Small Cyclic Peptide for Pyrophosphate Dependent Ligation in Prebiotic Environments
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. In Silico Design and Synthesis
- -
- a linear version of cyc3: lin3, H-RPDDHR-OH,
- -
- a partial alanine scan of cyc3: cyc6, c(RPDDAR); cyc7, c(RPADHR); cyc8, c(RPDAHR),
- -
- duplication of the putative active region of cyc3: cyc9, c(RHDRHD).
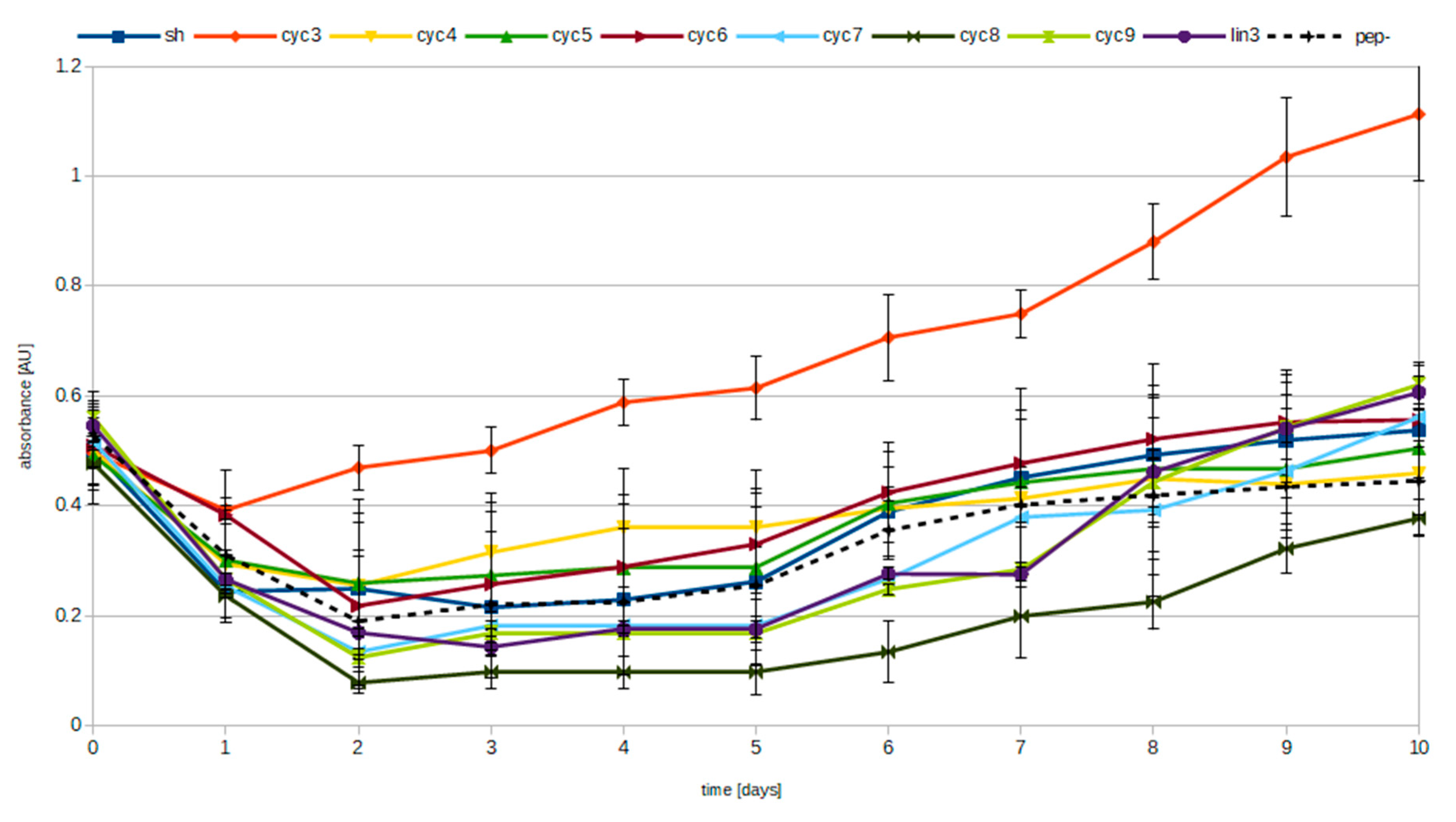
3.2. Pyrophosphate Hydrolysis Assay
3.3. Acylation of Peptide cyc3
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DIPEA | N,N-Diisopropylethylamine |
| DCM | Dichloromethane |
| ESI-TOF | Electrospray ionization time-of-flight |
| Fmoc | Fluorenylmethyloxycarbonyl protecting group |
| HOBT | Hydroxybenzotriazole |
| MALDI-TOF | matrix assisted laser desorption and ionization time-of-flight |
| MES | 2-(N-morpholino)ethanesulfonic acid |
| OtBu | tert-butyl ester group |
| Pbf | 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl group |
| Pi | Phosphate |
| PPi | Pyrophosphate |
| TBTU | 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethylaminium tetrafluoroborate |
| TCA | trichloroacetic acid |
| TFA | trifluoroacetic acid |
| Tris | tris(hydroxymethyl)aminomethane |
| Trt | trityl group |
| standard one-letter and three-letter codes for amino acids are used throughout the paper | |
References
- Damiano, L.; Luisi, P.L. Towards an autopoietic redefinition of life. Orig. Life Evol. Biosph. 2010, 40, 145–149. [Google Scholar] [CrossRef]
