Assessing Viral Abundance and Community Composition in Four Contrasting Regions of the Southern Ocean
Abstract
1. Introduction
2. Materials and Methods
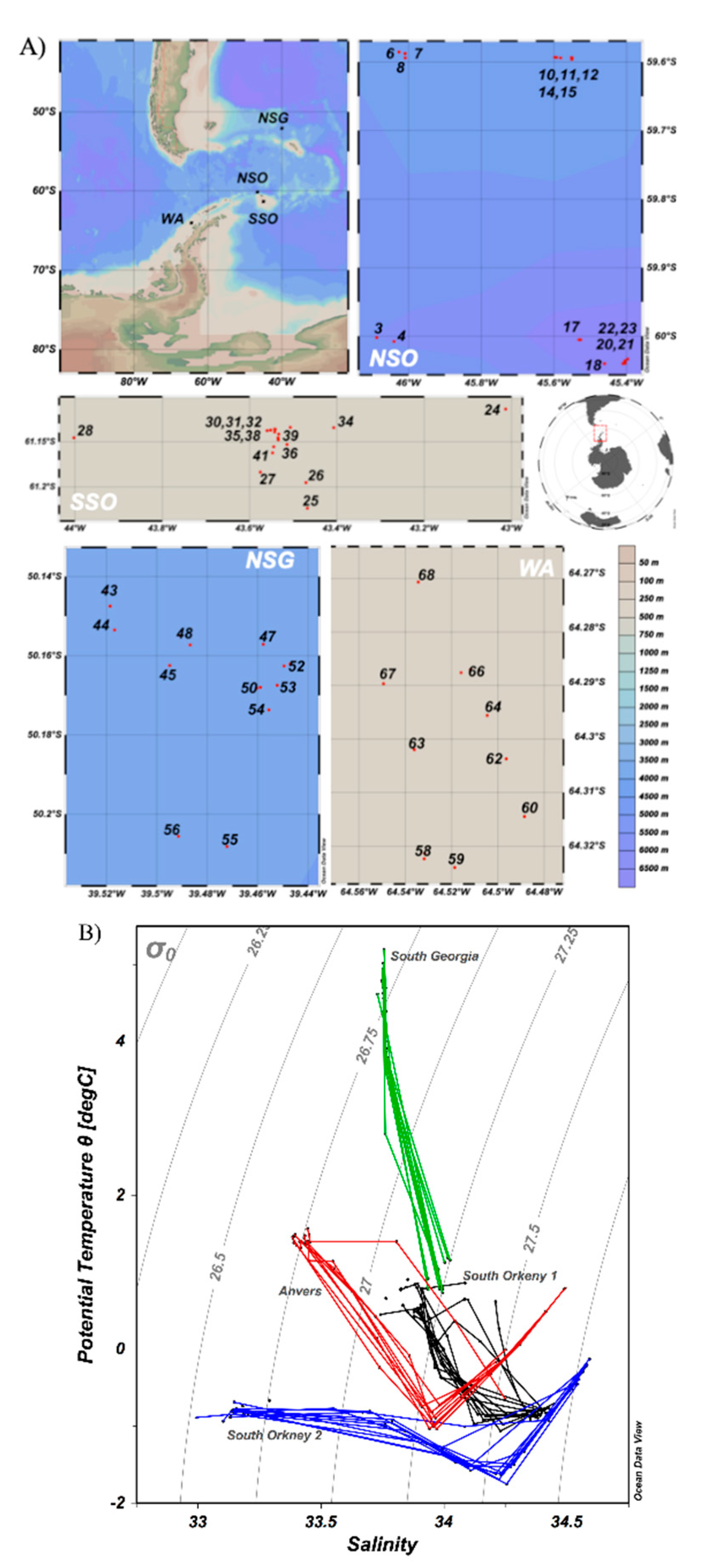
2.1. Sampling Regions and Strategy
2.2. Physicochemical Variables
2.3. Microbiological Variables
2.4. Viral Community Composition: Random Amplified Polymorphic DNA (RAPD)
2.5. Data Analyses
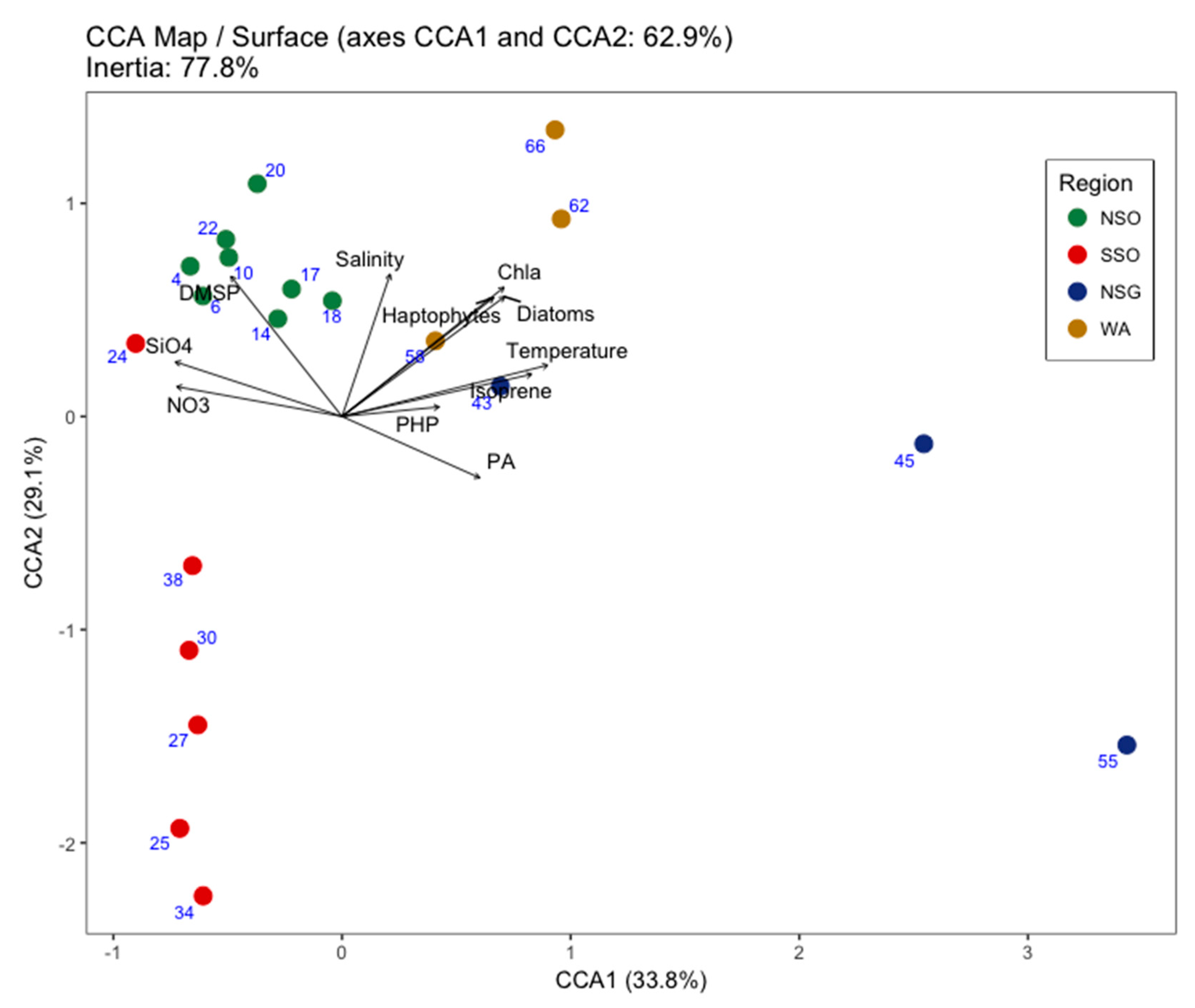
3. Results
3.1. Characterization of the Study Area: Environmental and Biological Variables
3.1.1. Temperature and Salinity
3.1.2. Inorganic Nutrients, FDOM and Secondary Compounds
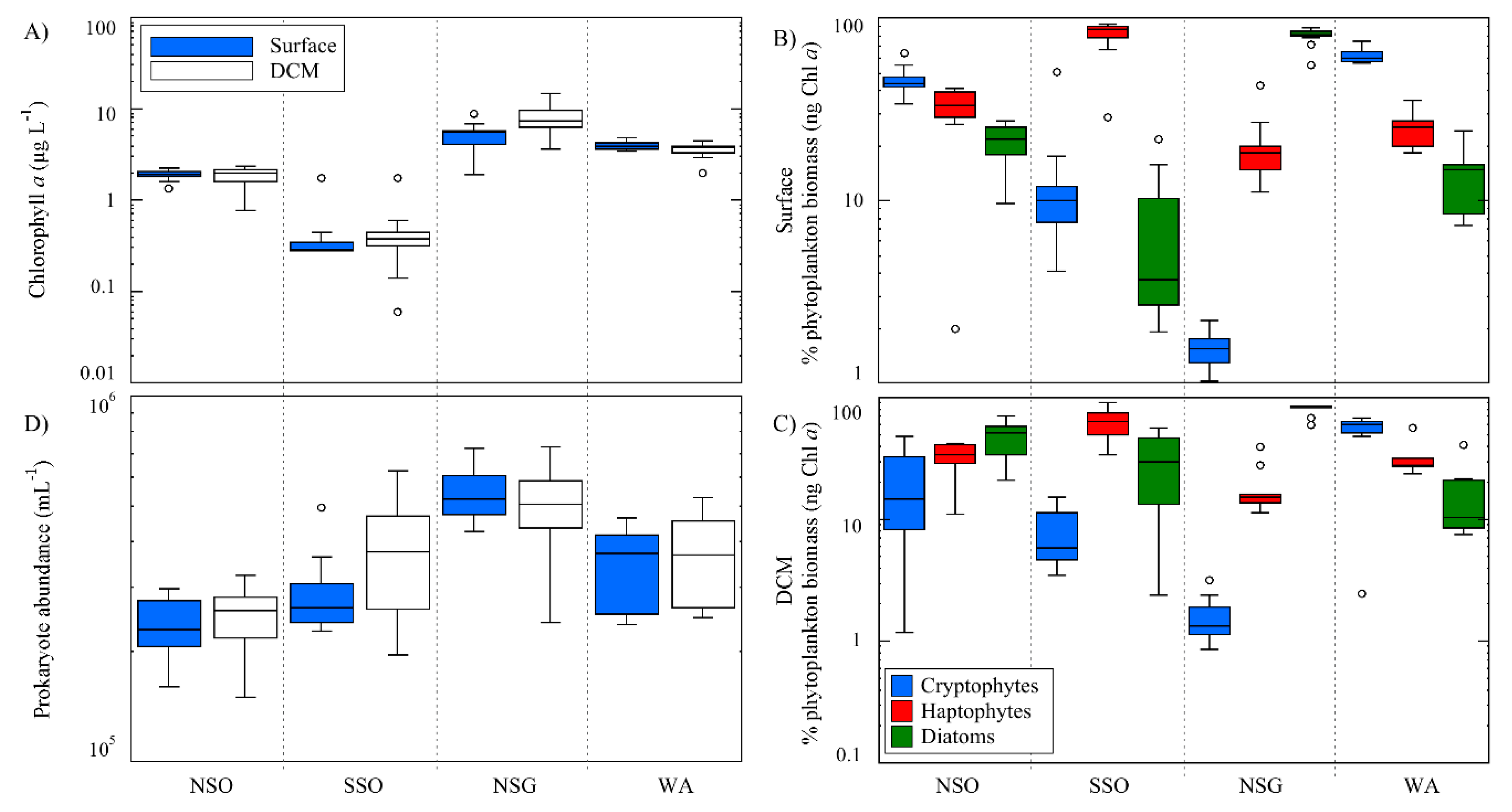
3.1.3. Chlorophyll a, Phytoplankton Taxa and Prokaryote Concentrations
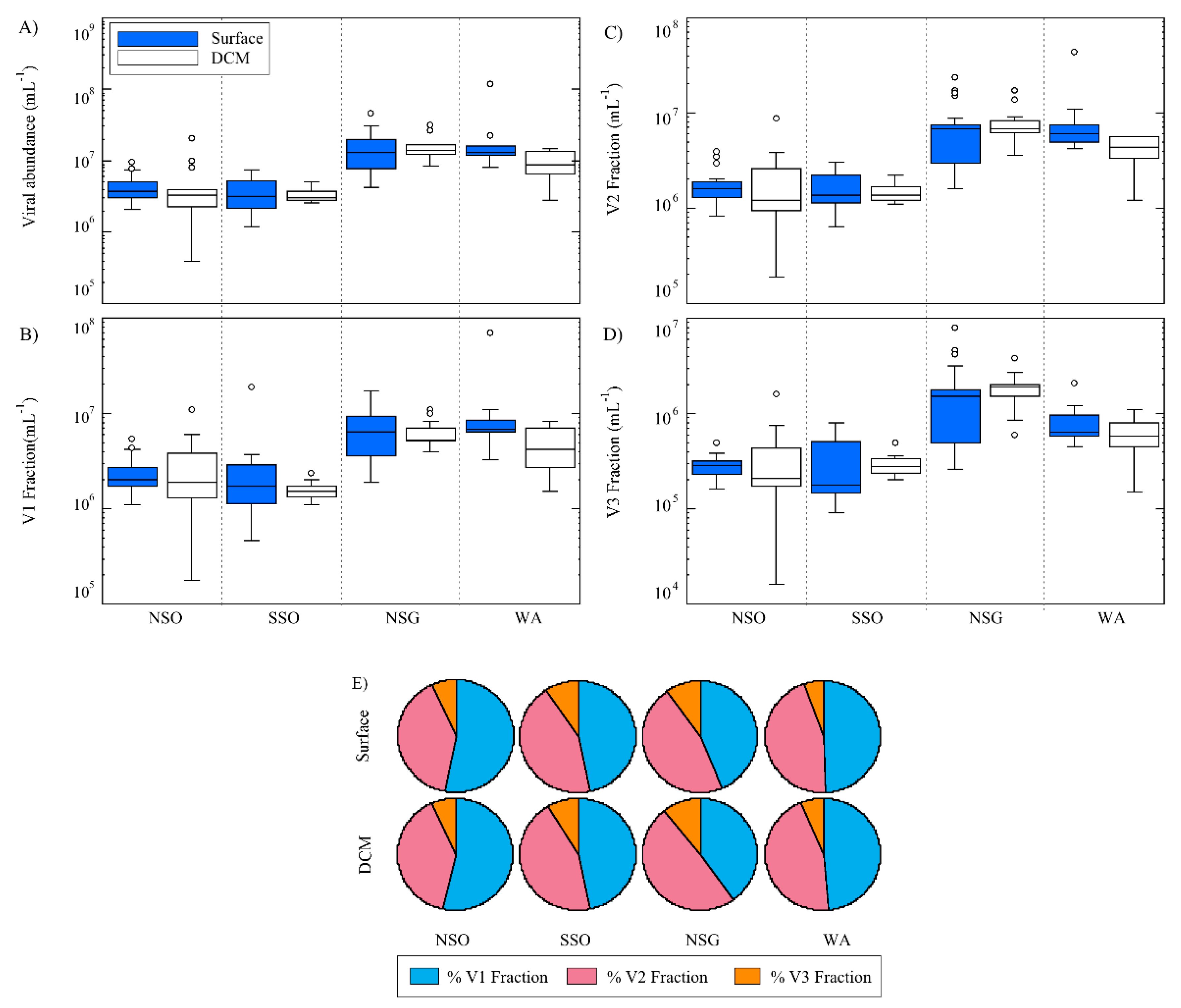
3.2. Viral Abundance
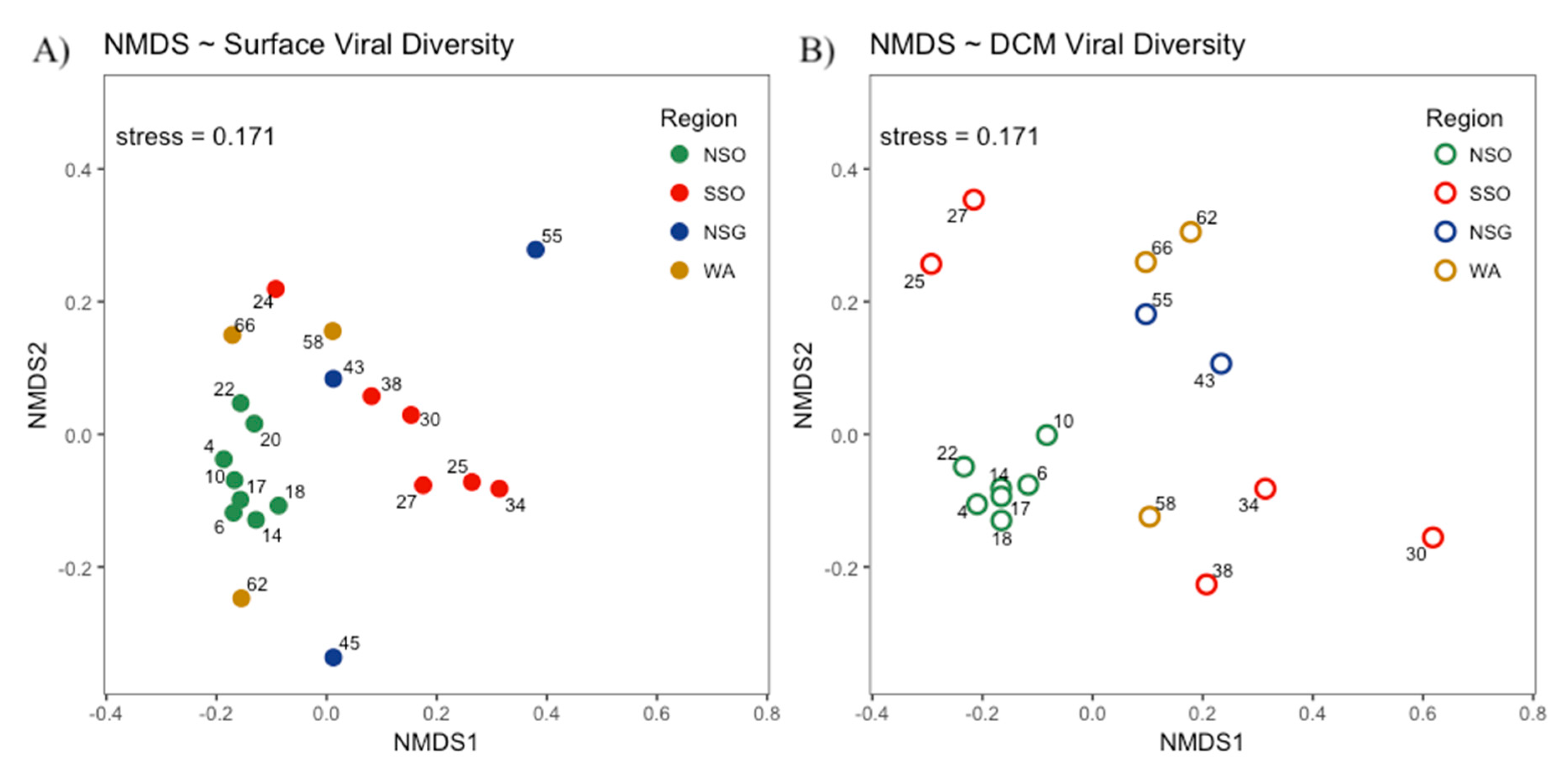
3.3. Viral Community Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suttle, C.A. Viruses in the sea. Nature. 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Fuhrman, J.A.; Middelburg, J.J. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef]
- Behrenfeld, M.J.; Kolber, Z.S. Widespread iron limitation of phytoplankton in the South Pacific Ocean. Science 1999, 283, 840–843. [Google Scholar] [CrossRef]
- Priddle, J.; Nedwell, D.B.; Whitehouse, M.; Reay, D.S.; Savidge, G.; Gilpin, L.; Murphy, E.; Ellis-Evansi, J.C. Re-examining the Antarctic Paradox: Speculation on the Southern Ocean as a nutrient-limited system. Ann. Glac. 1998, 27, 661–668. [Google Scholar] [CrossRef]
- Tréguer, P.; Jacques, G. Review Dynamics of nutrients and phytoplankton, and fluxes of carbon, nitrogen and silicon in the Antarctic Ocean. In Weddell Sea Ecology; Hempel, G., Ed.; Springer: Berlin/Heidelberg, Germany, 1992. [Google Scholar]
- Davison, B.; O’dowd, C.; Hewitt, C.N.; Smith, M.H.; Harrison, R.M.; Peel, D.A.; Wolf, E.; Mulvaney, R.; Schwikowski, M.; Baltenspergert, U. Dimethyl sulfide and its oxidation products in the atmosphere of the Atlantic and Southern Oceans. Atmosph. Environ. 1996, 30, 1895–1906. [Google Scholar] [CrossRef]
- Gowing, M.M.; Garrison, D.L.; Gibson, A.H.; Krupp, J.M.; Jeffries, M.O.; Fritsen, C.H. Bacterial and viral abundance in Ross Sea summer pack ice communities. Mar. Ecol. Prog. Ser. 2004, 279, 3–12. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Gobler, C.; Hutchins, A.; Fisher, N.S.; Cosper, E.M.; Sañudo-Wilhelmy, S.S. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 1997, 42, 1492–1504. [Google Scholar] [CrossRef]
