Dual RNase and β-lactamase Activity of a Single Enzyme Encoded in Archaea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Antibiotic Susceptibility Testing
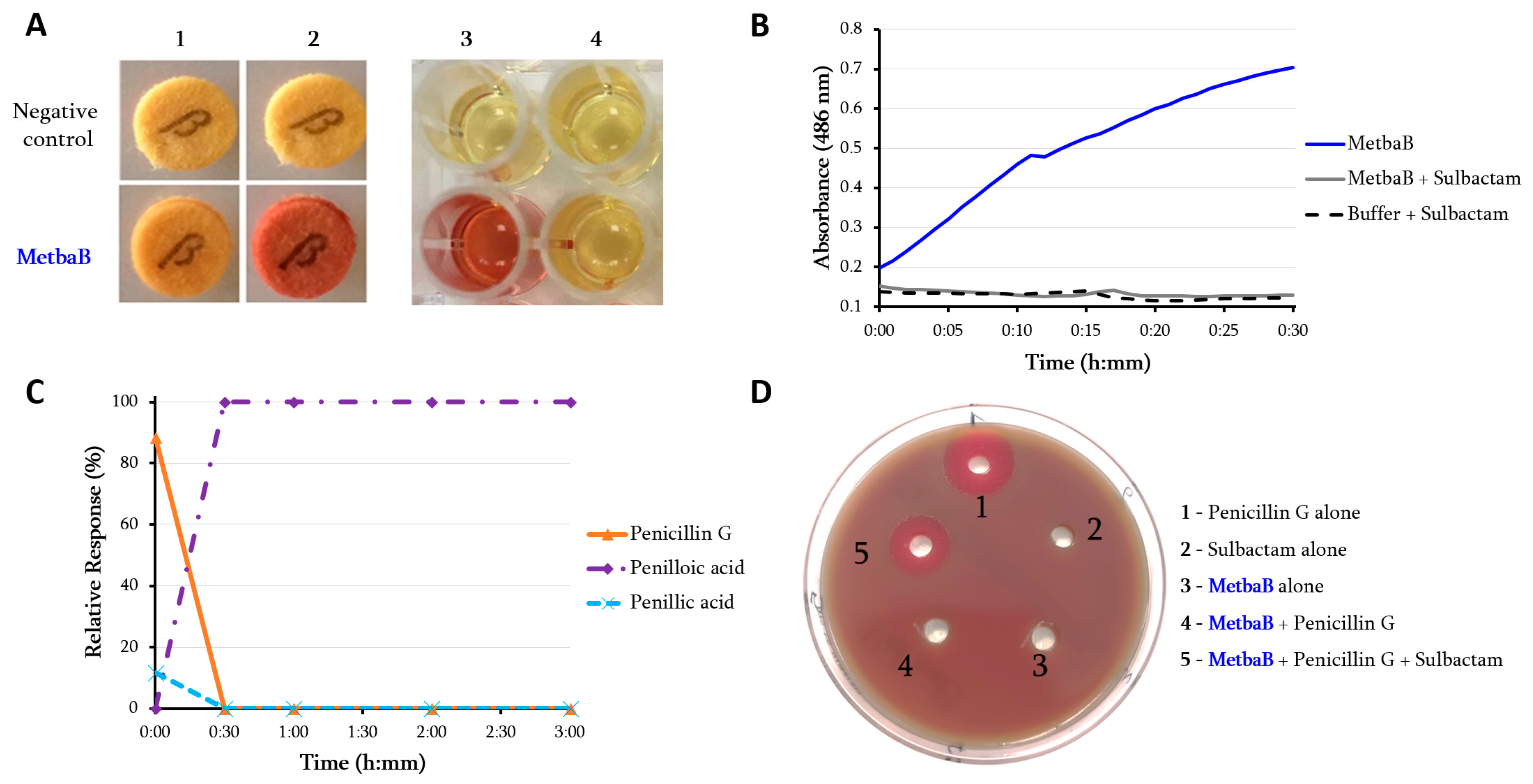
2.3. In Vitro Assessment of the β-Lactamase Activity
2.4. Imipenem Antibiotic Degradation Monitored by Liquid Chromatography-Mass
2.5. DNAse and RNAse Activity Evaluation
2.6. Glyoxalase II Activity Assay
3. Results
3.1. Archaeal Class B Metallo-β-Lactamase
3.2. Characterization of the DNAse and RNAse Activities
3.3. Glyoxalase Activity
3.4. Archaeal Class C-Like β-Lactamases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial Jungle. Natl. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Kim, K.M.; Caetano-Anollés, D.; Kim, M.; Caetano-anolle, G.; Caetano-anolle, D. The phylogenomic roots of modern biochemistry: Origins of proteins, cofactors and protein biosynthesis. J. Mol. Evol. 2012, 74, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, D.; Kim, K.M.; Mittenthal, J.E.; Caetano-Anollés, G.; Caetano-anolle, D.; Caetano-anolle, J.E.M.G. Proteome evolution and the metabolic origins of translation and cellular life. J. Mol. Evol. 2011, 72, 14–33. [Google Scholar] [CrossRef] [PubMed]
- Keshri, V.; Panda, A.; Levasseur, A.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. Phylogenomic Analysis of β-Lactamase in Archaea and Bacteria Enables the Identification of Putative New Members. Genome Biol. Evol. 2018, 10, 1106–1114. [Google Scholar] [CrossRef]
- Pettinati, I.; Brem, J.; Lee, S.Y.; McHugh, P.J.; Schofield, C.J. The Chemical Biology of Human Metallo-β-Lactamase Fold Proteins. Trends Biochem. Sci. 2016, 41, 338–355. [Google Scholar] [CrossRef]
- Aravind, L. An evolutionary classification of the metallo-beta-lactamase fold proteins. Silico Biol. 1999, 1, 69–91. [Google Scholar]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef]
- Horsfall, L.E.; Izougarhane, Y.; Lassaux, P.; Selevsek, N.; Lienard, B.M.R.; Poirel, L.; Kupper, M.B.; Hoffmann, K.M.; Frere, J.M.; Galleni, M.; et al. Broad antibiotic resistance profile of the subclass B3 metallo-β- lactamase GOB-1, a di-zinc enzyme. FEBS J. 2011, 278, 1252–1263. [Google Scholar] [CrossRef]