- Piast, R.W. Shannon’s information, Bernal’s biopoiesis and Bernoulli distribution as pillars for building a definition of life. J. Theor. Biol. 2019, 470, 101–107. [Google Scholar] [CrossRef]
- Franklin, M.H. The Vital Force: A Study of Bioenergetics; W.H. Freeman & Company: New York, NY, USA, 1987; p. 577. [Google Scholar]
- Piast, R.W.; Wieczorek, R.M. Origin of life and the phosphate transfer catalyst. Astrobiology 2017, 17, 277–285. [Google Scholar] [CrossRef]
- Liu, Z.; Rossi, J.C.; Pascal, R. How Prebiotic Chemistry and Early Life Chose Phosphate. Life 2019, 9, 26. [Google Scholar] [CrossRef]
- Handschuh, G.J.; Orgel, L.E. Struvite and prebiotic phosphorylation. Science 1973, 179, 483–484. [Google Scholar] [CrossRef] [PubMed]
- Pasek, M.A. Schreibersite on the early Earth: Scenarios for prebiotic phosphorylation. Geosci. Front. 2017, 8, 329–335. [Google Scholar] [CrossRef]
- Pasek, M.A.; Kee, T.P.; Bryant, D.E.; Pavlov, A.A.; Lunine, J.I. Production of potentially prebiotic condensed phosphates by phosphorus redox chemistry. Angew. Chem. Int. Ed. 2008, 47, 7918–7920. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Watanabe, H.; Saitoh, M.; Namba, T. Volcanic production of polyphosphates and its relevance to prebiotic evolution. Nature 1991, 352, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Coggins, A.J.; Powner, M.W. Prebiotic synthesis of phosphoenol pyruvate by α-phosphorylation-controlled triose glycolysis. Nat. Chem. 