- Poorvin, L.; Rinta-Kanto, J.M.; Hutchins, D.A.; Wilhelm, S. Viral release of Fe and its bioavailability to marine plankton. Limnol. Oceanog. 2004, 49, 1734–1741. [Google Scholar] [CrossRef]
- Mioni, C.E.; Poorvin, L.; Wilhelm, S.W. Virus and sidero- phore-mediated transfer of available Fe between hetero- trophic bacteria: Characterization using an Fe-specific bioreporter. Aquat. Microb. Ecol. 2005, 41, 233–245. [Google Scholar] [CrossRef]
- Evans, C.; Brussaard, C.P.D. Regional Variation in Lytic and Lysogenic Viral Infection in the Southern Ocean and Its Contribution to Biogeochemical Cycling. Appl. Environ. Microbiol. 2012, 78, 6741–6748. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.; Liss, P.S. Dimethylsulfide and climate. In Surface Ocean—Lower Atmosphere Processes; Le Quére, Ć., Saltzman, E.S., Eds.; AGU: Washington, DC, USA, 2009; Volume 187, pp. 197–232. [Google Scholar]
- Shaw, S.L.; Gantt, B.; Meskhidze, N. Production and emissions of marine isoprene and monoterpenes: A review. Adv. Meteor. 2010, 1–24. [Google Scholar] [CrossRef]
- Simó, R. Production of atmospheric sulfur by oceanic plankton: Biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 2001, 16, 287–294. [Google Scholar] [CrossRef]
- Stefels, J.; Steinke, M.; Turner, S.; Malin, G.; Belviso, S. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 2007, 83, 245–275. [Google Scholar] [CrossRef]
- Steiner, P.A.; Sintes, E.; Simó, R.; De Corte, D.; Pfannkuchen, D.M.; Ivancic, I.; Najdek, M.; Herndl, G.J. Seasonal dynamics of marine snow-associated and free-living demethylating bacterial communities in the coastal northern Adriatic Sea. Envion. Microb. Rep. 2019, 11, 699–707. [Google Scholar] [CrossRef]
- Shaw, S.L.; Chisholm, S.W.; Prinn, R.G. Isoprene production by Prochlorococcus, a marine cyanobacterium, and other phytoplankton. Mar. Chem. 2003, 80, 227–245. [Google Scholar] [CrossRef]
- Charlson, R.J.; Lovelock, J.E.; Andreae, M.O.; Warren, S.G. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 1987, 326, 655–661. [Google Scholar] [CrossRef]
- Archer, S.D.; Stelfox-Widdicombe, C.E.; Burkill, P.H.; Malin, G. A dilution approach to quantify the production of dissolved dimethylsulphoniopropionate and dimethyl sulphide due to microzooplankton herbivory. Aquat. Microb. Ecol. 2001, 23, 131–154. [Google Scholar] [CrossRef]
- Keller, M.D. Dimethyl sulfide production and marine phytoplankton: The importance of species composition and cell size. Biol. Oceanogr. 1989, 6, 375–382. [Google Scholar]
