Nuclear and Mitochondrial Data on Trichuris from Macaca fuscata Support Evidence of Host Specificity
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Molecular Methods
2.2. Sequencing, Evolutionary Distance, and Phylogenetic Analyses
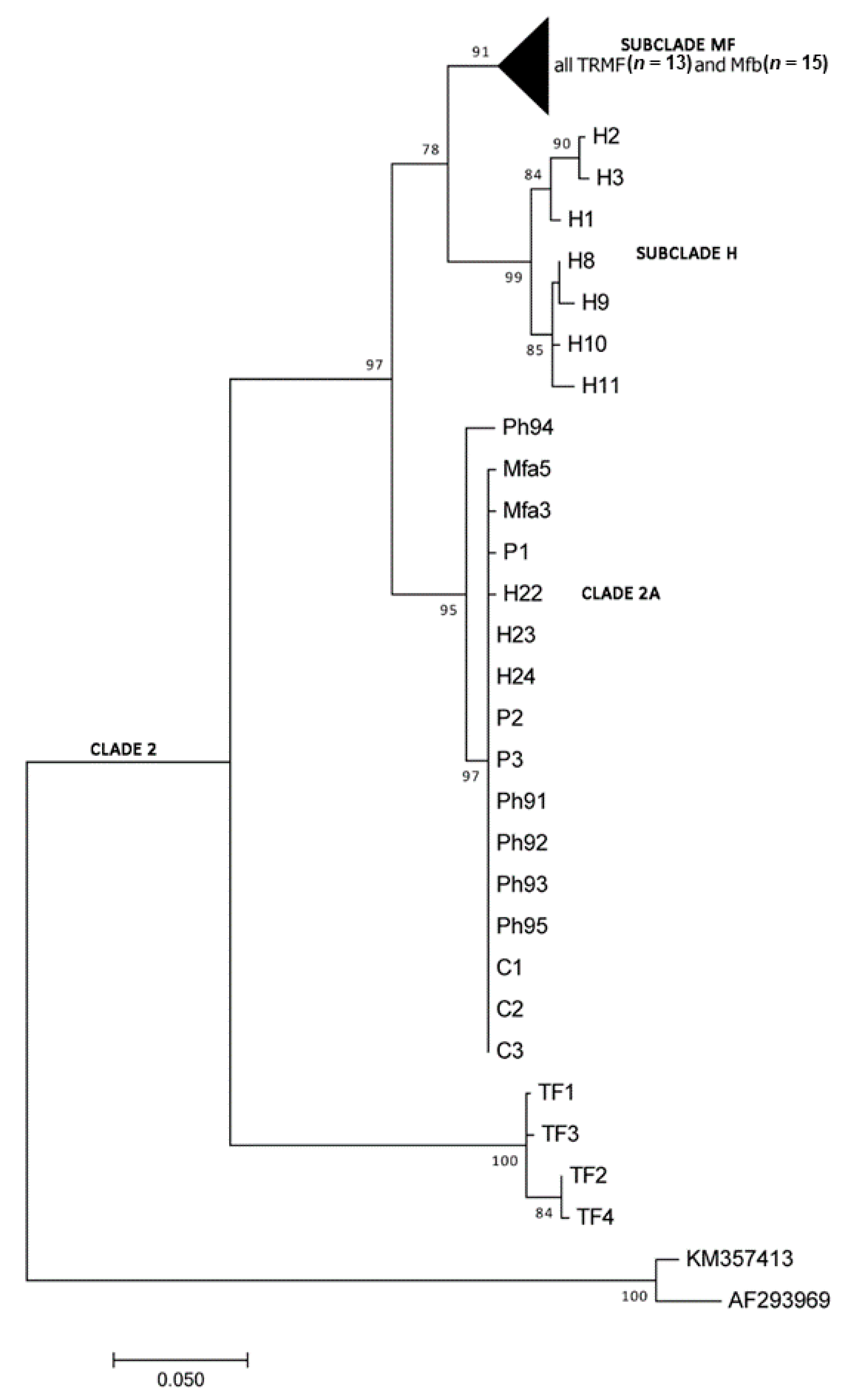
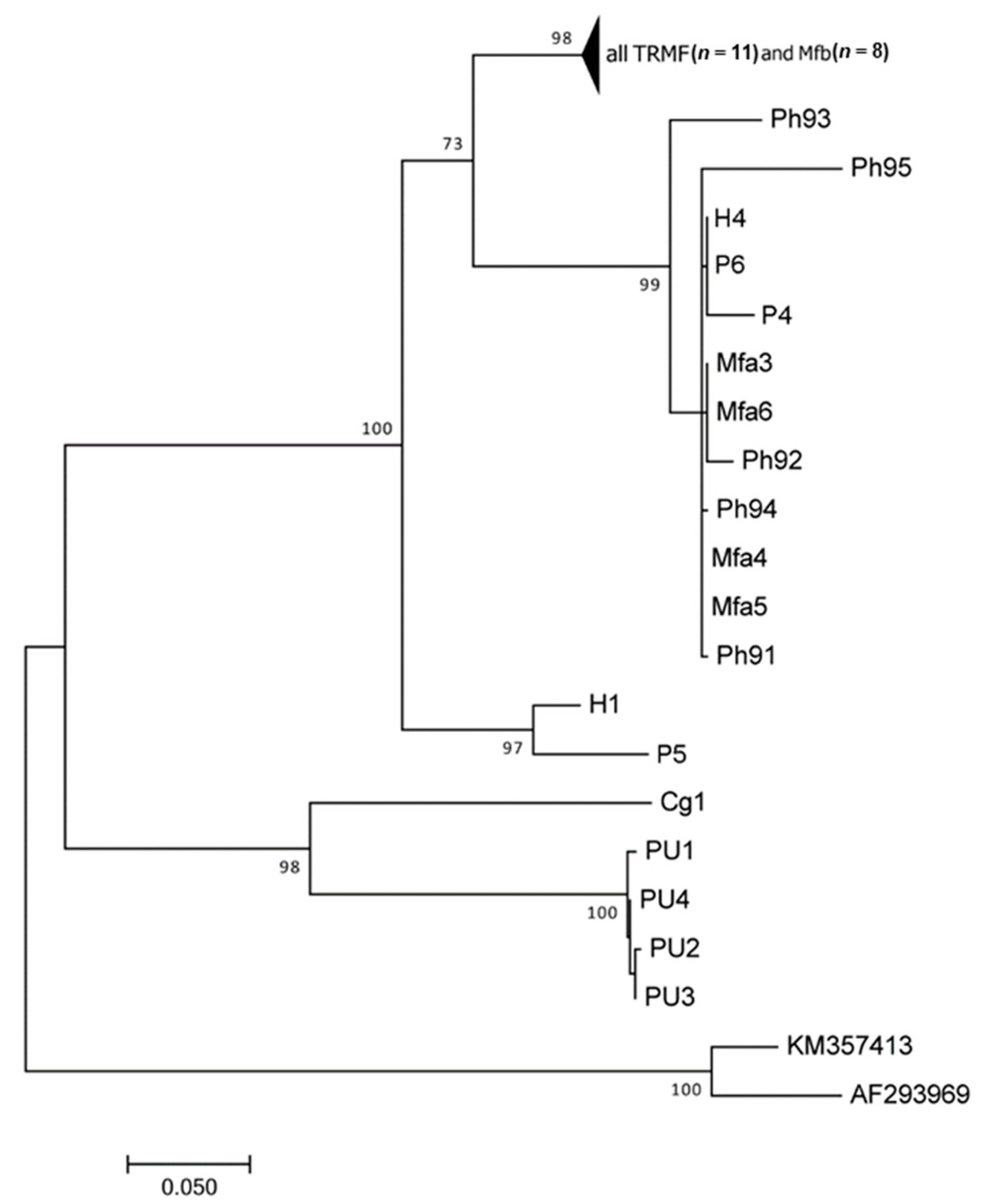
3. Results
Molecular and Phylogenetic Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Levecke, B.; Dorny, P.; Geurden, T.; Vercammen, F.; Vercruysse, J. Gastrointestinal protozoa in non-human primates of four zoological gardens in Belgium. Vet. Parasitol. 2007, 148, 236–246. [Google Scholar] [CrossRef]
- Calle, P.; Joslin, J. New world and old world monkeys. In Zoo and Wild Animal Medicine; Fowler, M., Miller, R., Eds.; Elsevier Saunders: St. Louis, MO, USA, 2014; pp. 301–335. [Google Scholar]
- Aviruppola, A.J.M.K.; Rajapakse, R.P.V.J.; Rajakaruna, R.S. Coprological survey of gastrointestinal parasites of mammals in Dehiwala National Zoological Gardens, Sri Lanka. Ceylon J. Sci. 2016, 45, 83. [Google Scholar] [CrossRef]
- Da Silva Barbosa, A.; Pinheiro, J.L.; dos Santos, C.R.; de Lima, C.S.C.C.; Dib, L.V.; Echarte, G.V.; Augusto, A.M.; Bastos, A.C.M.P.; Antunes Uchôa, C.M.; Bastos, O.M.P.; et al. Gastrointestinal parasites in captive animals at the Rio de Janeiro zoo. Acta Parasitol. 2020, 65, 237–249. [Google Scholar] [CrossRef]
- Li, M.; Zhao, B.; Li, B.; Wang, Q.; Niu, L.; Deng, J.; Gu, X.; Peng, X.; Wang, T.; Yang, G. Prevalence of gastrointestinal parasites in captive non-human primates of twenty-four zoological gardens in China. J. Med. Primatol. 2015, 44, 168–173. [Google Scholar] [CrossRef]
- Yamaguti, S. The nematodes of Vertebrates. In Systema Helminthum; Interscience (Wiley): New York, NY, USA, 1962. [Google Scholar]
- Bethony, J.; Brooker, S.; Albonico, M.; Geiger, S.M.; Loukas, A.; Diemert, D.; Hotez, P.J. Soil-Transmitted helminth infections: Ascariasis, trichuriasis, and hookworm. Lancet 2006, 367, 1521–1532. [Google Scholar] [CrossRef]
- Ravasi, D.F.; O’Riain, M.J.; Davids, F.; Illing, N. Phylogenetic evidence that two distinct Trichuris genotypes infect both humans and non-human primates. PLoS ONE 2012, 7, e44187. [Google Scholar] [CrossRef]