2017, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Kee, T.P.; Bryant, D.E.; Herschy, B.; Marriott, K.E.; Cosgrove, N.E.; Pasek, M.A.; Atlas, Z.D.; Cousins, C.R. Phosphate activation via reduced oxidation state phosphorus (P). Mild routes to condensed-P energy currency molecules. Life 2013, 3, 386–402. [Google Scholar] [CrossRef]
- Sousa-Silva, C.; Seager, S.; Ranjan, S.; Petkowski, J.J.; Zhan, Z.; Hu, R.; Bains, W. Phosphine as a Biosignature Gas in Exoplanet Atmospheres. Astrobiology 2020, 20, 235–268. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Urey, H.C. Organic compound synthesis on the primitive earth. Science 1959, 130, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.W.; Windsor, C.R. Synthesis of amino acids by the heating of formaldehyde and ammonia. Science 1970, 170, 984–986. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Tsuchiya, M.; Oshima, T.; Yanagawa, H. Abiotic synthesis of amino acids and imidazole by proton irradiation of simulated primitive earth atmospheres. Orig. Life Evol. Biosph. 1990, 20, 99–109. [Google Scholar] [CrossRef]
- Engel, M.H.; Macko, S.A. Isotopic evidence for extraterrestrial non-racemic amino acids in the Murchison meteorite. Nature 1997, 389, 265–268. [Google Scholar] [CrossRef]
- Bada, J.L. Amino acid cosmogeochemistry. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1991, 333, 349–358. [Google Scholar]
- Ehrenfreund, P.; Charnley, S.B. Organic molecules in the interstellar medium, comets, and meteorites: A voyage from dark clouds to the early Earth. Annu. Rev. Astron. Astrophys. 2000, 38, 427–483. [Google Scholar] [CrossRef]