- Matrai, P.A.; Keller, M.D. Total organic sulfur and dimethylsulfoniopropionate in marine phytoplankton: Intracellular variations. Mar. Biol. 1994, 119, 61–68. [Google Scholar] [CrossRef]
- Laroche, D.; Vezina, A.F.; Levasseur, M.; Gosselin, M.; Stefels, J.; Keller, M.D.; Matrai, P.A.; Kwint, R.L.J. DMSP synthesis and exudation in phytoplankton: A modeling approach. Mar. Ecol. Prog. Ser. 1999, 180, 37–49. [Google Scholar] [CrossRef]
- Simó, R.; Saló, V.; Almeda, R.; Movilla, J.; Trepat, I.; Saiz, E.; Calbet, A. The quantitative role of microzooplankton grazing in dimethylsulfide (DMS) production in the NW Mediterranean. Biogeochem. 2018, 141, 125–142. [Google Scholar] [CrossRef]
- Bratbak, G.; Levasseur, M.; Michaud, S.; Cantin, G.; Fernández, E.; Heimdal, B.R.; Heldal, M. Viral activity in relation to Emiliania huxleyi blooms: A mechanism of DMSP release? Mar. Ecol. Prog. Ser. 1995, 128, 133–142. [Google Scholar] [CrossRef]
- Hill, R.W.; White, B.A.; Cottrell, M.T.; Dacey, J.W. Virus-mediated total release of dimethylsulfoniopropionate from marine phytoplankton: A potential climate process. Aquat. Microb. Ecol. 1998, 14, 1–6. [Google Scholar] [CrossRef]
- Malin, G.; Wilson, W.H.; Bratbak, G.; Liss, P.S.; Mann, N.H. Elevated production of dimethylsulfide resulting from viral infection of cultures of Phaeocystis pouchetii. Limnol. Oceanogr. 1998, 43, 1389–1393. [Google Scholar] [CrossRef]
- Bonsang, B.; Polle, C.; Lambert, G. Evidence for marine production of isoprene. Geoph. Res. Lett. 1992, 19, 1129–1132. [Google Scholar] [CrossRef]
- Milne, P.J.; Riemer, D.D.; Zika, R.G.; Brand, L.E. Measurement of vertical distribution of isoprene in surface seawater, its chemical fate, and its emission from several phytoplankton monocultures. Mar. Chem. 1995, 48, 237–244. [Google Scholar] [CrossRef]
- Broadgate, W.J.; Liss, P.S.; Penkett, S.A. Seasonal emissions of isoprene and other reactive hydrocarbon gases from the ocean. Geoph. Res. Lett. 1997, 24, 2675–2678. [Google Scholar] [CrossRef]
- Yokouchi, Y.; Li, H.J.; Machida, T.; Aoki, S.; Akimoto, H. Isoprene in the marine boundary layer (Southeast Asian Sea, eastern Indian Ocean, and Southern Ocean): Comparison with dimethyl sulfide and bromoform. J. Geophys. Res. Atm. 1999, 104, 8067–8076. [Google Scholar] [CrossRef]
- Booge, D.; Marandino, C.A.; Schlundt, C.; Palmer, P.I.; Schlundt, M.; Atlas, E.L.; Bracher, A.; Saltzman, E.S.; Wallace, D.W.R. Can simple models predict large scale surface ocean isoprene concentrations? Atmos. Chem. Phys. 2016, 16, 11807–11821. [Google Scholar] [CrossRef]