- Zanzani, S.A.; Gazzonis, A.L.; Epis, S.; Manfredi, M.T. Study of the gastrointestinal parasitic fauna of captive non-human primates (Macaca fascicularis). Parasitol. Res. 2016, 115, 307–312. [Google Scholar] [CrossRef]
- Cavallero, S.; De Liberato, C.; Friedrich, K.G.; Di Cave, D.; Masella, V.; D’Amelio, S.; Berrilli, F. Genetic heterogeneity and phylogeny of Trichuris spp. from captive non-human primates based on ribosomal DNA sequence data. Infect. Genet. Evol. 2015, 34, 450–456. [Google Scholar] [CrossRef]
- Cavallero, S.; Nejsum, P.; Cutillas, C.; Callejón, R.; Doležalová, J.; Modrý, D.; D’Amelio, S. Insights into the molecular systematics of Trichuris infecting captive primates based on mitochondrial DNA analysis. Vet. Parasitol. 2019, 272, 23–30. [Google Scholar] [CrossRef]
- Reichard, M.V.; Wolf, R.F.; Carey, D.W.; Garrett, J.J.; Briscoe, H.A. Efficacy of fenbendazole and milbemycin oxime for treating baboons (Papio cynocephalus anubis) infected with Trichuris trichiura. J. Am. Assoc. Lab. Anim. Sci. 2007, 46, 42–45. [Google Scholar]
- Montalbano Di Filippo, M.; Berrilli, F.; De Liberato, C.; Di Giovanni, V.; D’Amelio, S.; Friedrich, K.G.; Cavallero, S. Molecular characterization of Trichuris spp. from captive animals based on mitochondrial markers. Parasitol. Int. 2020, 75. [Google Scholar] [CrossRef]
- Jenkins, T.A. Morphological and histochemical study of Trichuris suis (Schrank, 1788) with special reference to the host-parasite relationship. Parasitology 1970, 61, 357–374. [Google Scholar] [CrossRef]
- Ooi, H.; Tenora, F.; Itoh, K.; Kamiya, M. Comparative study of Trichuris trichiura from non-human primates and from man, and their difference with T. suis. J. Vet. Med. Sci. 1993, 55, 363–366. [Google Scholar] [CrossRef]
- Liu, G.H.; Gasser, R.B.; Su, A.; Nejsum, P.; Peng, L.; Lin, R.Q.; Li, M.W.; Xu, M.J.; Zhu, X.Q. Clear genetic distinctiveness between human- and pig-derived Trichuris based on analyses of mitochondrial datasets. PLoS Negl. Trop. Dis. 2012, 6, e1539. [Google Scholar] [CrossRef]