- Rode, B.M. Peptides and the origin of life. Peptides 1999, 20, 773–786. [Google Scholar] [CrossRef]
- Gorlero, M.; Wieczorek, R.; Adamala, K.; Giorgi, A.; Schininà, M.E.; Stano, P.; Luisi, P.L. Ser-His catalyses the formation of peptides and PNAs. FEBS Lett. 2009, 583, 153–156. [Google Scholar] [CrossRef]
- Erastova, V.; Degiacomi, M.T.; Fraser, D.G.; Greenwell, H.C. Mineral surface chemistry control for origin of prebiotic peptides. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Brack, A. From interstellar amino acids to prebiotic catalytic peptides: A review. Chem. Biodivers 2007, 4, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, R.; Adamala, K.; Gasperi, T.; Polticelli, F.; Stano, P. Small and random peptides: An unexplored reservoir of potentially functional primitive organocatalysts. The case of seryl-histidine. Life 2017, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Bonfio, C.; Godino, E.; Corsini, M.; de Biani, F.F.; Guella, G.; Mansy, S.S. Prebiotic iron–sulfur peptide catalysts generate a pH gradient across model membranes of late protocells. Nat. Catal. 2018, 1, 616–623. [Google Scholar] [CrossRef]
- Plankensteiner, K.; Righi, A.; Rode, B.M. Glycine and diglycine as possible catalytic factors in the prebiotic evolution of peptides. Orig. Life Evol. B 2002, 32, 225–236. [Google Scholar] [CrossRef]
- Wieczorek, R.; Dörr, M.; Chotera, A.; Luisi, P.L.; Monnard, P.A. Formation of RNA Phosphodiester Bond by Histidine-Containing Dipeptides. ChemBioChem 2013, 14, 217–223. [Google Scholar] [CrossRef]
