The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol
Abstract
1. Introduction
2. Results
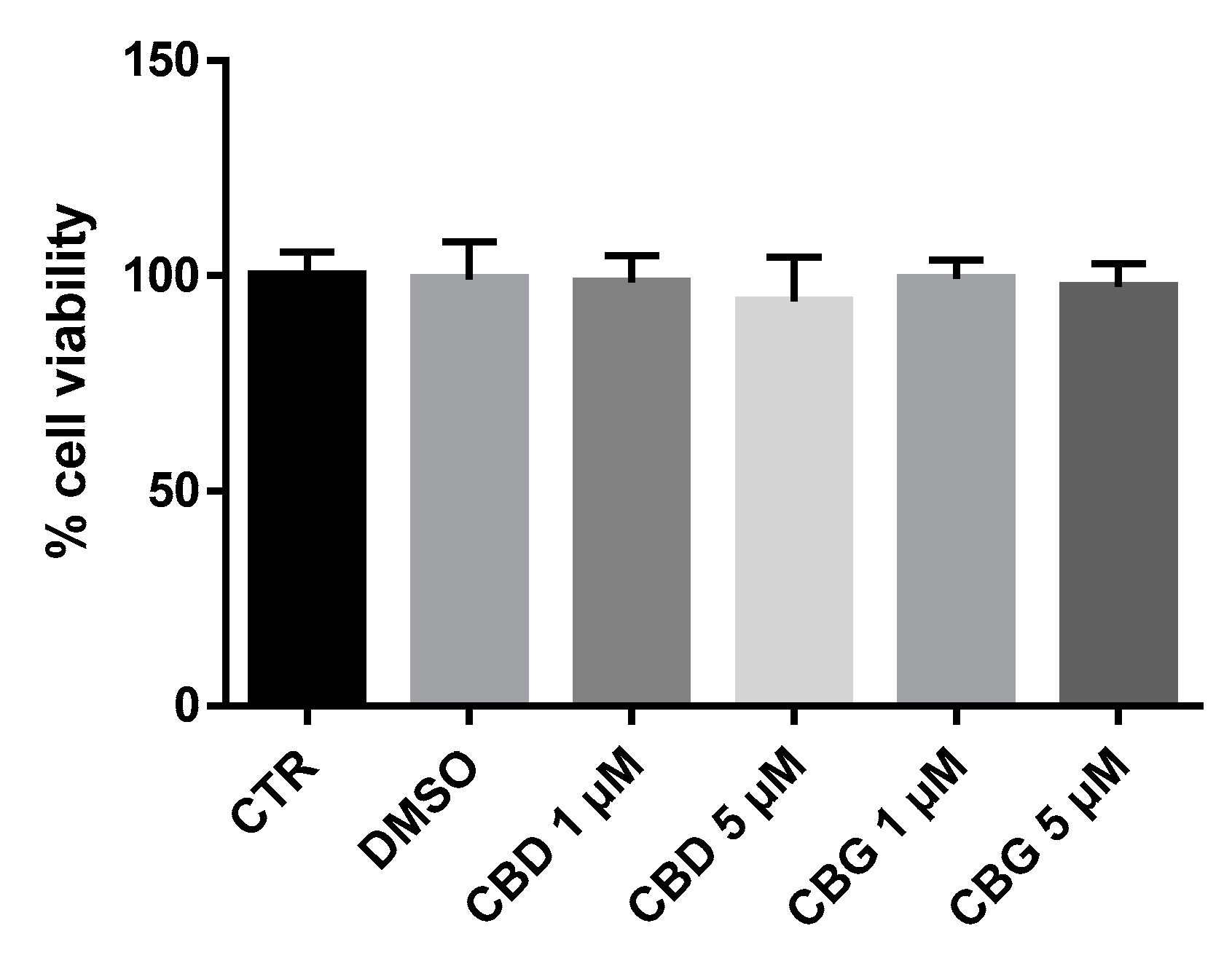
2.1. Cell Viability
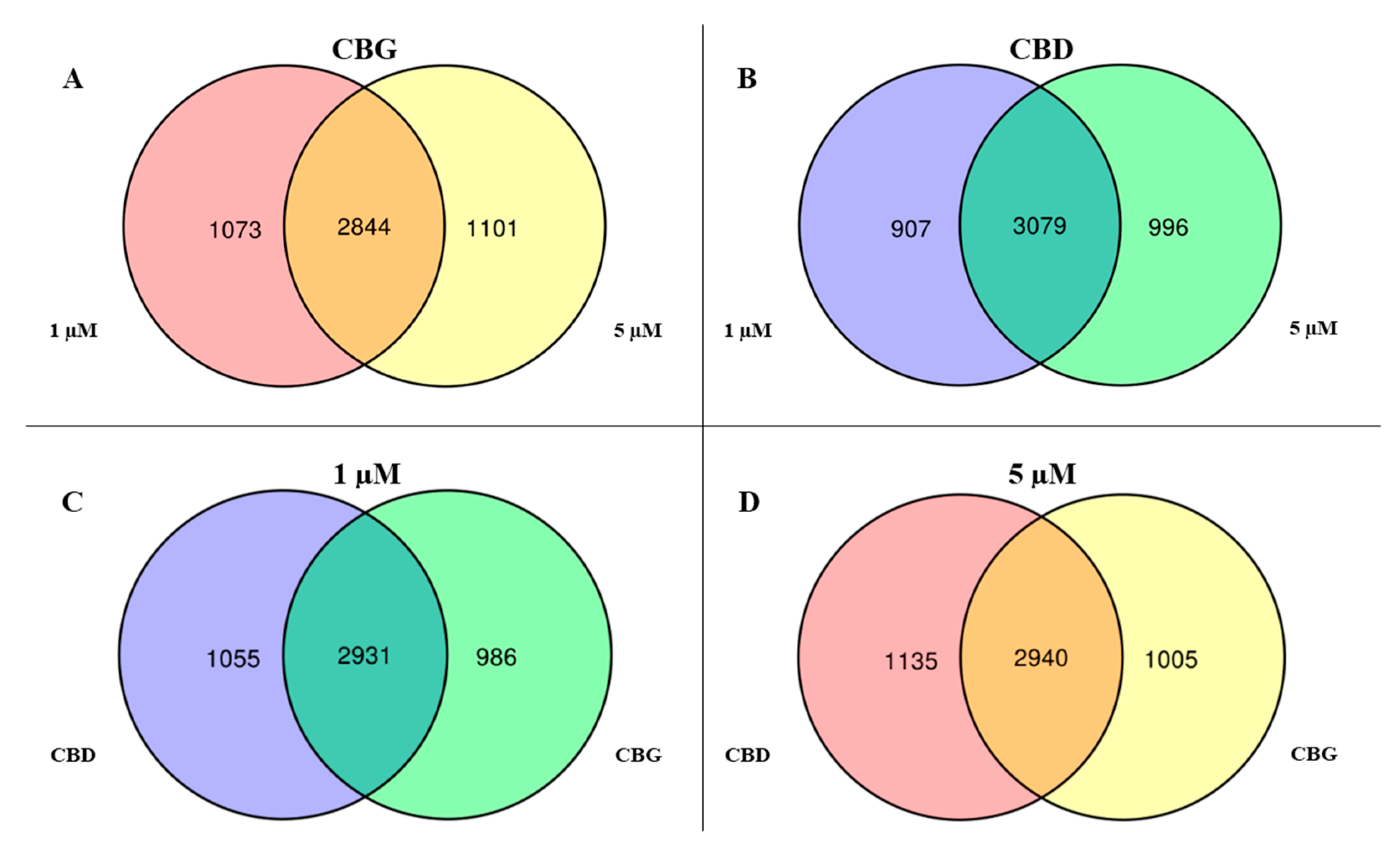
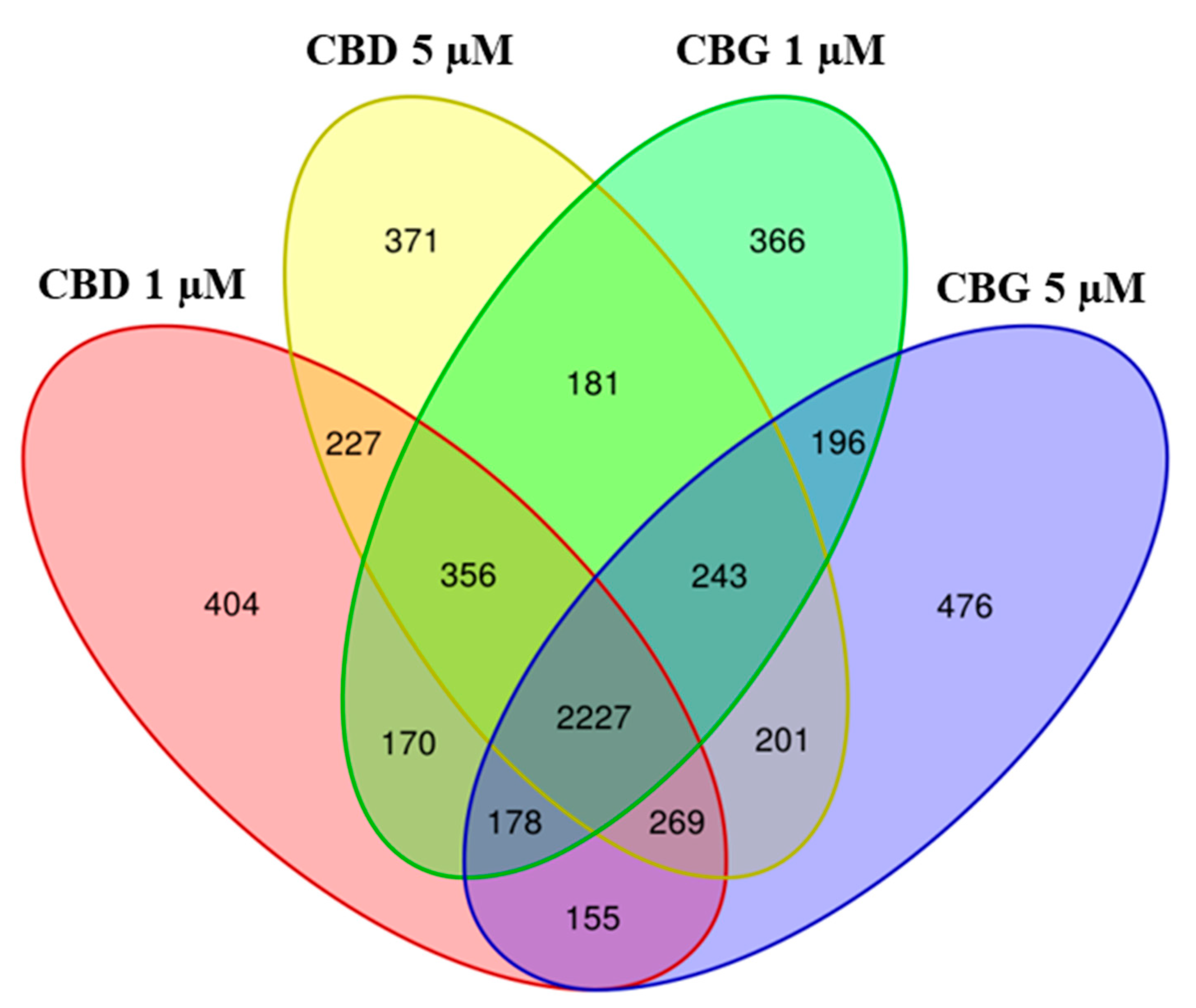
2.2. Differential Expressed Genes Inspection
2.3. KEGG Pathway Analysis
3. Discussion
4. Materials and Methods
4.1. Extraction and Isolation of CBG and CBD
4.2. Cell Culture and Cannabinoid Treatments
4.3. Thiazolyl Blue Tetrazolium Bromide (MTT) Assay
4.4. Statistical Analysis of Cell Viability
4.5. Total RNA Extraction and cDNA Library Preparation
4.6. Transcriptomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CBD | Cannabidiol |
| CBG | Cannabigerol |
| NGS | Next-generation sequencing |
| GABA | Gamma-aminobutyric acid |
| Δ9-THC | Delta-9-tetrahydrocannabinol |
| CBs | Cannabinoids recepors |
| GPCRs | G protein-coupled receptors |
| MTT | Thiazolyl Blue Tetrazolium Bromide |
| CTR-NSC-34 | Control cells |
| DMSO | Dimethyl sulfoxide |
| AKT1 | thymoma viral proto-oncogene 1 |
| ATF4 | activating transcription factor 4 |
| BCL2L1 | BCL2-like 1 |
| EIF2AK3 | eukaryotic translation initiation factor 2 alpha kinase 3 |
| ERN1 | endoplasmatic reticulum (ER) to nucleus signaling 1 |
| MAP3K5 | mitogen-activated protein kinase kinase kinase 5 |
| PARP4 | poly(ADP-ribose) polymerase family, member 4 |
| CASP6 | caspase 6 |
| CASP8 | caspase 8 |
| ADCY5 | adenylate cyclase 5 |
| DLGAP1 | DLG associated protein 1 |
| DRD4 | dopamine receptor D4 |
| GNAI2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| GNB4 | guanine nucleotide binding protein (G protein), beta 4 |
| HAP1 | huntingtin-associated protein 1 |
| PRKCA | protein kinase C, alpha |
| SLC32A1 | solute carrier family 32 (GABA vesicular transporter), member 1 |
| ADCY9 | adenylate cyclase 9 |
| CAMK2B | calcium/calmodulin-dependent protein kinase II, beta |
| CLOCK | circadian locomotor output cycles kaput |
| CREB1 | cAMP responsive element binding protein 1 |
| DRD2 | dopamine receptor D2 |
| GNAL | guanine nucleotide binding protein, alpha stimulating, olfactory type |
| PLD1 | phospholipase D1 |
| PPP3R1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) |
| PRKCB | protein kinase C, beta |
| SHANK1 | SH3 and multiple ankyrin repeat domains 1 |