- Nissen, S.; Al-Jubury, A.; Hansen, T.V.A.; Olsen, A.; Christensen, H.; Thamsborg, S.M.; Nejsum, P. Genetic analysis of Trichuris suis and Trichuris trichiura recovered from humans and pigs in a sympatric setting in Uganda. Vet. Parasitol. 2012, 188, 68–77. [Google Scholar] [CrossRef]
- Meekums, H.; Hawash, M.B.; Sparks, A.M.; Oviedo, Y.; Sandoval, C.; Chico, M.E.; Stothard, J.R.; Cooper, P.J.; Nejsum, P.; Betson, M.A. Genetic analysis of Trichuris trichiura and Trichuris suis from Ecuador. Parasites Vectors 2015, 8, 1–5. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hawash, M.; Betson, M.; Al-Jubury, A.; Ketzis, J.; LeeWillingham, A.; Bertelsen, M.; Cooper, P.; Littlewood, D.; Zhu, X.; Nejsum, P. Whipworms in humans and pigs: Origins and demography. Parasites Vectors 2016, 9. [Google Scholar] [CrossRef]
- Liu, G.; Gasser, R.; Nejsum, P.; Wang, Y.; Chen, Q.; Song, H.; Zhu, X. Mitochondrial and nuclear ribosomal DNA evidence supports the existence of a new Trichuris species in the endangered francois’ leaf-monkey. PLoS ONE 2013, 8, e66249. [Google Scholar] [CrossRef]
- Hawash, M.B.F.; Andersen, L.O.; Gasser, R.B.; Stensvold, C.R.; Nejsum, P. Mitochondrial genome analyses suggest multiple Trichuris species in humans, baboons, and pigs from different geographical regions. PLoS Negl. Trop. Dis. 2015, 9, e0004059. [Google Scholar] [CrossRef]
- Callejón, R.; Cutillas, C.; Nadler, S. Nuclear and mitochondrial genes for inferring Trichuris phylogeny. Parasitol. Res. 2015, 114, 4591–4599. [Google Scholar] [CrossRef]
- Rivero, J.; García-Sánchez, Á.M.; Zurita, A.; Cutillas, C.; Callejón, R. Trichuris trichiura isolated from Macaca sylvanus: Morphological, biometrical, and molecular study. BMC Vet. Res. 2020, 16, 445. [Google Scholar] [CrossRef]
- Mohandas, N.; Pozio, E.; La Rosa, G.; Korhonen, P.; Young, N.; Koehler, A.; Hall, R.; Sternberg, P.; Boag, P.; Jex, A.; et al. Mitochondrial genomes of Trichinella species and genotypes—A basis for diagnosis, and systematic and epidemiological explorations. Int. J. Parasitol. 2014, 44, 1073–1080. [Google Scholar] [CrossRef]
- Lavrov, D.; Brown, W. Trichinella spiralis mtDNA: A nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAS and has a gene arrangement relatable to those of coelomate metazoans. Genetics 2001, 157, 621–637. [Google Scholar]
