Effects of Diesel Exhaust Particles on Mouse Gastric Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of DEPs
2.2. Cell Culture
2.3. Cell Viability Assay
2.4. Glutathione Measurement
2.5. Thiobarbituric Acid Reactive Substances (TBARS) Measurement
2.6. NO Measurement
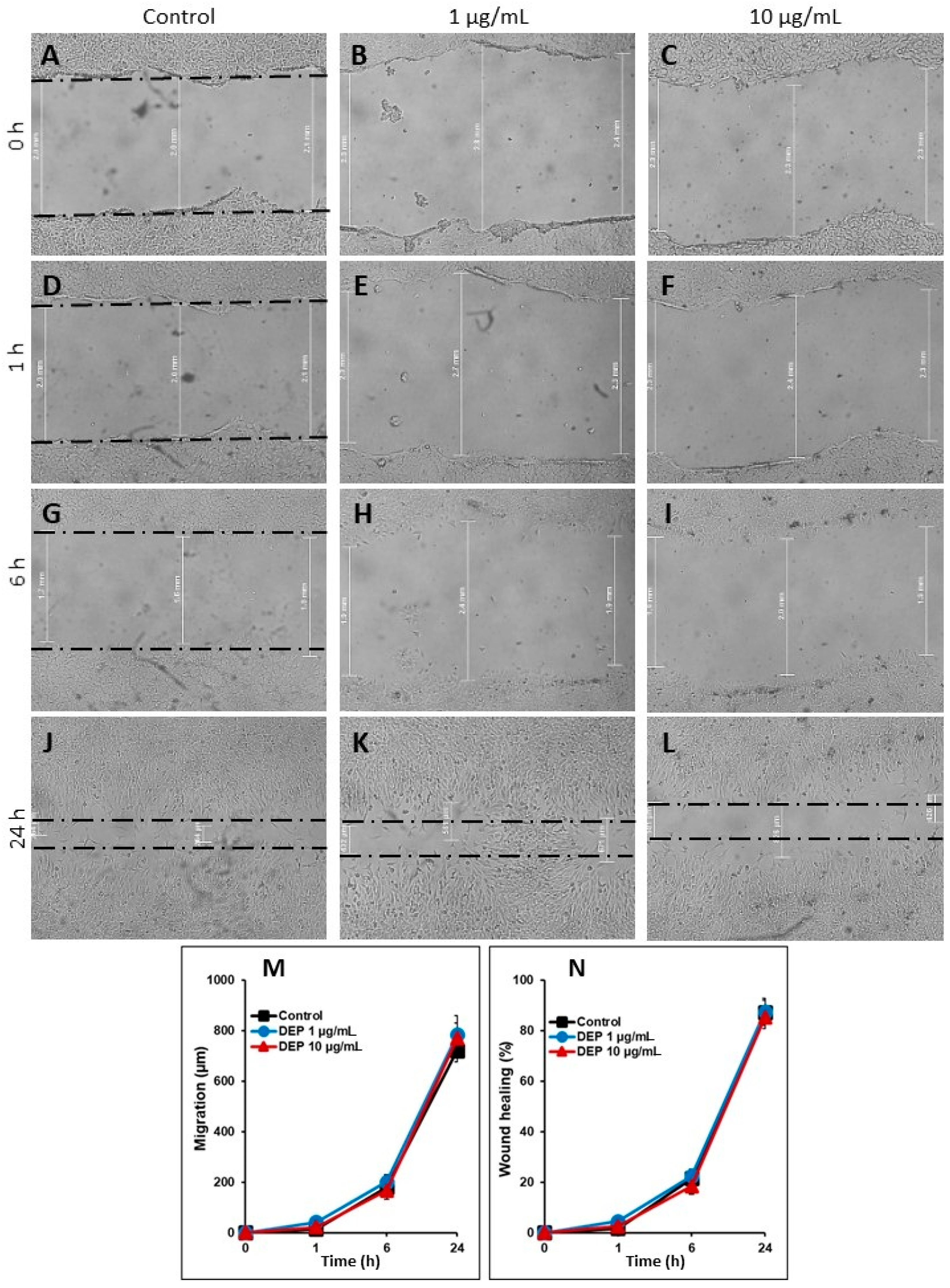
2.7. Cell Migration or Scratch Wound Healing Assay
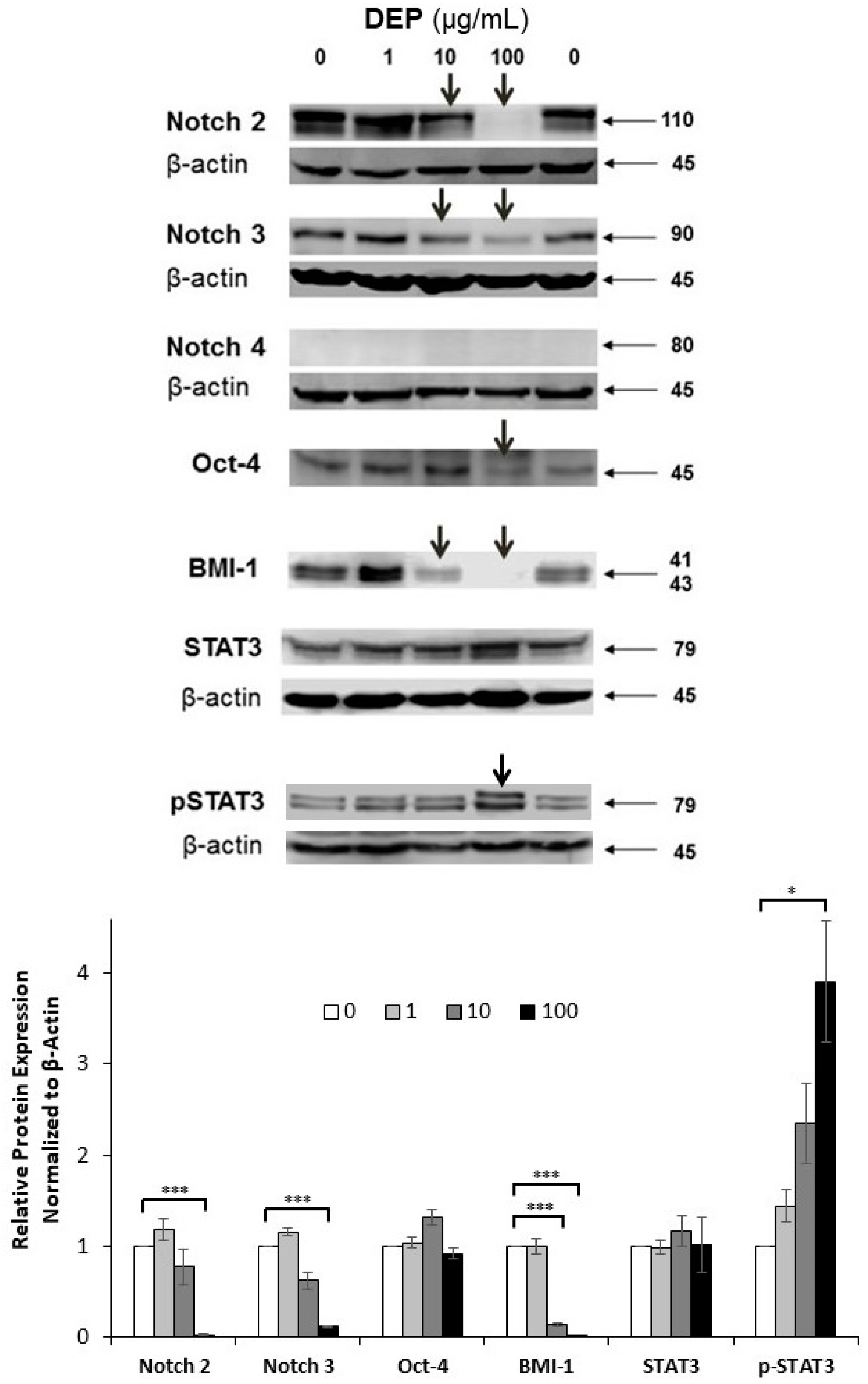
2.8. Western Blot Analysis
2.9. Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
2.10. Statistical Analysis
3. Results
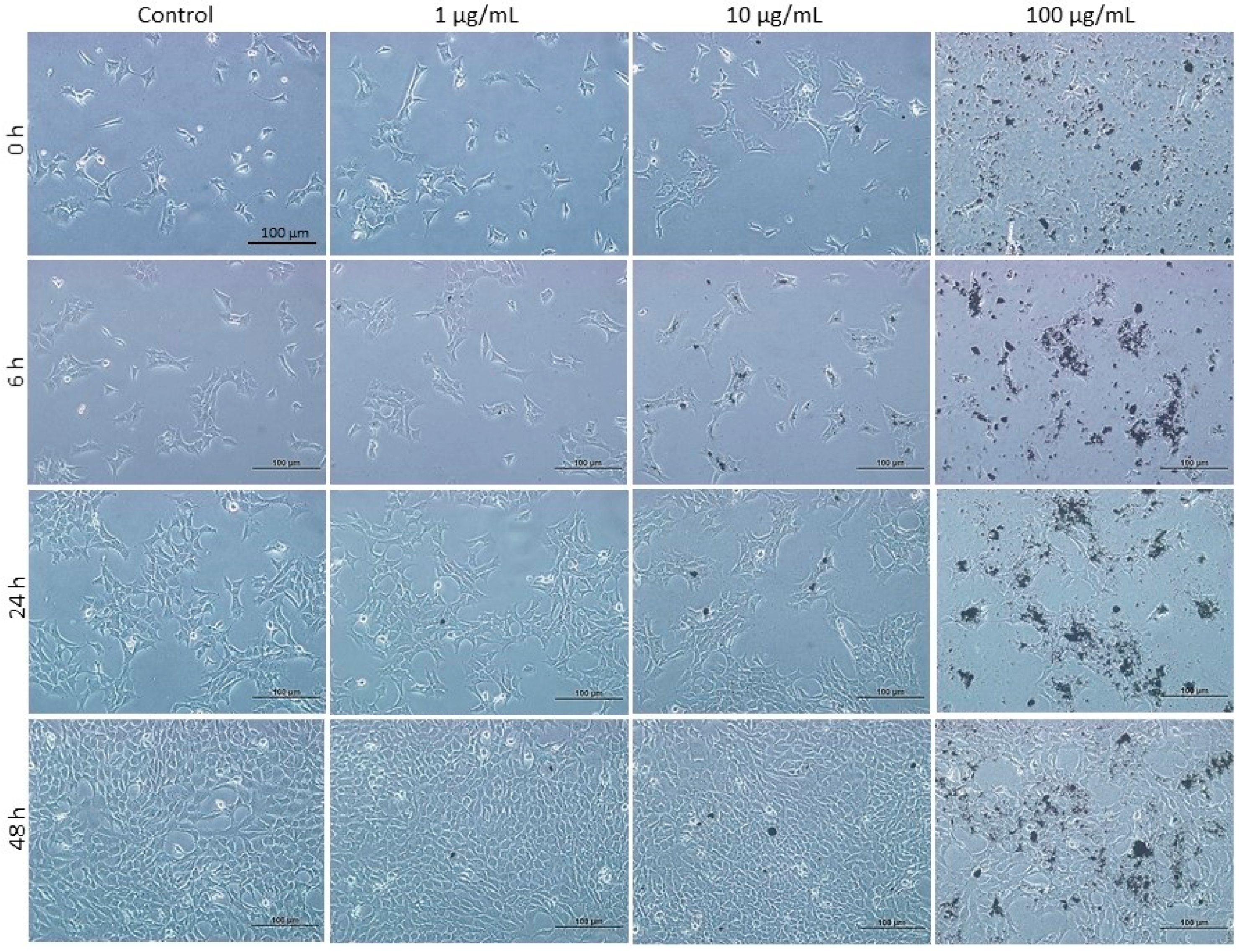
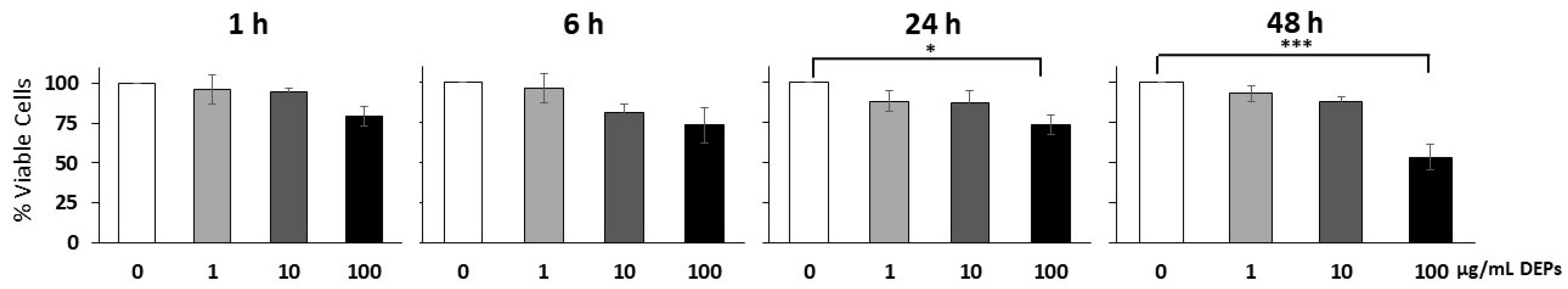
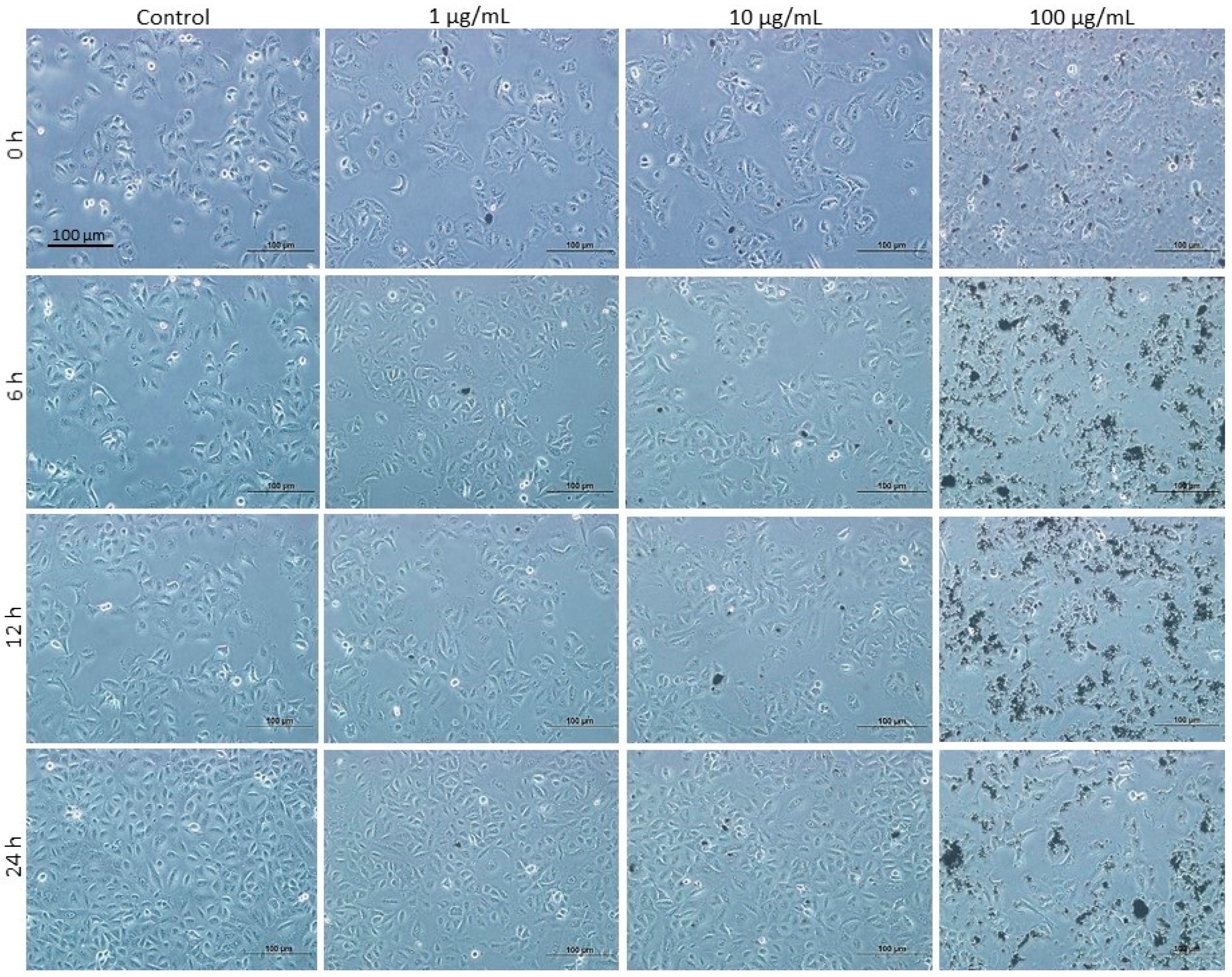
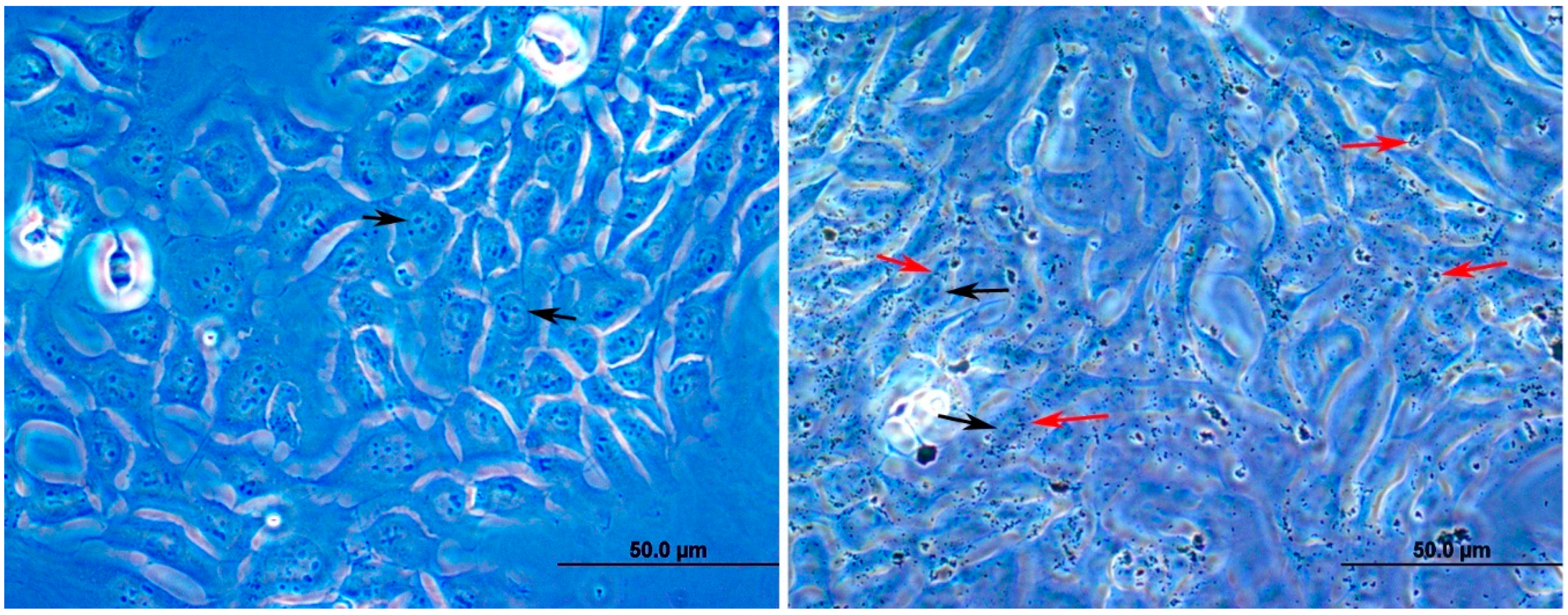
3.1. Effect of DEPs on the Viability of mGS Cells
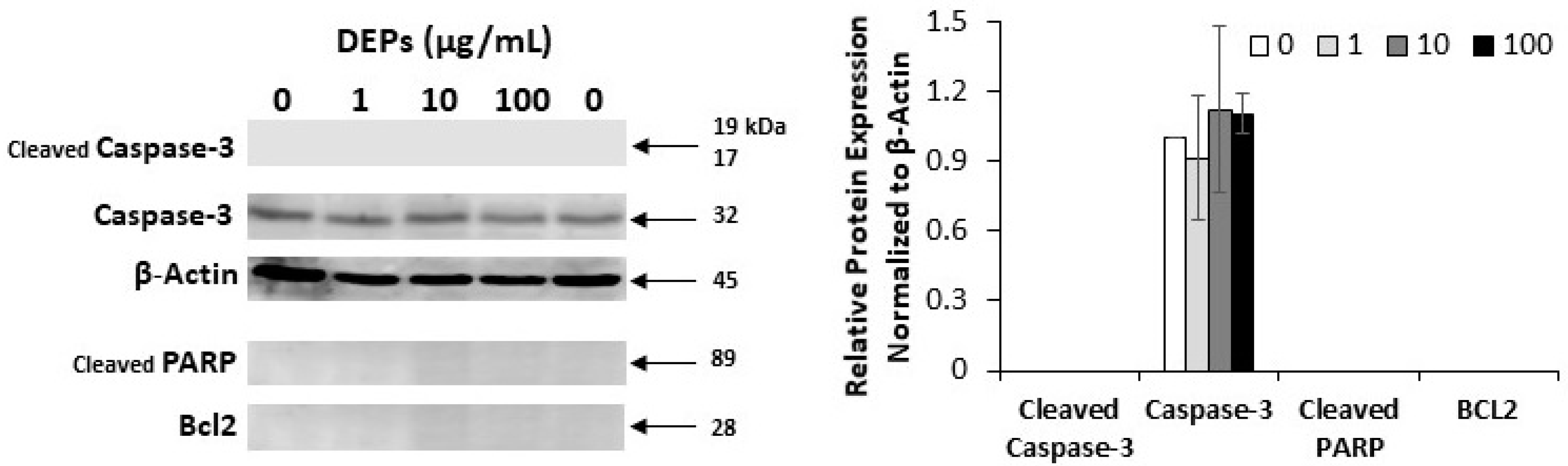
3.2. Effect of DEPs on the Expression of Apoptosis-related Proteins
3.3. Effect of DEPs on Oxidative Stress Markers
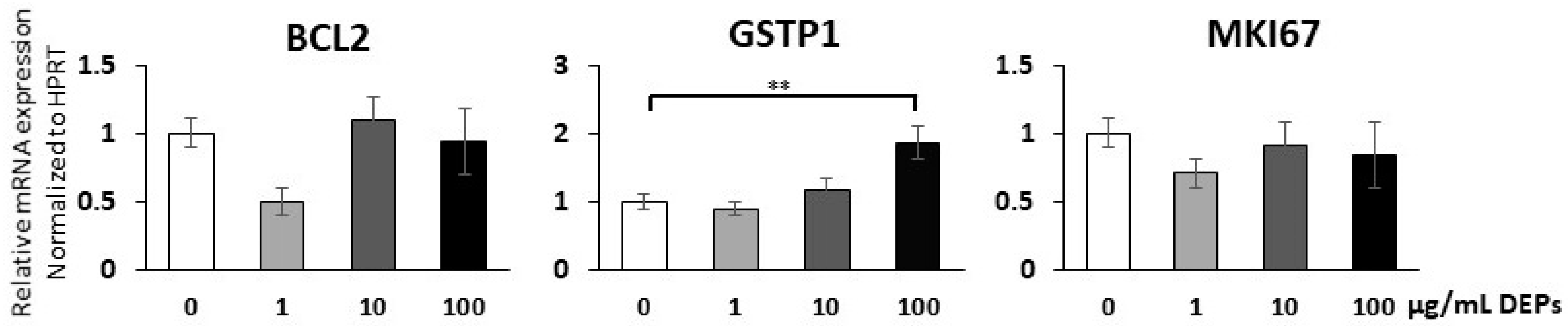
3.4. Gene Expression Analysis Using qRT-PCR
3.5. Effect of DEPs on the Migration of mGS Cells
3.6. Effects of DEPs on the Expression of Stem Cell-related Proteins in mGS Cells
3.7. Effects of DEPs on A549 Lung Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nemmar, A.; Beegam, S.; Yuvaraju, P.; Yasin, J.; Fahim, M.A.; Kazzam, E.E.; Alhaddabi, I.; Ali, B.H. Potentiation of cisplatin-induced nephrotoxicity by repeated exposure to diesel exhaust particles: An experimental study in rats. Exp. Biol. Med. 2014, 239, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Nemmar, A.; Al Dhaheri, R.; Alamiri, J.; Al Hefeiti, S.; Al Saedi, H.; Beegam, S.; Yuvaraju, P.; Yasin, J.; Ali, B.H. Diesel exhaust particles induce impairment of vascular and cardiac homeostasis in mice: Ameliorative effect of emodin. Cell. Physiol. Biochem. 