- Peloquin, J.A.; Smith, W.O. Phytoplankton blooms in the Ross Sea, Antarctica: Interannual variability in magnitude, temporal patterns, and composition. J. Geophys. Res. Ocean. 2007, 112, 1–12. [Google Scholar] [CrossRef]
- Liu, X.; Smith, W.O. Physiochemical controls on phytoplankton distributions in the Ross Sea, Antarctica. J. Mar. Syst. 2012, 94, 135–144. [Google Scholar] [CrossRef]
- Mangoni, O.; Saggiono, V.; Bolinesi, F.; Margiotta, F.; Budillon, G.; Cotroneo, Y.; Misic, C.; Rivaro, P.; Saggiorno, M. Phytoplankton blooms during austral summer in the Ross Sea, Antarctica: Driving factors and trophic implications. PLoS ONE 2017, 12, e0176033. [Google Scholar] [CrossRef]
- Nunes, S.; Latasa, M.; Delgado, M.; Simó, R.; Estrada, M. Phytoplankton community structure in contrasting ecosystems of the Southern Ocean and western Antarctic Peninsula. Deep-Sea-Res. I 2019, 151, 103059. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; González, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. 2014, 12, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Fandino, L.B.; Riemann, L.; Steward, G.F.; Long, R.A.; Azam, F. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 2001, 23, 119–130. [Google Scholar] [CrossRef]
- Fandino, L.B.; Riemann, L.; Steward, G.F.; Azam, F. Population dynamics of Cytophaga-Flavobacteria during marine phytoplankton blooms analyzed by real-time quantitative PCR. Aquat. Microb. Ecol. 2005, 40, 251–257. [Google Scholar] [CrossRef]
- Pinhassi, J.; Sala, M.M.; Havskum, H.; Peters, F.; Guadayol, O.; Malits, A.; Marrasé, C. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 2004, 70, 6753–6766. [Google Scholar] [CrossRef]
- Guixa-Boixereu, N.; Vaqué, D.; Gasol, J.M.; Sánchez-Cámara, J.; Pedrós-Alió, C. Viral distribution and activity in Antarctic waters. Deep Sea Res. II Trop. Stud. Ocean. 2002, 49, 827–845. [Google Scholar] [CrossRef]
- Evans, C.; Pearce, I.; Brussaard, C.P.D. Viral-mediated lysis of microbes and carbon release in the sub-Antarctic and Polar Frontal zones of the Australian Southern Ocean. Environ. Microbiol. 2009, 11, 2924–2934. [Google Scholar] [CrossRef]
- Christaki, U.; Lefèvre, D.; Georges, C.; Colombet, J.; Catala, P.; Courties, C.; Sime-Ngando, T.; Blain, S.; Obernosterer, I. Microbial food web dynamics during spring phytoplankton blooms in the naturally iron-fertilized Kerguelen area (Southern Ocean). Biogeosciences 2014, 11, 6739–6753. [Google Scholar] [CrossRef]