| SLC1A2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| SLC18A1 | solute carrier family 18 (vesicular monoamine), member 1 |
| SLC38A1 | solute carrier family 38, member 1 |
| Akt1 | serine-threonine protein kinase 1 |
| α | Alpha |
| β | Beta |
| γ | Gamma |
| G αi/o | Inhibiting G proteins |
| AC | Adenylyl cyclase |
| cAMP | Cyclic adenosine monophosphate |
| CREB | cAMP-response element-binding protein |
| CRE | cAMP response element |
| MAPK | mitogen-activated protein kinases |
| CaMK | calmodulin-dependent protein kinase |
| PKA | Protein kinase A |
| EAATs | Excitatory amino acid transporters |
| GlnT | Glutamine transporter |
| DLGAPs | Discs large associated proteins 1 |
| SHANK | Multiple ankyrin repeat domain proteins |
| PDZ | postsynaptic density protein-95, disk-large tumor suppressor protein, zonula occludens-1 |
| SAM | sterile alpha motif |
| mGluRs | Metabotropic glutamate receptors |
| NMDARs | N-Methyl-D-aspartate receptors |
| AMPARs | α-amino-3-hydroxyl-5-methyl-4-isoxazole propionic acid receptors |
| PKC | Protein kinase C |
| PLD1 | Phospholipase D1 |
| HAT-1 | Huntingtin-associated proteins-1 |
| STAR | Spliced Transcripts Alignment to a Reference |
References
- Pellati, F.; Borgonetti, V.; Brighenti, V.; Biagi, M.; Benvenuti, S.; Corsi, L. Cannabis sativa L. and nonpsychoactive cannabinoids: Their chemistry and role against oxidative stress, inflammation, and cancer. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Morales, P.; Reggio, P.H. An update on non-cb1, non-cb2 cannabinoid related g-protein-coupled receptors. Cannabis Cannabinoid Res. 2017, 2, 265–273. [Google Scholar] [CrossRef]
- Laprairie, R.; Bagher, A.; Kelly, M.; Denovan-Wright, E. Cannabidiol is a negative allosteric modulator of the cannabinoid cb1 receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. [Google Scholar] [CrossRef]
- Bih, C.I.; Chen, T.; Nunn, A.V.; Bazelot, M.; Dallas, M.; Whalley, B.J. Molecular targets of cannabidiol in neurological disorders. Neurotherapeutics 2015, 12, 699–730. [Google Scholar]
- Braun, D.A.; Wu, C.J. Antigen discovery and therapeutic targeting in hematologic malignancies. Cancer J. 2017, 23, 115–124. [Google Scholar] [CrossRef]