2015, 36, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Nemmar, A.; Al-Salam, S.; Beegam, S.; Yuvaraju, P.; Hamadi, N.; Ali, B.H. In vivo protective effects of nootkatone against particles-induced lung injury caused by diesel exhaust is mediated via the NF-κB pathway. Nutrients 2018, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Ehsanifar, M.; Jafari, A.J.; Nikzad, H.; Zavareh, M.S.; Atlasi, M.A.; Mohammadi, H.; Tameh, A.A. Prenatal exposure to diesel exhaust particles causes anxiety, spatial memory disorders with alters expression of hippocampal pro-inflammatory cytokines and NMDA receptor subunits in adult male mice offspring. Ecotoxicol. Environ. Saf. 2019, 176, 34–41. [Google Scholar] [CrossRef]
- Weitekamp, C.A.; Kerr, L.B.; Dishaw, L.; Nichols, J.; Lein, M.; Stewart, M.J. A systematic review of the health effects associated with the inhalation of particle-filtered and whole diesel exhaust. Inhal. Toxicol. 2020, 32, 1–13. [Google Scholar] [CrossRef]
- Pálková, L.; Vondráček, J.; Trilecová, L.; Ciganek, M.; Pěnčíková, K.; Neča, J.; Milcová, A.; Topinka, J.; Machala, M. The aryl hydrocarbon receptor-mediated and genotoxic effects of fractionated extract of standard reference diesel exhaust particle material in pulmonary, liver and prostate cells. Toxicol. Vitro 2015, 29, 438–448. [Google Scholar] [CrossRef]
- Gamble, J.F.; Nicolich, M.J.; Boffetta, P. Lung Cancer and Diesel Exhaust: An Updated Critical Review of the Occupational Epidemiology Literature. Crit. Rev. Toxicol. 2012, 42, 549–598. [Google Scholar] [CrossRef]
- Wichmann, H.E. Diesel exhaust particles. Inhal. Toxicol. 2017, 19 (Suppl. 1), 241–244. [Google Scholar] [CrossRef]
- Sjödahl, K.; Jansson, C.; Bergdahl, I.A.; Adami, J.; Boffetta, P.; Lagergren, J. Airborne exposures and risk of gastric cancer: A prospective cohort study. Int. J. Cancer 2007, 120, 2013–2018. [Google Scholar] [CrossRef]
