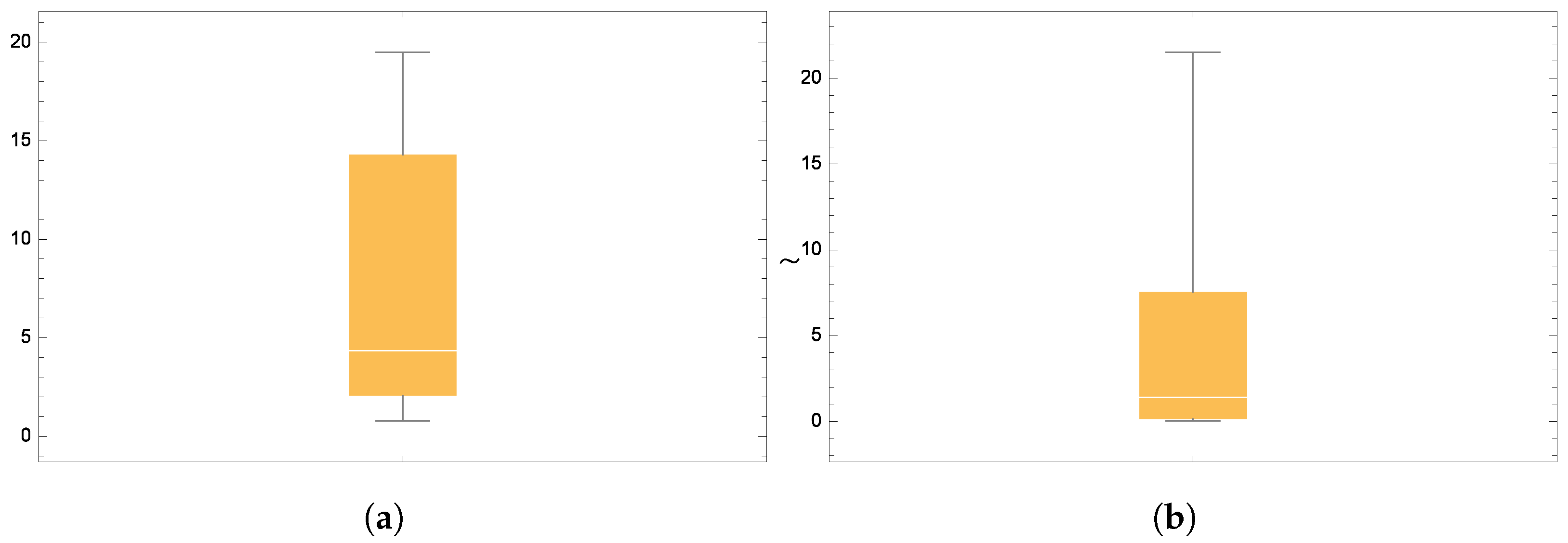