- Lassila, J.K.; Zalatan, J.G.; Herschlag, D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. [Google Scholar] [CrossRef]
- Storer, A.C.; Cornish-Bowden, A.T. Concentration of MgATP2- and other ions in solution. Calculation of the true concentrations of species present in mixtures of associating ions. Biochem. J. 1976, 159, 1–5. [Google Scholar] [CrossRef]
- Wieczorek, R. On prebiotic ecology, supramolecular selection and autopoiesis. Orig. Life Evol. Biosph. 2012, 42, 445–452. [Google Scholar] [CrossRef]
- Debiec, K.T.; Cerutti, D.S.; Baker, L.R.; Gronenborn, A.M.; Case, D.A.; Chong, L.T. Further along the road less traveled: AMBER ff15ipq, an original protein force field built on a self-consistent physical model. J. Chem. Theory Comput. 2016, 12, 3926–3947. [Google Scholar] [CrossRef]
- Barlos, K.L.; Chatzi, O.; Gatos, D.I.; Stavropoulos, G.E. 2-Chlorotrityl chloride resin. Int. J. Pept. Protein Res. 1991, 37, 513–520. [Google Scholar]
- Chen, P.S.; Toribara, T.T.; Warner, H. Microdetermination of phosphorus. Anal. Chem. 1956, 28, 1756–1758. [Google Scholar] [CrossRef]
- Rizo, J.; Gierasch, L.M. Constrained peptides: Models of bioactive peptides and protein substructures. Annu. Rev. Biochem. 1992, 61, 387–416. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Gill, H.S.; Pfluegl, G.M.; Rotstein, S.H. Structure–function relationships of glutamine synthetases. BBA Protein Struct. Mol. Enzym. 2000, 1477, 122–145. [Google Scholar] [CrossRef]