- Karam, S.M.; Leblond, C.P. Identifying and counting epithelial cell types in the “corpus” of the mouse stomach. Anat. Rec. 1992, 232, 231–246. [Google Scholar] [CrossRef]
- Karam, S.M.; Leblond, C.P. Dynamics of epithelial cells in the corpus of the mouse stomach. I. Identification of proliferative cell types and pinpointing of the stem cell. Anat. Rec. 1993, 236, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Karam, S.M. Stem Cells and Gastric Carcinogenesis: From Mouse to Human. In Stem Cells and Cancer Stem Cells; Hayat, M.A., Ed.; Springer: New York, NY, USA, 2012; Volume 3, pp. 221–229. [Google Scholar]
- Giannakis, M.; Chen, S.L.; Karam, S.M.; Engstrand, L.; Gordon, J.I. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 4358–4363. [Google Scholar] [CrossRef] [PubMed]
- Farook, V.S.; Alkhalaf, M.; Karam, S.M. Establishment of a gastric epithelial progenitor cell line from a transgenic mouse expressing the simian virus 40 large T antigen gene in the parietal cell lineage. Cell prolif. 2008, 41, 310–320. [Google Scholar] [CrossRef]
- Tai, M.H.; Chang, C.C.; Kiupel, M.; Webster, J.D.; Trosko, J.E. Oct-4 expression in adult stem cells: Evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 2005, 26, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Karam, S.M. Gastric stem cell biology: Proliferation kinetics, differentiation hierarchies, and role in carcinogenesis. In Stem Cells Handbook, 2nd ed.; Sell, S., Ed.; Humana Press: New Jersey, NJ, USA, 2013; Part III; pp. 255–271. [Google Scholar]
- Oh, J.D.; Karam, S.M.; Gordon, J.I. Intracellular Helicobacter pylori in gastric epithelial progenitors. PNAS 2005, 102, 5186–5191. [Google Scholar] [CrossRef] [PubMed]