- Hassel, B.; Tessler, S.; Faull, R.L.; Emson, P.C. Glutamate uptake is reduced in prefrontal cortex in huntington’s disease. Neurochem. Res. 2008, 33, 232–237. [Google Scholar] [CrossRef]
- Olsen, R.W.; DeLorey, T. Gaba and glycine. In Basic Neurochemistry: Molecular, Cellular and Medical Aspects; Lippincott-Raven: Philadelphia, PA, USA, 1999; p. 6. [Google Scholar]
- Moretto, E.; Murru, L.; Martano, G.; Sassone, J.; Passafaro, M. Glutamatergic synapses in neurodevelopmental disorders. Prog. Neuro Psychopharmacol. Biol. Psychiatry 2018, 84, 328–342. [Google Scholar] [CrossRef]
- Stagg, C.J.; Bachtiar, V.; Johansen-Berg, H. The role of gaba in human motor learning. Curr. Biol. 2011, 21, 480–484. [Google Scholar] [CrossRef]
- Nuss, P. Anxiety disorders and gaba neurotransmission: A disturbance of modulation. Neuropsychiatr. Dis. Treat. 2015, 11, 165. [Google Scholar]
- Ziemann, U.; Muellbacher, W.; Hallett, M.; Cohen, L.G. Modulation of practice-dependent plasticity in human motor cortex. Brain 2001, 124, 1171–1181. [Google Scholar] [CrossRef]
- Floyer-Lea, A.; Wylezinska, M.; Kincses, T.; Matthews, P.M. Rapid modulation of gaba concentration in human sensorimotor cortex during motor learning. J. Neurophysiol. 2006, 95, 1639–1644. [Google Scholar] [CrossRef]
- Musella, A.; De Chiara, V.; Rossi, S.; Prosperetti, C.; Bernardi, G.; Maccarrone, M.; Centonze, D. Trpv1 channels facilitate glutamate transmission in the striatum. Mol. Cell. Neurosci. 2009, 40, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.S.; Stella, N.; Catterall, W.A.; Westenbroek, R.E. Cannabidiol attenuates seizures and social deficits in a mouse model of dravet syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, 11229–11234. [Google Scholar] [CrossRef] [PubMed]
- Al-Hussaini, M.; Rettig, M.P.; Ritchey, J.K.; Karpova, D.; Uy, G.L.; Eissenberg, L.G.; Gao, F.; Eades, W.C.; Bonvini, E.; Chichili, G.R.; et al. Targeting cd123 in acute myeloid leukemia using a t-cell-directed dual-affinity retargeting platform. Blood 2016, 127, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, A.; Liu, Q.; Li, T.; Yuan, X.; Han, X.; Wu, K. Chimeric antigen receptor t cells: A novel therapy for solid tumors. J. Hematol. Oncol. 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Servier Medical Art by Servier. Available online: http://smart.servier.com/ (accessed on 27 August 2020).