- Vaqué, D.; Boras, J.A.; Torrent-Llagostera, F.; Agustí, S.; Arrieta, J.M.; Lara, E.; Castillo, Y.N.; Duarte, C.M.; Sala, M.M. Viruses and protists induced-mortality of prokaryotes around the antarctic peninsula during the austral summer. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D.; Yau, S.; Williams, T.J.; Allen, M.A.; Brown, M.V.; DeMaere, M.Z.; Lauro, F.M.; Cavicchioli, R. Key microbial drivers in Antarctic aquatic environments. FEMS Microbial. Rev. 2013, 37, 303–333. [Google Scholar] [CrossRef] [PubMed]
- López-Bueno, A.; Tamames, J.; Velázquez, D.; Moya, A.; Quesada, A.; Alcamí, A. High diversity of the viral community from an Antarctic lake. Science 2009, 326, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Bouvy, M.; Bettarel, Y.; Bouvier, C.; Domaizon, I.; Jacquet, S.; Le Floc’h, E.; Montanié, H.; Mostajir, B.; Sime-Ngando, T.; Torréton, J.P.; et al. Trophic interactions between viruses, bacteria and nanoflagellates under various nutrient conditions and simulated climate change. Environ. Microb. 2011, 13, 1842–1857. [Google Scholar] [CrossRef]
- Maranger, R.; Vaqué, D.; Nguyen, D.; Hébert, M.P.; Lara, E. Pan-Arctic patterns of planktonic heterotrophic microbial abundance and processes: Controlling factors and potential impacts of warming. Prog. Oceanogr. 2015, 139, 221–232. [Google Scholar] [CrossRef]
- Finke, J.F.; Hunt, B.P.V.; Winter, C.; Carmack, E.C.; Suttle, C.A. Nutrients and other environmental factors influence virus abundances across oxic and hypoxic marine environments. Viruses 2017, 9, 152. [Google Scholar] [CrossRef]
- Kukkaro, P.; Bamford, D.H. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. Rep. 2009, 1, 71–77. [Google Scholar] [CrossRef]
- Jover, L.F.; Effler, T.C.; Buchan, A.; Wilhelm, S.W.; Weitz, J.S. The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 2014, 12, 519–528. [Google Scholar] [CrossRef]
- Miranda, J.A.; Culley, A.I.; Schvarcz, C.R.; Steward, G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016, 18, 3714–3727. [Google Scholar] [CrossRef]
- Orsi, A.H.; Whitworth, T.; Nowlin, W.D., Jr. On the meridional extent and fronts of the Antarctic Circumpolar Current. Deep-Sea Res. I 1995, 42, 641–673. [Google Scholar] [CrossRef]
- Dall’Osto, M.; Ovadnevaite, J.; Paglione, M.; Beddows, D.C.O.; Ceburnis, D.; Cree, C.; Cortés, P.; Zamanillo, M.; Nunes, S.O.; Pérez, G.L.; et al. Antarctic sea ice region as a source of biogenic organic nitrogen in aerosols. Sci. Repp. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.P.; Koroleff, F. Determination of nutrients. In Methods of Seawater Analysis, 3rd ed.; Grasshoff, K., Kremling, K., Ehrhardt, M., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 1999. [Google Scholar]