- Ohkusa, T.; Fujiki, K.; Tamura, Y.; Yamamoto, M.; Kyoi, T. Freeze-fracture and immunohistochemical studies of gap junctions in human gastric mucosa with special reference to their relationship to gastric ulcer and gastric carcinoma. Microsc. Res. Tech. 1995, 31, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Nemmar, A.; Al-Maskari, S.; Ali, B.H.; Al-Amri, I.S. Cardiovascular and lung inflammatory effects induced by systemically administered diesel exhaust particles in rats. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007, 292, L664–L670. [Google Scholar] [CrossRef]
- Nemmar, A.; Zia, S.; Subramaniyan, D.; Al-Amri, I.; Al Kindi, M.A.; Ali, B.H. Interaction of diesel exhaust particles with human, rat and mouse erythrocytes in vitro. Cell. Physiol. Biochem. 2012, 29, 163–170. [Google Scholar] [CrossRef]
- Mohyeldin, A.; Garzon-Muvdi, T.; Quinones-Hinojosa, A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef]
- Linning, K.D.; Tai, M.H.; Madhukar, B.V.; Chang, C.C.; Reed, D.N.; Ferber, S.; Trosko, J.E.; Olson, L.K. Redox-mediated enrichment of self-renewing adult human pancreatic cells which possesses endocrine differentiation potential. Pancreas 2004, 29, e64–e76. [Google Scholar] [CrossRef] [PubMed]
- Nowak, E.; Kammerer, S.; Küpper, J.H. ATP-based cell viability assay is superior to trypan blue exclusion and XTT assay in measuring cytotoxicity of anticancer drugs Taxol and Imatinib, and proteasome inhibitor MG-132 on human hepatoma cell line HepG2. Clin. Hemorheol. Microcirc. 2018, 69, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.K.; Maulik, M.; Mancahanda, S.C.; Dinda, A.K.; Gupta, S.K.; Maulik, S.K. Dose-dependent induction of endogenous antioxidants in rat heart by chronic administration of garlic. Life Sci. 2002, 70, 1509–1518. [Google Scholar] [CrossRef]
- Janero, D.R. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radic. Biol. Med. 1990, 9, 515–540. [Google Scholar] [CrossRef]