- Creative Commons Attribution 3.0 Unported License. Available online: https://creativecommons.org/licenses/by/3.0 (accessed on 27 August 2020).
- Mirzaei, H.R.; Rodriguez, A.; Shepphird, J.; Brown, C.E.; Badie, B. Chimeric antigen receptors t cell therapy in solid tumor: Challenges and clinical applications. Front. Immunol. 2017, 8, 1850. [Google Scholar] [CrossRef]
- Walseng, E.; Koksal, H.; Sektioglu, I.M.; Fane, A.; Skorstad, G.; Kvalheim, G.; Gaudernack, G.; Inderberg, E.M.; Walchli, S. A tcr-based chimeric antigen receptor. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ilieva, K.M.; Cheung, A.; Mele, S.; Chiaruttini, G.; Crescioli, S.; Griffin, M.; Nakamura, M.; Spicer, J.F.; Tsoka, S.; Lacy, K.E.; et al. Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front. Immunol. 2017, 8, 1911. [Google Scholar] [CrossRef]
- Caruso, H.G.; Heimberger, A.B.; Cooper, L.J.N. Steering car t cells to distinguish friend from foe. Oncoimmunology 2019, 8, e1271857. [Google Scholar] [CrossRef]
- Van den Pol, A. Weighing the role of hypothalamic feeding neurotransmitters. Neuron 2003, 40, 1059–1061. [Google Scholar] [CrossRef]
- Rodriguez-Cueto, C.; Santos-Garcia, I.; Garcia-Toscano, L.; Espejo-Porras, F.; Bellido, M.; Fernandez-Ruiz, J.; Munoz, E.; de Lago, E. Neuroprotective effects of the cannabigerol quinone derivative vce-003.2 in sod1(g93a) transgenic mice, an experimental model of amyotrophic lateral sclerosis. Biochem. Pharmacol. 2018, 157, 217–226. [Google Scholar] [CrossRef]
- Pretzsch, C.M.; Freyberg, J.; Voinescu, B.; Lythgoe, D.; Horder, J.; Mendez, M.A.; Wichers, R.; Ajram, L.; Ivin, G.; Heasman, M. Effects of cannabidiol on brain excitation and inhibition systems; a randomised placebo-controlled single dose trial during magnetic resonance spectroscopy in adults with and without autism spectrum disorder. Neuropsychopharmacology 2019, 44, 1398–1405. [Google Scholar] [CrossRef]
- Cifelli, P.; Ruffolo, G.; De Felice, E.; Alfano, V.; van Vliet, E.A.; Aronica, E.; Palma, E. Phytocannabinoids in neurological diseases: Could they restore a physiological gabaergic transmission? Int. J. Mol. Sci. 2020, 21, 723. [Google Scholar] [CrossRef]
- Guo, Y.; He, X.; Zhao, L.; Liu, L.; Song, H.; Wang, X.; Xu, J.; Ju, X.; Guo, W.; Zhu, X. Gβ2 regulates the multipolar-bipolar transition of newborn neurons in the developing neocortex. Cereb. Cortex 2017, 27, 3414–3426. [Google Scholar] [CrossRef]
- Schwindinger, W.F.; Robishaw, J.D. Heterotrimeric g-protein βγ-dimers in growth and differentiation. Oncogene 2001, 20, 1653–1660. [Google Scholar] [CrossRef]