- Rossolini, G.; Franceschini, N.; Riccio, M.; Mercuri, P.; Perilli, M.; Galleni, M.; Frere, J.; Amicosante, G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: A new molecular class B β-lactamase showing a broad substrate profile. Biochem. J. 1998, 332, 145–152. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, R.; Diene, S.M.; Lopez-rojas, R.; Kempf, M.; Landraud, L. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Khelaifia, S.; Raoult, D.; Drancourt, M. A Versatile Medium for Cultivating Methanogenic Archaea. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Pinault, L.; Keshri, V.; Armstrong, N.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Colson, P.; La Scola, B.; Rolain, J.-M.; et al. Human metallo-β-lactamase enzymes degrade penicillin. Sci. Rep. 2019, 9, 12173. [Google Scholar] [CrossRef]
- Sharma, V.; Colson, P.; Giorgi, R.; Pontarotti, P.; Raoult, D. DNA-Dependent RNA Polymerase Detects Hidden Giant Viruses in Published Databanks. Genome Biol. Evol. 2014, 6, 1603–1610. [Google Scholar] [CrossRef]
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef]
- Opota, O.; Diene, S.M.; Bertelli, C.; Prod’hom, G.; Eckert, P.; Greub, G. Genome of the carbapenemase-producing clinical isolate Elizabethkingia miricola EM_CHUV and comparative genomics with Elizabethkingia meningoseptica and Elizabethkingia anophelis: Evidence for intrinsic multidrug resistance trait of emerging pathogens. Int. J. Antimicrob. Agents. 2016, 1, 93–97. [Google Scholar] [CrossRef]
- Crofts, T.S.; Wang, B.; Spivak, A.; Gianoulis, T.A.; Forsberg, K.J.; Gibson, M.K.; Johnsky, L.A.; Broomall, S.M.; Rosenzweig, C.N.; Skowronski, E.W.; et al. Shared strategies for β-lactam catabolism in the soil microbiome. Nat. Chem. Biol. 2018, 14, 556–564. [Google Scholar] [CrossRef]
- Peimbert, M.; Segovia, L. Evolutionary engineering of β-lactamase activity on a D-Ala D-Ala transpeptidase fold. Protein Eng. 2003, 16, 27–35. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deppenmeier, U.; Johann, A.; Hartsch, T.; Merkl, R.; Schmitz, R.A.; Martinez-Arias, R.; Henne, A.; Wiezer, A.; Bäumer, S.; Jacobi, C.; et al. The Genome of Methanosarcina mazei: Evidence for Lateral Gene Transfer Between Bacteria and Archaea JMMB Research Article. J. Mol. Microbiol. Biotechnol. 2002, 4, 453–461. [Google Scholar] [PubMed]
- Dominski, Z. Nucleases of the Metallo-β-lactamase Family and Their Role in DNA and RNA Metabolism. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; John von Freyend, S.; Sabag-Daigle, A.; Daniels, C.J.; Allers, T.; Marchfelder, A. Assigning a function to a conserved archaeal metallo-β-lactamase from Haloferax volcanii. Extremophiles 2012, 16, 333–343. [Google Scholar] [CrossRef][Green Version]
- Baier, F.; Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014, 426, 2442–2456. [Google Scholar] [CrossRef]