- Guevara, I.; Iwanejko, J.; Dembińska-Kieć, A.; Pankiewicz, J.; Wanat, A.; Anna, P.; Gołabek, I.; Bartuś, S.; Malczewska-Malec, M.; Szczudlik, A. Determination of nitrite/nitrate in human biological material by the simple Griess reaction. Clin. Chim. Acta 1998, 274, 177–188. [Google Scholar] [CrossRef]
- Al-Marzoqee, F.Y.; Khoder, G.; Al-Awadhi, H.; John, R.; Beg, A.; Vincze, A.; Branicki, F.; Karam, S.M. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int. J. Oncol. 2012, 41, 1733–1743. [Google Scholar] [CrossRef]
- Reinisch, C.; Kandutsch, S.; Uthman, A.; Pammer, J. BMI-1: A protein expressed in stem cells, specialized cells and tumors of the gastrointestinal tract. Histol. Histopathol. 2006, 21, 1143–1149. [Google Scholar]
- Chen, C.; Zhang, W. Itraconazole Alters the Stem Cell Characteristics of A549 and NCI-H460 Human Lung Cancer Cells by Suppressing Wnt Signaling. Med. Sci. Monit. 2019, 25, 9509–9516. [Google Scholar] [CrossRef]
- Upham, B.L.; Weis, L.M.; Trosko, J.E. Modulated gap junctional intercellular communication as a biomarker of PAH epigenetic toxicity: Structure-function relationship. Environ. Health Perspect. 1998, 106 (Suppl. 4), 975–981. [Google Scholar]
- Rummel, A.M.; Trosko, J.E.; Wilson, M.R.; Upham, B.L. Polycyclic aromatic hydrocarbons with bay-like regions inhibited gap junctional intercellular communication and stimulated MAPK activity. Toxicol. Sci. 1999, 49, 232–240. [Google Scholar] [CrossRef]
- Romo, D.; Velmurugan, K.; Upham, B.L.; Dwyer-Nield, L.D.; Bauer, A.K. Dysregulation of gap junction function and cytokine production in response to non-genotoxic polycyclic aromatic hydrocarbons in an in vitro lung cell model. Cancers 2019, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Osgood, R.S.; Upham, B.L.; Hill, T., 3rd; Helms, K.L.; Velmurugan, K.; Babica, P.; Bauer, A.K. Polycyclic aromatic hydrocarbon-induced signaling events relevant to inflammation and tumorigenesis in lung cells are dependent on molecular structure. PLoS ONE 2013, 8, e65150. [Google Scholar] [CrossRef] [PubMed]