- Rosskopf, D.; Nikula, C.; Manthey, I.; Joisten, M.; Frey, U.; Kohnen, S.; Siffert, W. The human g protein beta4 subunit: Gene structure, expression, ggamma and effector interaction. FEBS Lett. 2003, 544, 27–32. [Google Scholar] [CrossRef][Green Version]
- Piniella, D.; Martinez-Blanco, E.; Ibanez, I.; Bartolome-Martin, D.; Porlan, E.; Diez-Guerra, J.; Gimenez, C.; Zafra, F. Identification of novel regulatory partners of the glutamate transporter glt-1. Glia 2018, 66, 2737–2755. [Google Scholar] [CrossRef]
- Lok, K.-C.; Fu, A.K.Y.; Ip, F.C.F.; Wong, Y.H.; Ip, N. Expression of g protein β subunits in rat skeletal muscle after nerve injury: Implication in the regulation of neuregulin signaling. Neuroscience 2007, 146, 594–603. [Google Scholar] [CrossRef]
- Seino, S.; Shibasaki, T. Pka-dependent and pka-independent pathways for camp-regulated exocytosis. Physiol. Rev. 2005, 85, 1303–1342. [Google Scholar] [CrossRef]
- Fahlke, C.; Kortzak, D.; Machtens, J.-P. Molecular physiology of eaat anion channels. Pflügers Arch. Eur. J. Physiol. 2016, 468, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Rothstein, J.D.; Dykes-Hoberg, M.; Pardo, C.A.; Bristol, L.A.; Jin, L.; Kuncl, R.W.; Kanai, Y.; Hediger, M.A.; Wang, Y.; Schielke, J.P. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron 1996, 16, 675–686. [Google Scholar] [CrossRef]
- Varoqui, H.; Zhu, H.; Yao, D.; Ming, H.; Erickson, J.D. Cloning and functional identification of a neuronal glutamine transporter. J. Biol. Chem. 2000, 275, 4049–4054. [Google Scholar] [CrossRef] [PubMed]
- Solbu, T.T.; Bjørkmo, M.; Berghuis, P.; Harkany, T.; Chaudhry, F.A. Sat1, a glutamine transporter, is preferentially expressed in gabaergic neurons. Front. Neuroanat. 2010, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, E.C.; Bariselli, S.; Bellone, C. Synaptic basis of social dysfunction: A focus on postsynaptic proteins linking group-i m g lu r s with ampar s and nmdar s. Eur. J. Neurosci. 2014, 39, 1114–1129. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, A.H.; Rasmussen, H.B.; Silahtaroglu, A. The dlgap family: Neuronal expression, function and role in brain disorders. Mol. Brain 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Naisbitt, S.; Kim, E.; Tu, J.C.; Xiao, B.; Sala, C.; Valtschanoff, J.; Weinberg, R.J.; Worley, P.F.; Sheng, M. Shank, a novel family of postsynaptic density proteins that binds to the nmda receptor/psd-95/gkap complex and cortactin. Neuron 1999, 23, 569–582. [Google Scholar] [CrossRef]
- Sheng, M.; Kim, E. The shank family of scaffold proteins. J. Cell Sci. 2000, 113, 1851–1856. [Google Scholar]
- Grabrucker, A.M.; Schmeisser, M.J.; Schoen, M.; Boeckers, T.M. Postsynaptic prosap/shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011, 21, 594–603. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Ehlers, M.D. Modeling autism by shank gene mutations in mice. Neuron 2013, 78, 8–27. [Google Scholar] [CrossRef]