- Bennet, A.; Barker, G.; Bundy, D.A. Beta-Tubulin gene from Trichuris trichiura. Mol. Biochem. Parasitol. 1999, 103, 111–116. [Google Scholar]
- Hansen, T.; Thamsborg, S.; Olsen, A.; Nejsum, P. Genetic variations in the beta-tubulin gene and the internal transcribed spacer (ITS-2) region of Trichuris species from man and baboons. Parasites Vectors 2013, 236. [Google Scholar] [CrossRef]
- Mitreva, M.; Jasmer, D.P.; Zarlenga, D.S.; Wang, Z.; Abubucker, S.; Martin, J.; Taylor, C.M.; Yin, Y.; Fulton, L.; Minx, P.; et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 2011, 43, 228–235. [Google Scholar] [CrossRef]
- Arizono, N.; Yamada, M.; Tegoshi, T.; Onishi, K. Molecular identification of Oesophagostomum and Trichuris eggs isolated from wild Japanese macaques. Korean J. Parasitol. 2012, 50, 253–257. [Google Scholar] [CrossRef]
- Putaporntip, C.; Jongwutiwes, S. Detection of Trichuris trichiura and T. vulpis in human and dog stool samples by nested PCR. Asian Biomed. 2010, 4. [Google Scholar] [CrossRef]
- Phosuk, I.; Sanpool, O.; Thanchomnang, T.; Sadaow, L.; Rodpai, R.; Anamnart, W.; Janwan, P.; Wijit, A.; Laymanivong, S.; Aung, W.; et al. Molecular identification of Trichuris suis and Trichuris trichiura eggs in human populations from Thailand, Lao PDR, and Myanmar. Am. J. Trop. Med. Hyg. 2018, 98, 39–44. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Song, M. Cloning and Sequence Analysis of the 18S Ribosomal RNA Gene from Five Isolates of Trichinella. Unpublished. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AY497012 (accessed on 30 December 2020).
- Doležalová, J.; Oborník, M.; Hajdušková, E.; Jirku, M.; Petrželková, K.J.; Bolechová, P.; Cutillas, C.; Callejón, R.; Jaroš, J.; Beránková, Z.; et al. How many species of whipworms do we share? Whipworms from man and other primates form two phylogenetic lineages. Folia Parasitol. 2015, 62, 1–12. [Google Scholar] [CrossRef]
| Parasite Species | Host Species | GenBank Accession Number | Specimen Code | Authors and References |
|---|---|---|---|---|
| Dataset_16S | ||||