- Pöss, J.; Lorenz, D.; Werner, C.; Pavlikova, V.; Gensch, C.; Speer, T.; Alessandrini, F.; Berezowski, V.; Kuntz, M.; Mempel, M.; et al. Diesel exhaust particles impair endothelial progenitor cells, compromise endothelial integrity, reduce neoangiogenesis, and increase atherogenesis in mice. Cardiovasc. Toxicol. 2013, 13, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Lucariello, A.; Savarese, L.; Cinelli, M.P.; Ferraraccio, F.; Bianco, A.; De Luca, A.; Mazzarella, G. Morphology changes in human lung epithelial cells after exposure to diesel exhaust micron sub particles (PM 1.0) and pollen allergens. Environ. Pollut. 2012, 171, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Turnquist, H.; Jackson, J.; Sgagias, M.; Yan, Y.; Gong, M.; Dean, M.; Sharp, J.G.; Cowan, K. The multidrug resistance transporter ABCG2 (breast cancer resistance protein 1) effluxes Hoechst 33342 and is overexpressed in hematopoietic stem cells. Clin. Cancer Res. 2002, 8, 22–28. [Google Scholar]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Abdelmawgoud, H.; Saleh, A. Anti-inflammatory and antioxidant effects of mesenchymal and hematopoietic stem cells in a rheumatoid arthritis rat model. Adv. Clin. Exp. Med. 2018, 27, 873–880. [Google Scholar] [CrossRef]
- Nemmar, A.; Yuvaraju, P.; Beegam, S.; John, A.; Raza, H.; Ali, B.H. Cardiovascular effects of nose-only water-pipe smoking exposure in mice. Am. J. Physiol.-Heart Circ. Physiol. 2013, 305, H740–H746. [Google Scholar] [CrossRef]
- Kim, Y.-D.; Lantz-McPeak, S.M.; Ali, S.F.; Kleinman, M.T.; Choi, Y.-S.; Kim, H. Effects of ultrafine diesel exhaust particles on oxidative stress generation and dopamine metabolism in PC-12 cells. Environ. Toxicol. Pharmacol. 2014, 37, 954–959. [Google Scholar] [CrossRef]