- Adnan, S.; Paterson, D.L.; Lipman, J.; Roberts, J.A. Ampicillin/sulbactam: Its potential use in treating infections in critically ill patients. Int. J. Antimicrob. Agents. 2013, 42, 384–389. [Google Scholar] [CrossRef]
- Keshri, V.; Chabrière, E.; Pinault, L.; Colson, P.; Diene, S.M.; Rolain, J.M.; Raoult, D.; Pontarotti, P. Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons from the β-Lactamases. Int. J. Mol. Sci. 2020, 21, 6260. [Google Scholar] [CrossRef]
- Kato, Y.; Takahashi, M.; Seki, M.; Nashimoto, M.; Shimizu-Ibuka, A. RNA-hydrolyzing activity of metallo-βlactamase IMP-1. PLoS ONE 2020, 15, e0241557. [Google Scholar] [CrossRef]
- Woappi, Y.; Gabani, P.; Singh, A.; Singh, O.V. Antibiotrophs: The complexity of antibiotic-subsisting and antibiotic-resistant microorganisms. Crit. Rev. Microbiol. 2016, 42, 17–30. [Google Scholar] [CrossRef]
- González, T.d.J.B.; Zuidema, T.; Bor, G.; Smidt, H.; van Passel, M.W.J. Study of the aminoglycoside subsistence phenotype of bacteria residing in the gut of humans and zoo animals. Front. Microbiol. 2016, 6, 1–7. [Google Scholar] [CrossRef]
- Xin, Z.; Fengwei, T.; Gang, W.; Xiaoming, L.; Qiuxiang, Z.; Hao, Z.; Wei, C. Isolation, identification and characterization of human intestinal bacteria with the ability to utilize chloramphenicol as the sole source of carbon and energy. FEMS Microbiol. Ecol. 2012, 82, 703–712. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diene, S.M.; Pinault, L.; Armstrong, N.; Azza, S.; Keshri, V.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Rolain, J.-M.; Pontarotti, P.; et al. Dual RNase and β-lactamase Activity of a Single Enzyme Encoded in Archaea. Life 2020, 10, 280. https://doi.org/10.3390/life10110280
Diene SM, Pinault L, Armstrong N, Azza S, Keshri V, Khelaifia S, Chabrière E, Caetano-Anolles G, Rolain J-M, Pontarotti P, et al. Dual RNase and β-lactamase Activity of a Single Enzyme Encoded in Archaea. Life. 2020; 10(11):280. https://doi.org/10.3390/life10110280
Chicago/Turabian StyleDiene, Seydina M., Lucile Pinault, Nicholas Armstrong, Said Azza, Vivek Keshri, Saber Khelaifia, Eric Chabrière, Gustavo Caetano-Anolles, Jean-Marc Rolain, Pierre Pontarotti, and et al. 2020. "Dual RNase and β-lactamase Activity of a Single Enzyme Encoded in Archaea" Life 10, no. 11: 280. https://doi.org/10.3390/life10110280
APA StyleDiene, S. M., Pinault, L., Armstrong, N., Azza, S., Keshri, V., Khelaifia, S., Chabrière, E., Caetano-Anolles, G., Rolain, J.-M., Pontarotti, P., & Raoult, D. (2020). Dual RNase and β-lactamase Activity of a Single Enzyme Encoded in Archaea. Life, 10(11), 280. https://doi.org/10.3390/life10110280