| T. trichiura | Homo sapiens | GU385218 AM993017-18 | H1 H2-3 | Liu et al. (2012) [16] |
| T. trichiura | Homo sapiens | KP781898-KP781901 | H8–11 | Meekums et al. (2015) [10] |
| T. trichiura | Homo sapiens | KU524541-43 | H22–24 | Hawash et al. (2016) [21] |
| Trichuris sp. | Macaca fuscata | MW403712-16 | TRMF4,34,48,61,72 plus 8 undeposited * | Present study (representative specimens) |
| Trichuris sp. | Macaca fuscata | MN088542-43MN088544-58 | Mfa3,5 Mfb2–4,6–8, 10–14,16–19 | Cavallero et al. (2019) [11] |
| Trichuris sp. | Papio sp. | KU524558-60 | P1–3 | Hawash et al. (2016) [21] |
| Trichuris sp. | Papio hamadryas | MN088578-82 | Ph92–96 | Cavallero et al. (2019) [11] |
| Trichuris sp. | Chlorocebus sabaeus | KU524595-97 | C1–3 | Hawash et al. (2016) [21] |
| Trichuris sp. | Trachypithecus francoisi | KC481232-35 | TF 1–4 | Liu et al. (2013) [22] |
| Dataset_cytb | ||||
| T. trichiura | Homo sapiens | GU385218 | H1 | Liu et al. (2012) [16] |
| T. trichiura | Homo sapiens | KT449826 | H4 | Hawash et al. (2015) [23] |
| T. colobae | Colobus guereza | LM994704 | Cg1 | Callejón et al. (2015) [24] |
| Trichuris sp. | Macaca fuscata | MW403707-11 | TRMF4,34,44,58,61 plus 6 undeposited | Present study (representative specimens) |
| Trichuris sp. | Macaca fuscata | MK914550-53MK914554-61 | Mfa3–5 Mfb2–4,7,10–13 | Cavallero et al. (2019) [11] |
| T. ursinus | Papio ursinus | LT627357-60 | PU1–4 | Rivero et al. (2020) [25] |
| Trichuris sp. | Papio hamadryas | KT449824 | P4 | Hawash et al. (2015) [23] |
| Trichuris sp. | Papio hamadryas | MK914573-77 | Ph91–95 | Cavallero et al. (2019) [11] |
| Trichuris sp. | Papio anubis | KT449825 | P5 | Hawash et al. (2015) [23] |
| Trichuris sp. | Papio sp. | LM994703 | P6 | Callejón et al. (2015) [24] |
| Outgroup species | ||||
| Trichinella britovi | KM357413 | Mohandas et al. (2014) [26] | ||
| Trichinella spiralis | AF293969 | Lavrov and Brown (2001) [27] | ||
| Parasite Species | Host Species | GenBank Accession Number | Specimen Code | Authors |
|---|---|---|---|---|
| Dataset_βtub | ||||
| T. trichiura | Homo sapiens | AF034219 | H4 | Bennett et al. (1999) [28] |
| T. trichiura | Homo sapiens | KF410623-24 | H5-H6 | Hansen et al. (Unpublished) [29] |