- Xiao, G.G.; Wang, M.; Li, N.; Loo, J.A.; Nel, A.E. Use of proteomics to demonstrate a hierarchical oxidative stress response to diesel exhaust particle chemicals in a macrophage cell line. J. Biol. Chem. 2003, 278, 50781–50790. [Google Scholar] [CrossRef]
- Trosko, J.E. The Role of the Mitochondria in the Evolution of Stem Cells, Including MUSE Stem Cells and Their Biology. Adv. Exp. Med. Biol. 2018, 1103, 131–152. [Google Scholar] [PubMed]
- Ebert, M.N.; Klinder, A.; Peters, W.H.M.; Schäferhenrich, A.; Sendt, W.; Scheele, J.; Pool-Zobel, B.L. Expression of glutathione S-transferases (GSTs) in human colon cells and inducibility of GSTM2 by butyrate. Carcinogenesis 2003, 24, 1637–1644. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Previs, R.A.; Coleman, R.L.; Harris, A.L.; Sood, A.K. Molecular pathways: Translational and therapeutic implications of the Notch signaling pathway in cancer. Clin. Cancer Res. 2015, 21, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Alketbi, A.; Attoub, A. Notch Signaling in Cancer: Rationale and Strategies for Targeting. Curr. Cancer Drug Targets 2015, 15, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E. Review paper: Cancer stem cells and cancer nonstem cells: From adult stem cells or from reprogramming of differentiated somatic cells. Vet. Pathol. 2009, 46, 176–193. [Google Scholar] [CrossRef]
- Darnell, J.E. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef]
- Chang, C.C. Recent translational research: Stem cells as the roots of breast cancer. Breast Cancer Res. 2006, 8, 103. [Google Scholar] [CrossRef]
- Galoczova, M.; Coates, P.; Vojtesek, B. STAT3, stem cells, cancer stem cells and p63. Cell. Mol. Biol. Lett. 2018, 23, 12. [Google Scholar] [CrossRef]