- Kreienkamp, H.J. Scaffolding proteins at the postsynaptic density: Shank as the architectural framework. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 365–380. [Google Scholar]
- Kornau, H.-C.; Schenker, L.T.; Kennedy, M.B.; Seeburg, P.H. Domain interaction between nmda receptor subunits and the postsynaptic density protein psd-95. Science 1995, 269, 1737–1740. [Google Scholar] [CrossRef]
- Chen, L.; Chetkovich, D.M.; Petralia, R.S.; Sweeney, N.T.; Kawasaki, Y.; Wenthold, R.J.; Bredt, D.S.; Nicoll, R.A. Stargazin regulates synaptic targeting of ampa receptors by two distinct mechanisms. Nature 2000, 408, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Sala, C.; Piëch, V.; Wilson, N.R.; Passafaro, M.; Liu, G.; Sheng, M. Regulation of dendritic spine morphology and synaptic function by shank and homer. Neuron 2001, 31, 115–130. [Google Scholar] [CrossRef]
- Mei, Y.; Monteiro, P.; Zhou, Y.; Kim, J.-A.; Gao, X.; Fu, Z.; Feng, G. Adult restoration of shank3 expression rescues selective autistic-like phenotypes. Nature 2016, 530, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Peça, J.; Feliciano, C.; Ting, J.T.; Wang, W.; Wells, M.F.; Venkatraman, T.N.; Lascola, C.D.; Fu, Z.; Feng, G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 2011, 472, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Roussignol, G.; Ango, F.; Romorini, S.; Tu, J.C.; Sala, C.; Worley, P.F.; Bockaert, J.; Fagni, L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J. Neurosci. 2005, 25, 3560–3570. [Google Scholar] [CrossRef] [PubMed]
- Mansuy, I.M. Calcineurin in memory and bidirectional plasticity. Biochem. Biophys. Res. Commun. 2003, 311, 1195–1208. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Tomizawa, K.; Oda, Y.; Wei, F.-Y.; Lu, Y.-F.; Matsushita, M.; Li, S.-T.; Moriwaki, A.; Matsui, H. Critical role of calpain-mediated cleavage of calcineurin in excitotoxic neurodegeneration. J. Biol. Chem. 2004, 279, 4929–4940. [Google Scholar] [CrossRef]
- Kittler, J.T.; Thomas, P.; Tretter, V.; Bogdanov, Y.D.; Haucke, V.; Smart, T.G.; Moss, S.J. Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating γ-aminobutyric acid type a receptor membrane trafficking. Proc. Natl. Acad. Sci. USA 2004, 101, 12736–12741. [Google Scholar] [CrossRef]
- Vithlani, M.; Terunuma, M.; Moss, S.J. The dynamic modulation of gabaa receptor trafficking and its role in regulating the plasticity of inhibitory synapses. Physiol. Rev. 2011, 91, 1009–1022. [Google Scholar] [CrossRef]
- Song, M.; Messing, R. Protein kinase c regulation of gaba a receptors. Cell. Mol. Life Sci. CMLS 2005, 62, 119–127. [Google Scholar] [CrossRef]