- Coble, P.G. Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy. Mar. Chem. 1996, 51, 325–346. [Google Scholar] [CrossRef]
- Yamashita, Y.; Tanoue, E. Chemical characterization of protein-like fluorophores in DOM in relation to aromatic amino acids. Mar. Chem. 2003, 82, 255–271. [Google Scholar] [CrossRef]
- Lawaetz, A.J.; Stedmon, C.A. Fluorescence Intensity Calibration Using the Raman Scatter Peak of Water. App. Spectr. 2009, 63, 936–940. [Google Scholar] [CrossRef]
- Strickland, J.D.H.; Parsons, T.R. A Practical Handbook of Seawater Analysis, 2nd ed.; Fisheries Research Board of Canada: Ottawa, ON, Canada, 1968. [Google Scholar]
- Latasa, M. A simple method to increase sensitivity for RP-HPLC phytoplankton pigment analysis. Limnol. Oceanogr. Methods 2014, 12, 46–53. [Google Scholar] [CrossRef]
- Mackey, M.D.; Mackey, D.J.; Higgins, H.W.; Wright, S.W. CHEMTAX—A program for estimating class abundances from chemical markers: Application to HPLC measurements of phytoplankton. Mar. Ecol. Prog. Ser. 1996, 144, 265–283. [Google Scholar] [CrossRef]
- Brussaard, C.P. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004, 70, 1506–1513. [Google Scholar] [CrossRef]
- Gasol, J.M.; Del Giorgio, P.A. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 2000, 64, 197–224. [Google Scholar] [CrossRef]
- Kirchman, D.; K’nees, E.; Hodson, R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 1985, 49, 599–607. [Google Scholar] [CrossRef]
- Smith, D.C.; Azam, F.A. Simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 1992, 6, 107–114. [Google Scholar]
- Kirchman, D.L. Incorporation of thymidine and leucine in the subarctic Pacific: Application to estimating bacterial production. Mar. Ecol. Prog. Ser. 1992, 1, 301–309. [Google Scholar] [CrossRef]
- Winget, D.M.; Wommack, K.E. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 2008, 74, 2612–2618. [Google Scholar] [CrossRef] [PubMed]
- Lara, E. Viruses in the Marine Environment: Community Dynamics, Phage-Host Interactions and Genomic Structure. Ph.D. Thesis, University of Las Palmas of Canary Island (ULPG), Las Palmas de Gran Canaria, Spain, 2014. [Google Scholar]
- Schlitzer, R. Ocean Data View. 2017. Available online: http://odv.awi.de (accessed on 15 January 2017).
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: Heidelberg, Germany; Dublin, Ireland, 2018. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 20 January 2017).