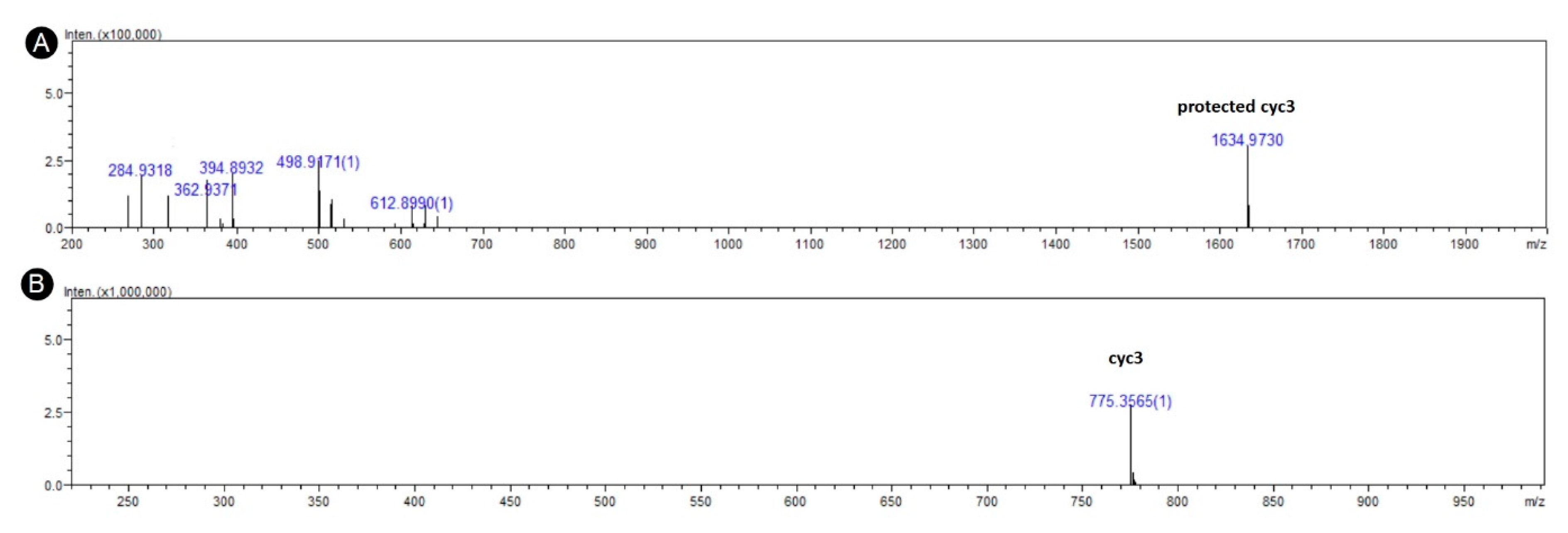

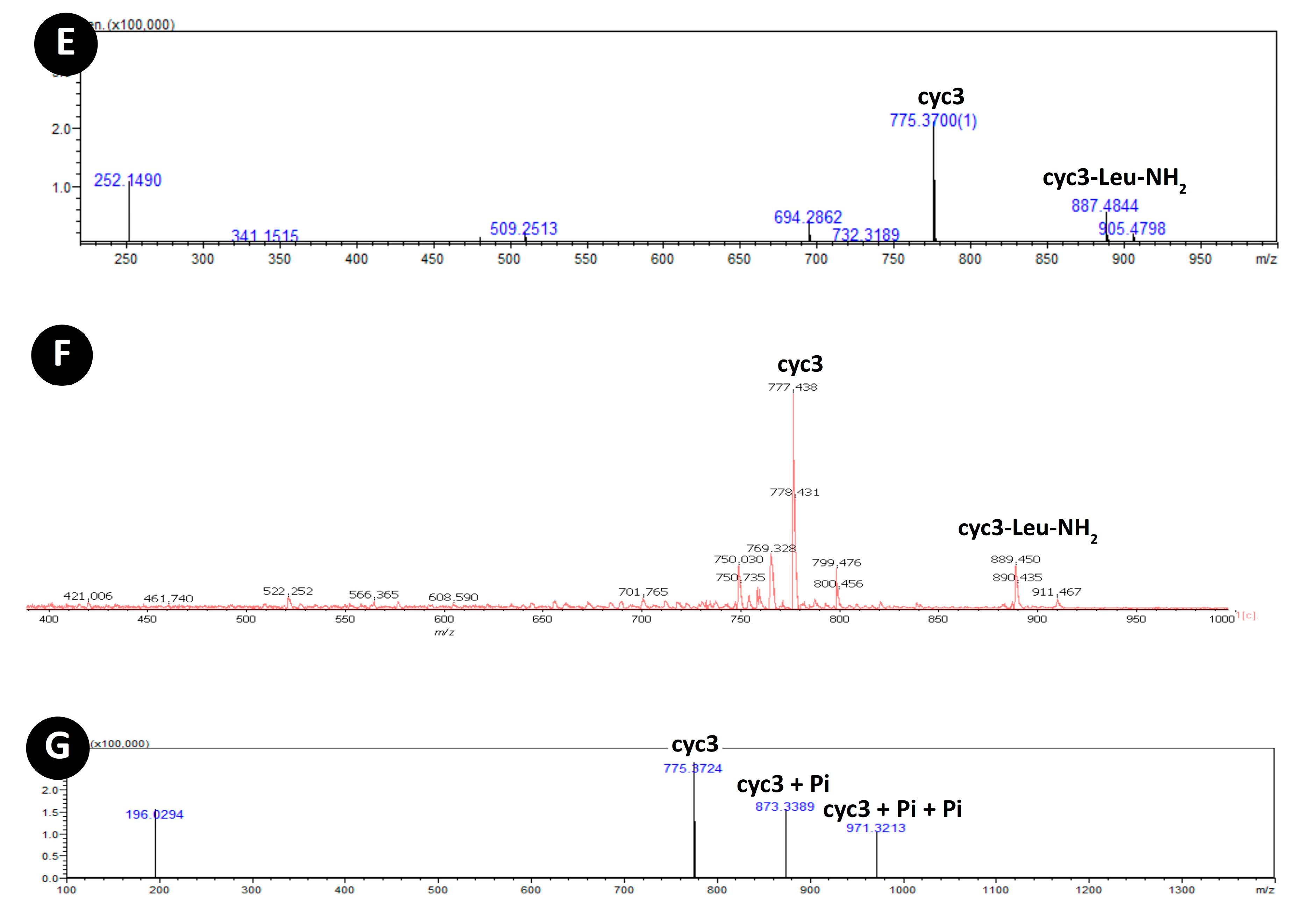
| Peptide | Sequence | Mass (g/mol) | ||
|---|---|---|---|---|
| Calc (g/mol) | TOF (g/mol) | MALDI (g/mol) | ||
| SH | H-SH-OH | 242.23 | 242.1 | 242.26 |
| cyc2 | c(DPDPDR) | 695.69 | -------- | -------- |
| cyc3 | c(RPDDHR) | 776.82 | 776.38 | 776.49 |
| cyc4 | c(RPDDPR) | 736.76 | 736.37 | 736.71 |
| cyc5 | c(DPDDPR) | 695.69 | 695.29 | 695.71 |
| cyc6 | c(RPDDAR) | 710.75 | 710.39 | 710.69 |
| cyc7 | c(RPADHR) | 732.81 | 732.38 | 732.75 |
| cyc8 | c(RPDAHR) | 732.81 | 732.38 | 732.74 |
| cyc9 | c(RHDRHD) | 816.84 | 816.37 | 816.77 |
| lin3 | H-RPDDHR-OH | 794.82 | 794.39 | 794.54 |
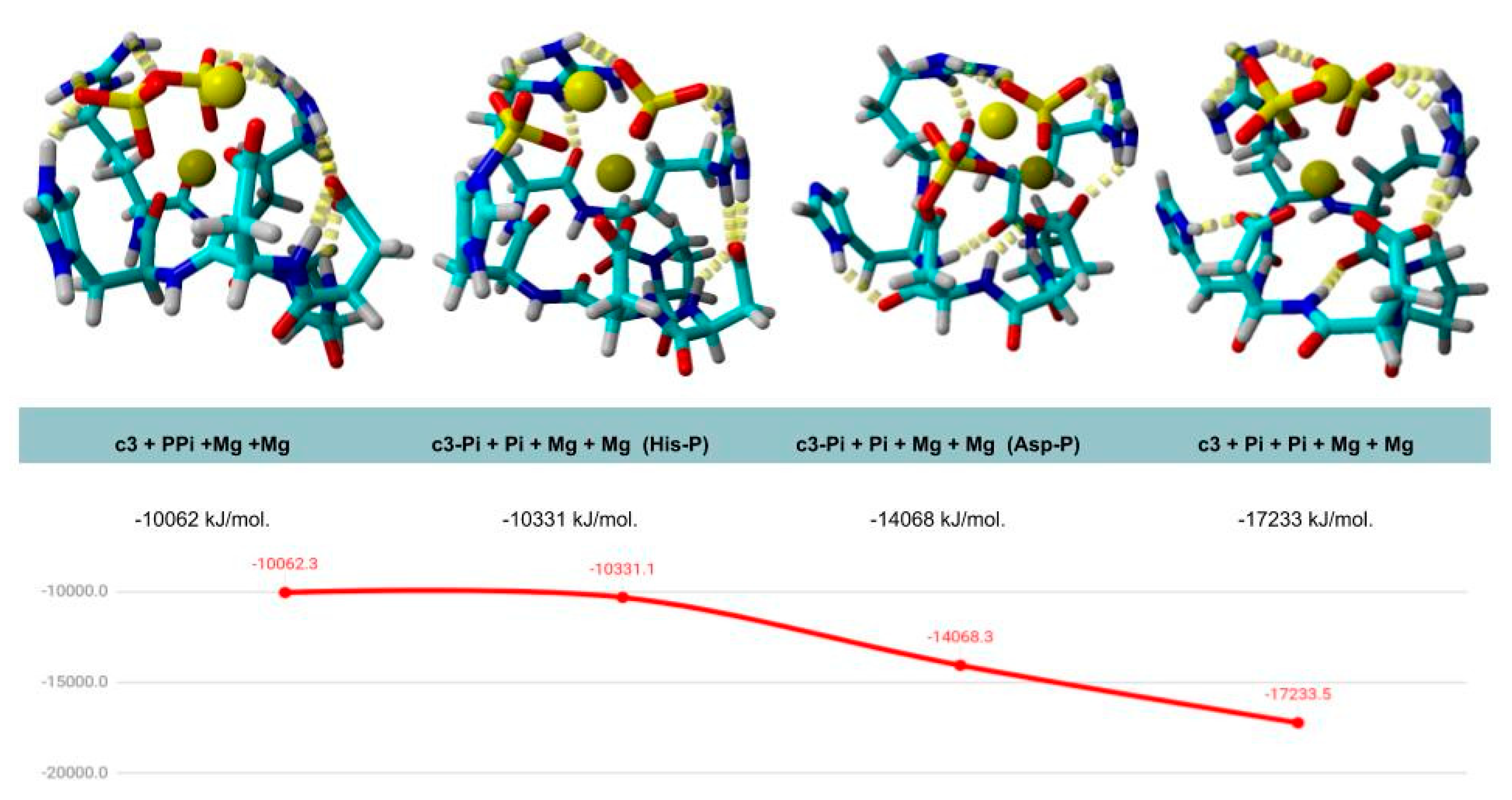
| Peptide Name | Sequence | Energy of Complex Formation with PPi and 2 × Mg2+ |
|---|---|---|
| cyc2 | c(DPDPDR) | −9331 kJ/mol |
| cyc3 | c(RPDDHR) | −10,062 kJ/mol |
| cyc4 | c(RPDDPR) | −10,210 kJ/mol |
| cyc5 | c(DPDDPR) | −9529 kJ/mol |
| Elution Step | SH | cyc3 | cyc4 | cyc5 | cyc6 | cyc7 | cyc8 | cyc9 | lin3 | -pep | -pep -Mg | cyc3 -Mg | cyc3 EDTA | Pi | PPi |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 H2O | 0.0904 | 0.0887 | 0.0538 | 0.0796 | 0.1001 | 0.0689 | 0.0562 | 0.1124 | 0.1015 | 0.1031 | 0.0987 | 0.0833 | 0.2038 | 2.2260 | 0.6642 |
| 2 HCl 0.05 mM | 0.0794 | 0.0999 | 0.0661 | 0.2219 | 0.0889 | 0.0767 | 0.0738 | 0.0885 | 0.1216 | 0.0935 | 0.0905 | 0.0749 | 0.0691 | 1.1816 | 0.3797 |