- Cao, D.; Tal, T.L.; Graves, L.M.; Gilmour, I.; Linak, W.; Reed, W.; Bromberg, P.A.; Samet, J.M. Diesel exhaust particulate-induced activation of Stat3 requires activities of EGFR and Src in airway epithelial cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007, 292, L422–L429. [Google Scholar] [CrossRef]
- Clifford, R.L.; Jones, M.J.; MacIsaac, J.L.; McEwen, L.M.; Goodman, S.J.; Mostafavi, S.; Kobor, M.S.; Carlsten, C. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J. Allergy Clin. Immunol. 2017, 139, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Nemmar, A.; Al-Salam, S.; Beegam, S.; Yuvaraju, P.; Ali, B.H. Thrombosis and systemic and cardiac oxidative stress and DNA damage induced by pulmonary exposure to diesel exhaust particles and the effect of nootkatone thereon. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H917–H927. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Sadik, H.; Sugathan, S.; Saseedharan, P.; Sulaiman, S.; Beegam, S.; Nemmar, A.; Attoub, S.; Karam, S.M. Effects of Diesel Exhaust Particles on Mouse Gastric Stem Cells. Life 2020, 10, 149. https://doi.org/10.3390/life10080149
Al-Sadik H, Sugathan S, Saseedharan P, Sulaiman S, Beegam S, Nemmar A, Attoub S, Karam SM. Effects of Diesel Exhaust Particles on Mouse Gastric Stem Cells. Life. 2020; 10(8):149. https://doi.org/10.3390/life10080149
Chicago/Turabian StyleAl-Sadik, Heba, Subi Sugathan, Prashanth Saseedharan, Shahrazad Sulaiman, Sumaya Beegam, Abderrahim Nemmar, Samir Attoub, and Sherif M. Karam. 2020. "Effects of Diesel Exhaust Particles on Mouse Gastric Stem Cells" Life 10, no. 8: 149. https://doi.org/10.3390/life10080149
APA StyleAl-Sadik, H., Sugathan, S., Saseedharan, P., Sulaiman, S., Beegam, S., Nemmar, A., Attoub, S., & Karam, S. M. (2020). Effects of Diesel Exhaust Particles on Mouse Gastric Stem Cells. Life, 10(8), 149. https://doi.org/10.3390/life10080149