- Martin, J.; Fitzwater, S.; Gordon, R. Iron Deficiency Limits Phytoplankton Growth in Antarctic Waters. Glob. Biogeochem. Cycles 1990, 4, 5–12. [Google Scholar] [CrossRef]
- Nielsdóttir, M.C.; Bibby, T.S.; Moore, C.M.; Hinz, D.J.; Sanders, R.; Whitehouse, M.; Korb, R.; Achterberg, E.P. Seasonal and spatial dynamics of iron availability in the Scotia Sea. Mar. Chem. 2012, 130, 62–72. [Google Scholar] [CrossRef]
- Short, S.M.; Suttle, C.A. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 2002, 68, 1290–1296. [Google Scholar] [CrossRef]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, 11. [Google Scholar] [CrossRef]
- Brum, J.R.; Hurwitz, B.L.; Schofield, O.; Ducklow, H.W.; Sullivan, M.B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016, 10, 437–449. [Google Scholar] [CrossRef]
- Moreno-Pino, M.; De la Iglesia, R.; Valdivia, N.; Henríquez-Castilo, C.; Galán, A.; Díez, B.; Trefault, N. Variation in coastal Antarctic microbial community composition at sub-mesoscale: Spatial distance or environmental filtering? FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zayed, A.A.; Conceição-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1109–1123. [Google Scholar] [CrossRef]
- De Corte, D.; Sintes, E.; Yokokawa, T.; Reinthaler, T.; Herndl, G.J. Links between viruses and prokaryotes throughout the water column along a North Atlantic latitudinal transect. ISME J. 2012, 6, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.H.; Garcia, J.A.; Herndl, G.J.; Reinthaler, T. Connectivity between surface and deep waters determines prokaryotic diversity in the North Atlantic Deep Water. Envion. Microb. 2016, 18, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, B.L.; Brum, J.R.; Sullivan, M.B. Depth-stratified functional and taxonomic niche specialization in the ‘core’ and ‘flexible’ Pacific Ocean Virome. ISME J. 2015, 9, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef]
- Ruiz-González, C.; Galí, M.; Gasol, J.M.; Simó, R. Sunlight effects on the DMSP-sulfur and leucine assimilation activities of polar heterotrophic bacterioplankton. Biogeochemistry 2012, 110, 57–74. [Google Scholar] [CrossRef]
- Johnson, W.M.; Kido-Soule, M.C.; Kujawinski, E.B. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 2016, 10, 2304–2316. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Suttle, C.A. Viruses and Nutrient Cycles in the Sea—Viruses play critical roles in the structure and function of aquatic food webs. Bioscience 1999, 49, 781–788. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microb. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Gabor, R.S.; Baker, A.; Mcknight, D.M.; Miller, M. Fluorescence indices and their interpretation. In Aquatic Organic Matter Fluorescence; Coble, P., Lead, J., Baker, A., Reynolds, D., Spencer, R., Eds.; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Kiene, R.P.; Service, S.K. Decomposition of dissolved DMSP and DMS in estuarine waters: Dependence on temperature and substrate concentration. Mar. Ecol. Prog. Ser. 1991, 76, 1–11. [Google Scholar] [CrossRef]
- Belviso, S.; Kim, S.K.; Rassoulzadegan, F.; Krajka, B.; Nguyen, B.C.; Mihalopoulos, N.; Buat-Menard, P. Production of dimethylsulfonium propionate (DMSP) and dimethylsulfide (DMS) by a microbial food web. Limnol. Oceanogr. 1990, 35, 1810–1821. [Google Scholar] [CrossRef]
- Kiene, R.P.; Linn, L.J. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulphide in the Gulf of Mexico. Limnol. Oceanogr. 2000, 45, 849–861. [Google Scholar] [CrossRef]
- Tomaru, Y.; Nagasaki, K. Flow Cytometric Detection and Enumeration of DNA and RNA Viruses Infecting Marine Eukaryotic Microalgae. J. Oceanogr. 2007, 63, 215–221. [Google Scholar] [CrossRef]
- Stefels, J.; van Leeuwe, M.A. Effects of iron and light stress on the biochemical composition of Antarctic Phaeocystis sp. (prymnesiophyceae). i. intracellular dmsp concentrations. J. Phycol. Phycol. 1998, 34, 486–495. [Google Scholar] [CrossRef]
- Moore, R.M.; Oram, D.E.; Penkett, S.A. Production of isoprene by marine phytoplankton cultures. Geophys. Res. Lett. 1994, 21, 2507–2510. [Google Scholar] [CrossRef]
- McKay, W.A.; Turner, M.F.; Jones, B.M.R.; Halliwell, C.M. Emissions of hydrocarbons from marine phytoplankton—Some results from controlled laboratory experiments. Atmos. Envion. 1996, 30, 2583–2593. [Google Scholar] [CrossRef]
| AREAS | NSO | SSO | NSG | WA | |||||
|---|---|---|---|---|---|---|---|---|---|
| VARIABLES | Depths | Mean | SD | Mean | SD | Mean | SD | Mean | SD |
| NO3− * | Surface | 27.76 | 1.89 | 28.44 | 3.42 | 18.35 | 5.47 | 18.63 | 0.89 |