- Taglialatela-Scafati, O.; Pagani, A.; Scala, F.; De Petrocellis, L.; Di Marzo, V.; Grassi, G.; Appendino, G. Cannabimovone, a cannabinoid with a rearranged terpenoid skeleton from hemp. Eur. J. Org. Chem. 2010, 2010, 2067–2072. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Pollastro, F.; Grassi, G.; Bramanti, P.; Mazzon, E. In vitro model of neuroinflammation: Efficacy of cannabigerol, a non-psychoactive cannabinoid. Int. J. Mol. Sci. 2018, 19, 1992. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Pizzicannella, J.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Extracellular vesicles derived from human gingival mesenchymal stem cells: A transcriptomic analysis. Genes 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal rna-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
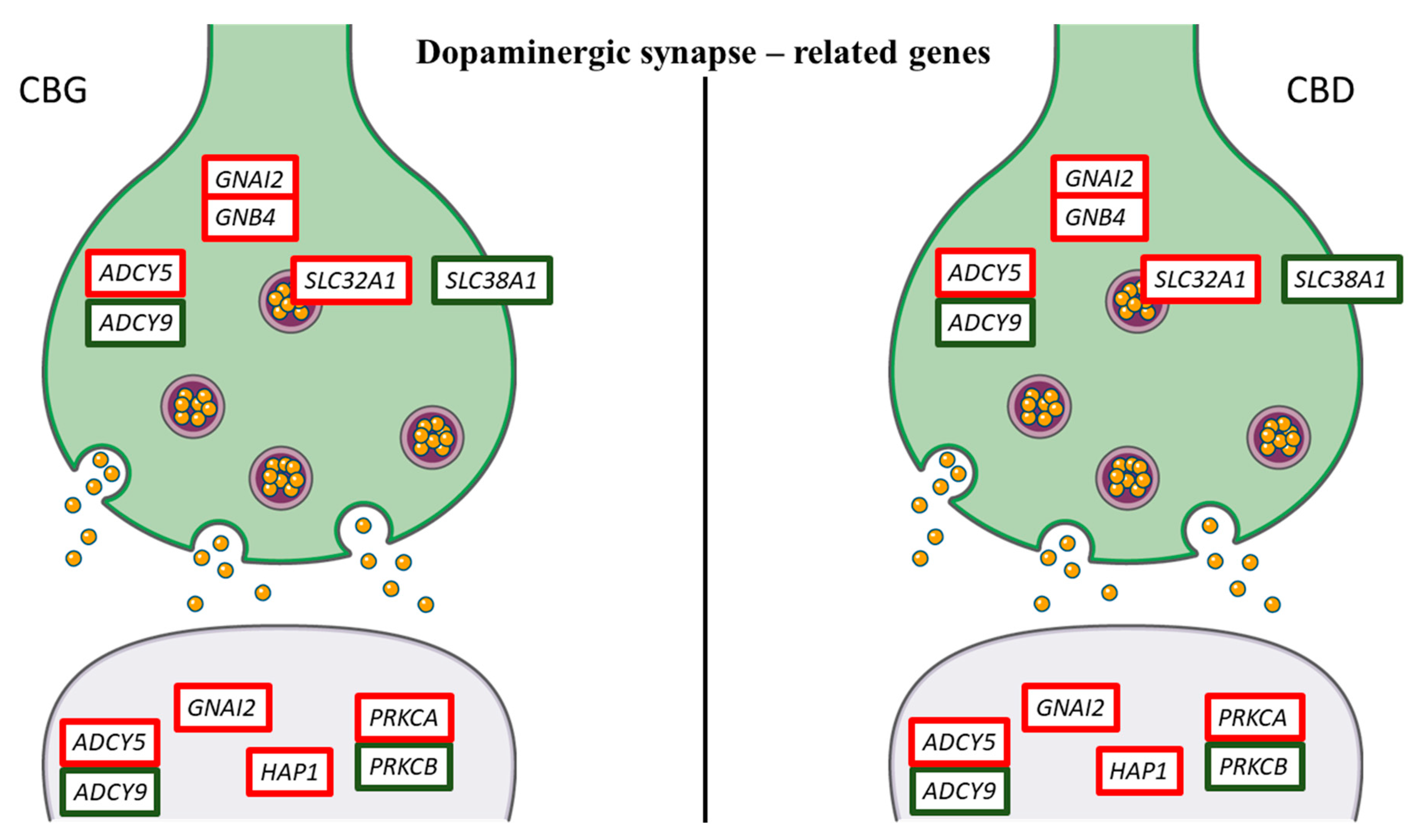
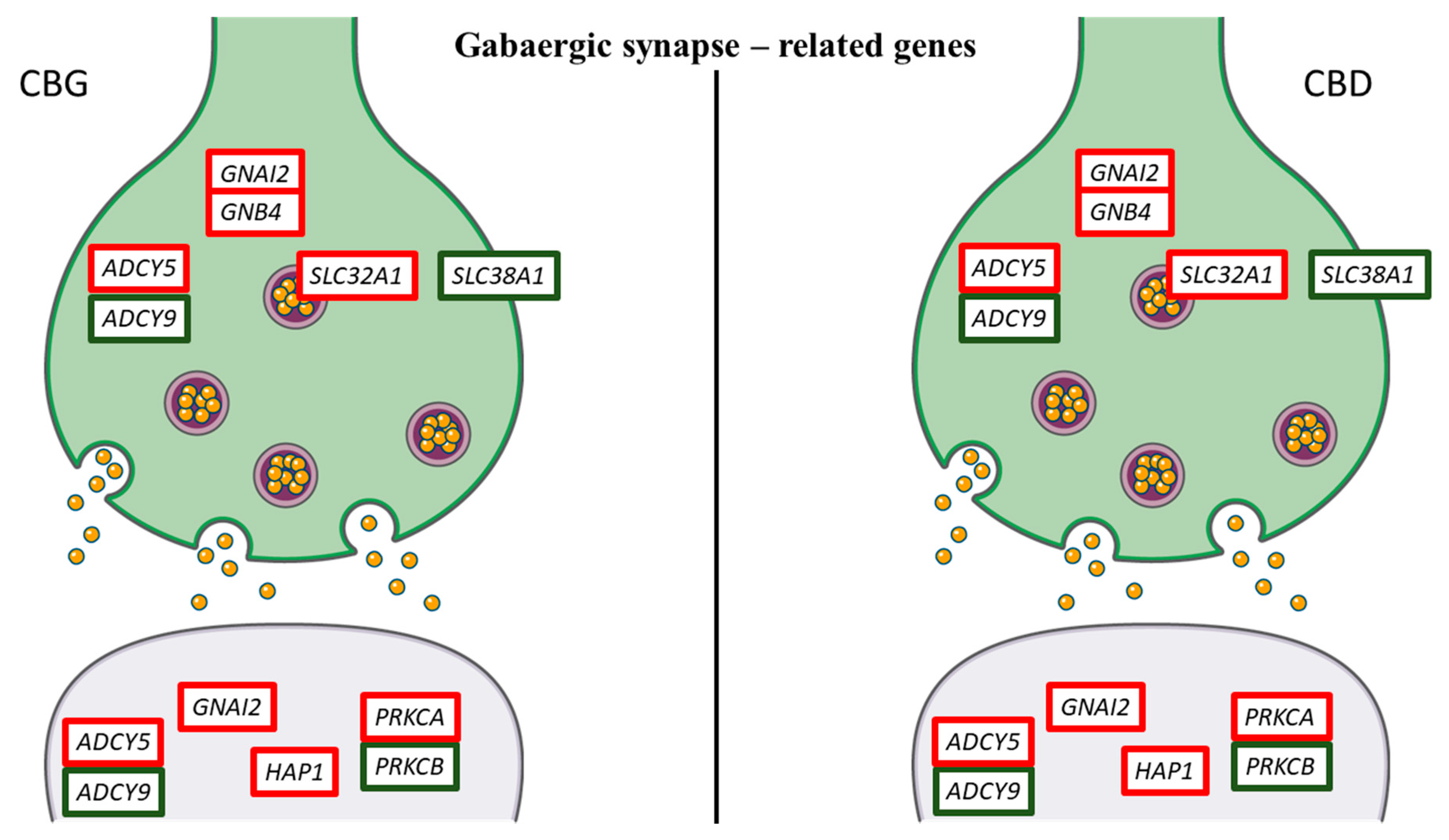
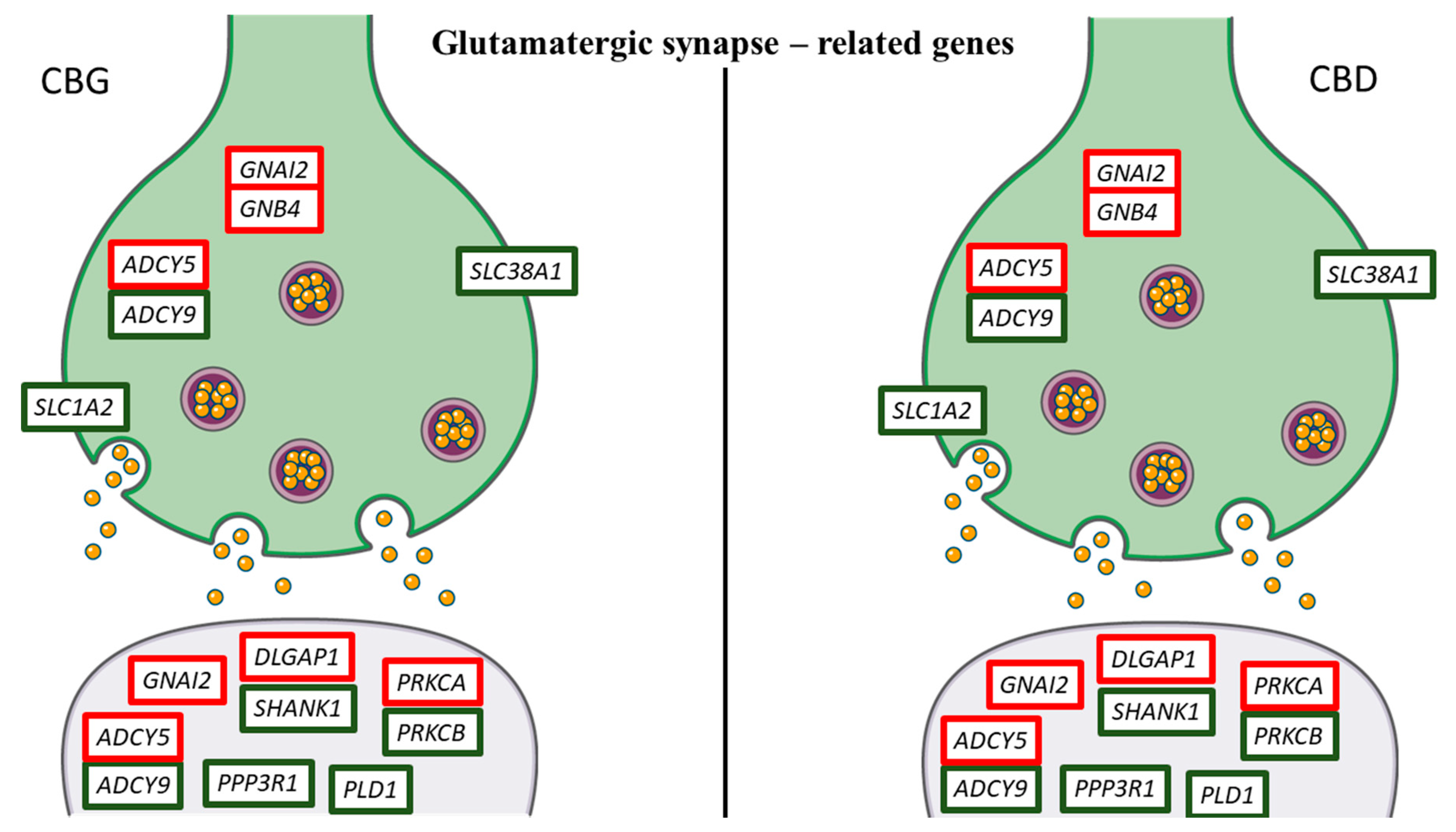
| Gene | Name | Fold Change CBG 1 µM | Fold Change CBG 5 µM | Fold Change CBD 1 µM | Fold Change CBD 5 µM |
|---|---|---|---|---|---|
| AKT1 | thymoma viral proto-oncogene 1 | 0.80 | 0.74 | 0.84 | 0.96 |
| ATF4 | activating transcription factor 4 | 0.71 | 0.74 | 0.80 | 0.76 |
| BCL2L1 | BCL2-like 1 | 0.97 | 1.20 | 0.96 | 1.29 |
| EIF2AK3 | eukaryotic translation initiation factor 2 alpha kinase 3 | −1.84 | −1.13 | −1.82 | −1.60 |
| ERN1 | endoplasmatic reticulum (ER) to nucleus signaling 1 | −0.91 | −0.91 | −0.71 | −0.84 |
| MAP3K5 | mitogen-activated protein kinase kinase kinase 5 | −1.30 | −0.97 | −0.98 | −0.82 |
| PARP4 | poly(ADP-ribose) polymerase family, member 4 | −1.60 | −1.59 | −1.28 | −1.50 |
| CASP6 | caspase 6 | - | - | - | −3.58 |
| CASP8 | caspase 8 | −1.39 | - | - | −1.81 |
| Gene | Name | Fold Change CBG 1 µM | Fold Change CBG 5 µM | Fold Change CBD 1 µM | Fold Change CBD 5 µM | Pathway |
|---|---|---|---|---|---|---|
| ADCY5 | adenylate cyclase 5 | 5.86 | 4.74 | 5.25 | 6.03 | Dopaminergic, GABAergic, Glutamatergic synapse |
| ADCY9 | adenylate cyclase 9 | −1.15 | −1.28 | −1.1 | −1.31 | GABAergic, Glutamatergic synapse |
| AKT1 | thymoma viral proto-oncogene 1 | 0.80 | 0.74 | 0.84 | 0.96 | Dopaminergic synapse |
| ATF4 | activating transcription factor 4 | 0.71 | 0.74 | 0.80 | 0.76 | Dopaminergic synapse |
| CAMK2B | calcium/calmodulin-dependent protein kinase II, beta | −1.33 | −1.25 | −1.02 | −1.29 | Dopaminergic synapse |
| CLOCK | circadian locomotor output cycles kaput | −1.59 | −1.53 | −1.41 | −1.42 | Dopaminergic synapse |
| CREB1 | cAMP responsive element binding protein 1 | −1.08 | −0.98 | −0.95 | −1.23 | Dopaminergic synapse |