| Trichuris sp. | Papio hamadryas | KF410632-34 | P7-P9 | Hansen et al. (Unpublished) [29] |
| Trichuris sp. | Macaca fuscata | MW403705-06 | TRMF4,48 plus 10 undeposited * | Present study (representative specimens) |
| Outgroup species | ||||
| Trichinella spiralis | Rattus norvegicus | XM_003369432 | Tspi | Mitreva et al. (2011) [30] |
| Dataset_18S | ||||
| T. trichiura | Macaca fuscata | AB699092 | TtMF1 | Arizono et al. (2012) [31] |
| Trichuris sp. | Macaca fuscata | MW396470-71 | TRMF4,6 plus 10 undeposited * | Present study (representative specimens) |
| T. trichiura | Homo sapiens | AB699090 | TtHS1 | Arizono et al. (2012) [31] |
| T. trichiura | Homo sapiens | GQ352553 | TH1 | Putaporntip et al. (2010) [32] |
| T. trichiura | Homo sapiens | MF288632 | TZY | Phosuk et al. (2017) [33] |
| Outgroup species | ||||
| Trichinella spiralis | Sus scrofa | AY497012 | Tspi | Li et al. (Unpublished) [34] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavallero, S.; Montalbano Di Filippo, M.; Rondón, S.; Liberato, C.D.; D’Amelio, S.; Friedrich, K.G.; Berrilli, F. Nuclear and Mitochondrial Data on Trichuris from Macaca fuscata Support Evidence of Host Specificity. Life 2021, 11, 18. https://doi.org/10.3390/life11010018
Cavallero S, Montalbano Di Filippo M, Rondón S, Liberato CD, D’Amelio S, Friedrich KG, Berrilli F. Nuclear and Mitochondrial Data on Trichuris from Macaca fuscata Support Evidence of Host Specificity. Life. 2021; 11(1):18. https://doi.org/10.3390/life11010018
Chicago/Turabian StyleCavallero, Serena, Margherita Montalbano Di Filippo, Silvia Rondón, Claudio De Liberato, Stefano D’Amelio, Klaus G. Friedrich, and Federica Berrilli. 2021. "Nuclear and Mitochondrial Data on Trichuris from Macaca fuscata Support Evidence of Host Specificity" Life 11, no. 1: 18. https://doi.org/10.3390/life11010018
APA StyleCavallero, S., Montalbano Di Filippo, M., Rondón, S., Liberato, C. D., D’Amelio, S., Friedrich, K. G., & Berrilli, F. (2021). Nuclear and Mitochondrial Data on Trichuris from Macaca fuscata Support Evidence of Host Specificity. Life, 11(1), 18. https://doi.org/10.3390/life11010018