| NO2− * | Surface | 0.23 | 0.06 | 0.17 | 0.04 | 0.27 | 0.05 | 0.19 | 0.03 |
| NH4+ * | Surface | 1.11 | 3.51 | 1.03 | 1.03 | 0.78 | 2.02 | 2.48 | 1.87 |
| SiO42− | Surface | 47.21 | 4.08 | 45.72 | 11.96 | 2.02 | 20.73 | 49.75 | 3.66 |
| PO43− * | Surface | 2.04 | 0.21 | 2.15 | 0.28 | 1.42 | 0.43 | 1.81 | 0.16 |
| FDOM (R.U.) | Surface | 18.5 | 4.9 | 7.9 | 1.3 | 15.4 | 3.1 | 40.8 | 6.3 |
| DMSP ** | Surface | 249.1 | 101.2 | 103.5 | 57.2 | 84.4 | 28.0 | 115.7 | 14.61 |
| DMS ** | Surface | 7.0 | 2.6 | 7.8 | 1.6 | 5.8 | 1.0 | 2.1 | 0.6 |
| Isoprene *** | Surface | 9.6 | 1.7 | 4.7 | 1.6 | 76.5 | 8.3 | 12.0 | 0.6 |
| Temperature | Surface | 0.5 | 0.3 | −0.8 | 0.1 | 4.8 | 0.5 | 1.4 | 0.1 |
| DCM | 0.0 | 0.3 | −1.5 | 0.4 | 4.3 | 0.4 | 1.7 | 0.6 | |
| Salinity | Surface | 33.88 | 0.073 | 33.14 | 0.064 | 33.74 | 0.007 | 33.41 | 0.002 |
| DCM | 34.02 | 0.068 | 34.20 | 0.148 | 33.77 | 0.004 | 33.42 | 0.053 | |
| Chl-a | Surface | 1.82 | 0.33 | 0.31 | 0.05 | 5.05 | 1.88 | 4.45 | 0.46 |
| DCM | 1.61 | 0.38 | 0.37 | 0.18 | 7.51 | 3.27 | 3.53 | 0.63 | |
| Cryptophytes | Surface | 288.1 | 110.5 | 11.1 | 4.16 | 28.27 | 14.21 | 898.7 | 268.2 |
| DCM | 117.8 | 112.4 | 9.43 | 4.80 | 34.47 | 19.67 | 676.5 | 203.0 | |
| Diatoms | Surface | 116.9 | 58.17 | 10.9 | 4.94 | 1983.9 | 1451 | 109.9 | 31.81 |
| DCM | 272.2 | 102.2 | 56.82 | 50.46 | 2402 | 1321.2 | 139.4 | 33.33 | |
| Haptophytes | Surface | 274.2 | 80.75 | 80.76 | 18.15 | 373.5 | 127.4 | 629.5 | 199.5 |
| DCM | 225.7 | 50.45 | 85.85 | 41.23 | 369.5 | 150.8 | 583.1 | 151.1 | |
| PA | Surface | 2.4 | 0.5 | 2.9 | 0.6 | 5.4 | 0.9 | 3.4 | 0.9 |
| DCM | 2.6 | 0.5 | 3.6 | 1.4 | 5.2 | 1.0 | 3.3 | 0.5 | |
| PHP | Surface | 0.46 | 0.17 | 0.31 | 0.18 | 1.04 | 0.44 | 0.40 | 0.13 |
| DCM | 0.30 | 0.10 | 0.27 | 0.13 | 1.24 | 0.41 | 0.29 | 0.05 | |
| VA | Surface | 4.5 | 2.0 | 3.0 | 1.5 | 21.5 | 10.7 | 13.5 | 4.1 |
| DCM | 3.1 | 1.7 | 4.0 | 1.5 | 17.9 | 6.6 | 12.0 | 7.2 | |
| V1 | Surface | 2.4 | 1.1 | 1.6 | 0.8 | 8.7 | 4.1 | 6.8 | 2.1 |
| DCM | 1.6 | 0.8 | 1.7 | 0.3 | 6.9 | 2.7 | 6.0 | 3.9 | |
| V2 | Surface | 1.8 | 0.8 | 1.2 | 0.7 | 10.8 | 5.9 | 6.1 | 2.0 |
| DCM | 1.3 | 0.7 | 1.6 | 0.3 | 8.3 | 3.9 | 5.3 | 3.0 | |
| V3 | Surface | 0.3 | 0.1 | 0.2 | 0.1 | 2.2 | 1.2 | 0.7 | 0.3 |
| DCM | 0.2 | 0.2 | 0.3 | 0.1 | 1.7 | 0.8 | 0.7 | 0.3 | |
| Temp | Salinity | PA | Chl-a | PHP | Crypto. | Diatoms | Hapto. | NO3− | NO2− | NH4+ | SiO42− | PO43− | FDOM | DMSP | DMS | Isoprene | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VA | 0.78 | 0.14 | 0.50 | 0.78 | 0.53 | 0.43 | 0.78 | 0.65 | −0.82 | 0.38 | −0.05 | −0.50 | −0.70 | 0.47 | −0.18 | −0.53 | 0.78 |
| V1 | 0.74 | 0.14 | 0.47 | 0.69 | 0.41 | 0.38 | 0.70 | 0.52 | −0.67 | 0.24 | −0.08 | −0.51 | −0.54 | 0.39 | −0.19 | −0.50 | 0.64 |
| V2 | 0.78 | 0.14 | 0.54 | 0.78 | 0.50 | 0.40 | 0.77 | 0.64 | −0.80 | 0.35 | −0.07 | −0.49 | −0.70 | 0.44 | −0.21 | −0.51 | 0.74 |
| V3 | 0.88 | 0.31 | 0.58 | 0.83 | 0.54 | 0.41 | 0.85 | 0.70 | −0.82 | 0.50 | −0.11 | −0.50 | −0.69 | 0.43 | −0.17 | −0.52 | 0.78 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotomayor-Garcia, A.; Montserrat Sala, M.; Ferrera, I.; Estrada, M.; Vázquez-Domínguez, E.; Emelianov, M.; Cortés, P.; Marrasé, C.; Ortega-Retuerta, E.; Nunes, S.; et al. Assessing Viral Abundance and Community Composition in Four Contrasting Regions of the Southern Ocean. Life 2020, 10, 107. https://doi.org/10.3390/life10070107
Sotomayor-Garcia A, Montserrat Sala M, Ferrera I, Estrada M, Vázquez-Domínguez E, Emelianov M, Cortés P, Marrasé C, Ortega-Retuerta E, Nunes S, et al. Assessing Viral Abundance and Community Composition in Four Contrasting Regions of the Southern Ocean. Life. 2020; 10(7):107. https://doi.org/10.3390/life10070107
Chicago/Turabian StyleSotomayor-Garcia, Ana, Maria Montserrat Sala, Isabel Ferrera, Marta Estrada, Evaristo Vázquez-Domínguez, Mikhail Emelianov, Pau Cortés, Cèlia Marrasé, Eva Ortega-Retuerta, Sdena Nunes, and et al. 2020. "Assessing Viral Abundance and Community Composition in Four Contrasting Regions of the Southern Ocean" Life 10, no. 7: 107. https://doi.org/10.3390/life10070107
APA StyleSotomayor-Garcia, A., Montserrat Sala, M., Ferrera, I., Estrada, M., Vázquez-Domínguez, E., Emelianov, M., Cortés, P., Marrasé, C., Ortega-Retuerta, E., Nunes, S., M. Castillo, Y., Serrano Cuerva, M., Sebastián, M., Dall’Osto, M., Simó, R., & Vaqué, D. (2020). Assessing Viral Abundance and Community Composition in Four Contrasting Regions of the Southern Ocean. Life, 10(7), 107. https://doi.org/10.3390/life10070107