| 3 HCl 0.05 mM | 0.0452 | 0.0716 | 0.0477 | 0.0882 | 0.0917 | 0.0472 | 0.0529 | 0.0696 | 0.0864 | 0.0397 | 0.0130 | 0.0601 | 0.0616 | 0.3230 | 0.1460 |
| 4 HCl 0.05 mM | 0.0787 | 0.0490 | 0.0698 | 0.0763 | 0.0693 | 0.0586 | 0.0769 | 0.1082 | 0.0946 | 0.0370 | 0.0103 | 0.0654 | 0.0767 | 0.0983 | 0.1314 |
| 5 HCl 0.5 mM | 1.1244 | 0.6759 | 0.9259 | 1.1511 | 0.9625 | 1.3295 | 1.0540 | 1.1386 | 1.0551 | 1.3751 | 0.0000 | 0.2761 | 1.0652 | 0.1386 | 1.4420 |
| 6 HCl 0.5 mM | 0.0133 | 0.0589 | 0.0224 | 0.0608 | 0.0145 | 0.0042 | 0.0208 | 0.0182 | 0.0822 | 0.0357 | 0.0208 | 0.0643 | 0.0158 | 0.0555 | 0.4817 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piast, R.W.; Garstka, M.; Misicka, A.; Wieczorek, R.M. Small Cyclic Peptide for Pyrophosphate Dependent Ligation in Prebiotic Environments. Life 2020, 10, 103. https://doi.org/10.3390/life10070103
Piast RW, Garstka M, Misicka A, Wieczorek RM. Small Cyclic Peptide for Pyrophosphate Dependent Ligation in Prebiotic Environments. Life. 2020; 10(7):103. https://doi.org/10.3390/life10070103
Chicago/Turabian StylePiast, Radosław W., Maciej Garstka, Aleksandra Misicka, and Rafał M. Wieczorek. 2020. "Small Cyclic Peptide for Pyrophosphate Dependent Ligation in Prebiotic Environments" Life 10, no. 7: 103. https://doi.org/10.3390/life10070103
APA StylePiast, R. W., Garstka, M., Misicka, A., & Wieczorek, R. M. (2020). Small Cyclic Peptide for Pyrophosphate Dependent Ligation in Prebiotic Environments. Life, 10(7), 103. https://doi.org/10.3390/life10070103