| DLGAP1 | DLG associated protein 1 | 1.61 | 1.56 | 1.32 | 1.26 | Glutamatergic synapse |
| DRD2 | dopamine receptor D2 | −0.71 | −0.74 | −0.81 | −1.08 | Dopaminergic synapse |
| DRD4 | dopamine receptor D4 | 1.61 | 2.03 | 1.36 | 1.84 | Dopaminergic synapse |
| GNAI2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | 0.86 | 0.95 | 0.76 | 0.77 | Dopaminergic, GABAergic, Glutamatergic synapse |
| GNAL | guanine nucleotide binding protein, alpha stimulating, olfactory type | −1.64 | −5.79 | −2.32 | −1.74 | Dopaminergic synapse |
| GNB4 | guanine nucleotide binding protein (G protein), beta 4 | 1.46 | 1.14 | 0.74 | 1.16 | Dopaminergic, GABAergic, Glutamatergic synapse |
| HAP1 | huntingtin-associated protein 1 | 1.14 | 0.93 | 0.86 | 1.41 | GABAergic synapse |
| PLD1 | phospholipase D1 | −1.12 | −0.83 | −2.39 | −1.22 | Glutamatergic synapse |
| PPP3R1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | −1.75 | −1.45 | −0.97 | −1.7 | Glutamatergic synapse |
| PRKCA | protein kinase C, alpha | 0.97 | 1.09 | 0.86 | 0.95 | Dopaminergic, GABAergic, Glutamatergic synapse |
| PRKCB | protein kinase C, beta | −2.71 | −2.83 | −4.81 | −1.49 | Dopaminergic, GABAergic, Glutamatergic synapse |
| SHANK1 | SH3 and multiple ankyrin repeat domains 1 | −1.22 | −0.76 | −0.86 | −1.42 | Glutamatergic synapse |
| SLC1A2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | −0.84 | −0.75 | −0.72 | −0.93 | Glutamatergic synapse |
| SLC18A1 | solute carrier family 18 (vesicular monoamine), member 1 | −1.49 | −3.61 | −6.03 | −1.58 | Dopaminergic synapse |
| SLC32A1 | solute carrier family 32 (GABA vesicular transporter), member 1 | 1.1 | 0.85 | 0.92 | 0.96 | GABAergic synapse |
| SLC38A1 | solute carrier family 38, member 1 | −1.12 | −0.91 | −1.64 | −1.33 | GABAergic and Glutamatergic synapse |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gugliandolo, A.; Silvestro, S.; Chiricosta, L.; Pollastro, F.; Bramanti, P.; Mazzon, E. The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol. Life 2020, 10, 227. https://doi.org/10.3390/life10100227
Gugliandolo A, Silvestro S, Chiricosta L, Pollastro F, Bramanti P, Mazzon E. The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol. Life. 2020; 10(10):227. https://doi.org/10.3390/life10100227
Chicago/Turabian StyleGugliandolo, Agnese, Serena Silvestro, Luigi Chiricosta, Federica Pollastro, Placido Bramanti, and Emanuela Mazzon. 2020. "The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol" Life 10, no. 10: 227. https://doi.org/10.3390/life10100227
APA StyleGugliandolo, A., Silvestro, S., Chiricosta, L., Pollastro, F., Bramanti, P., & Mazzon, E. (2020). The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol. Life, 10(10), 227. https://doi.org/10.3390/life10100227