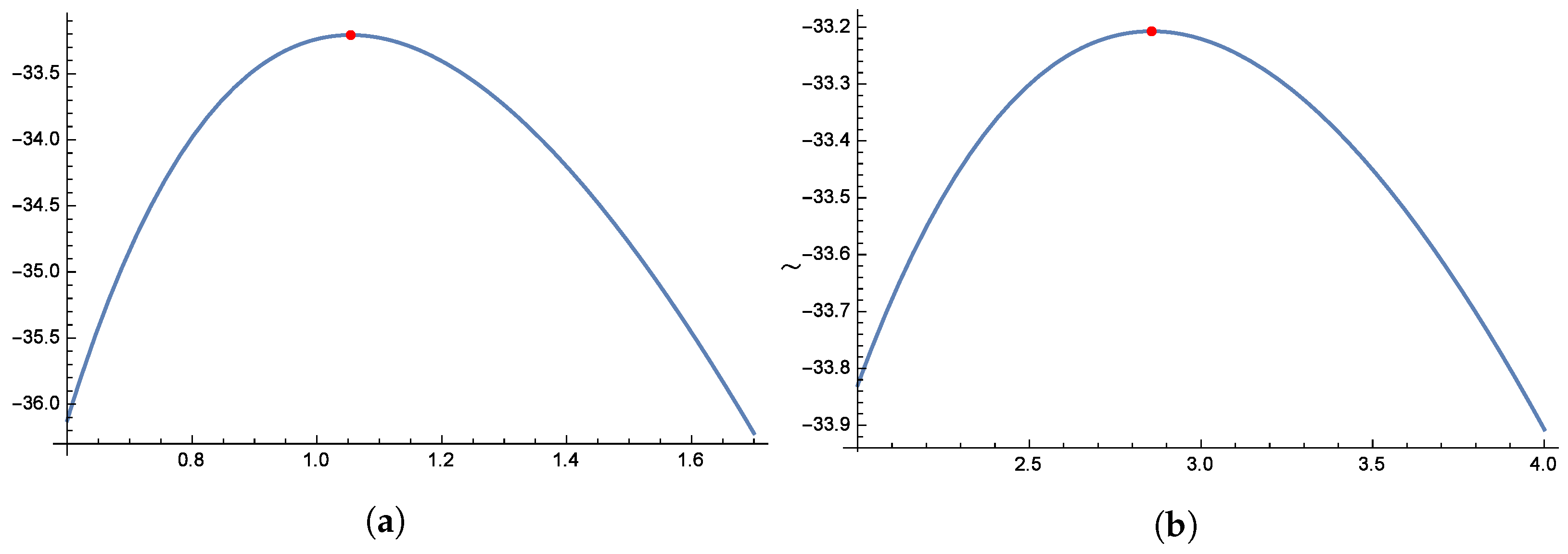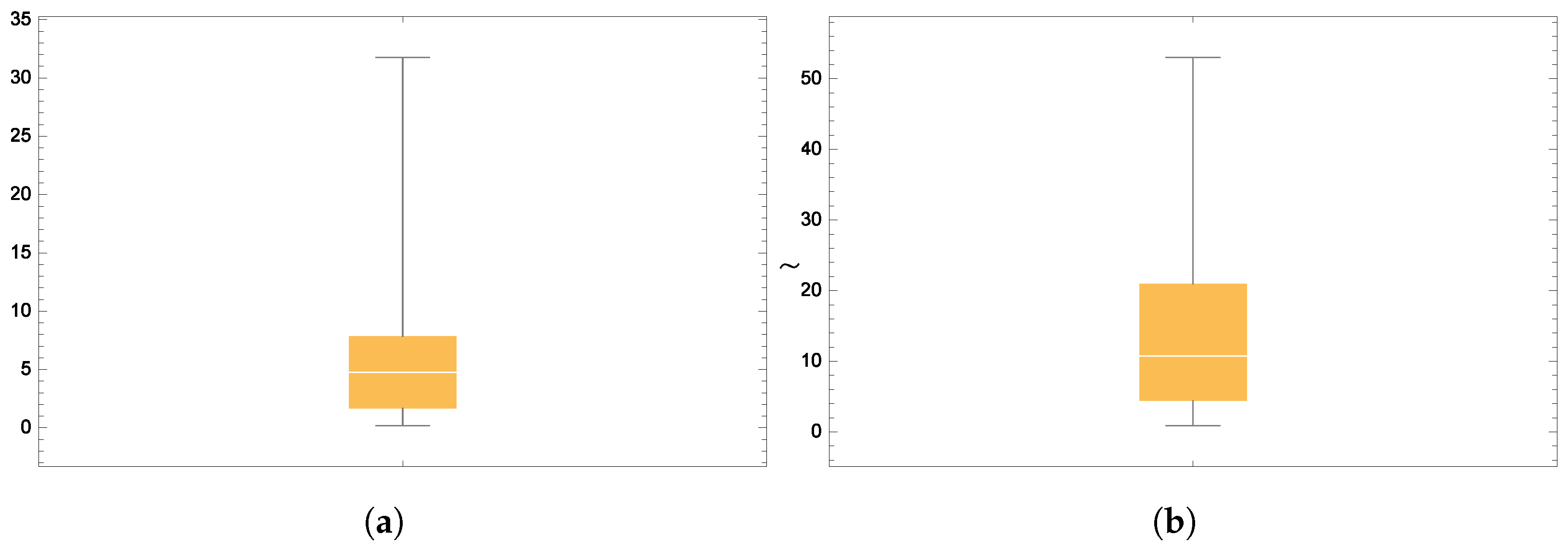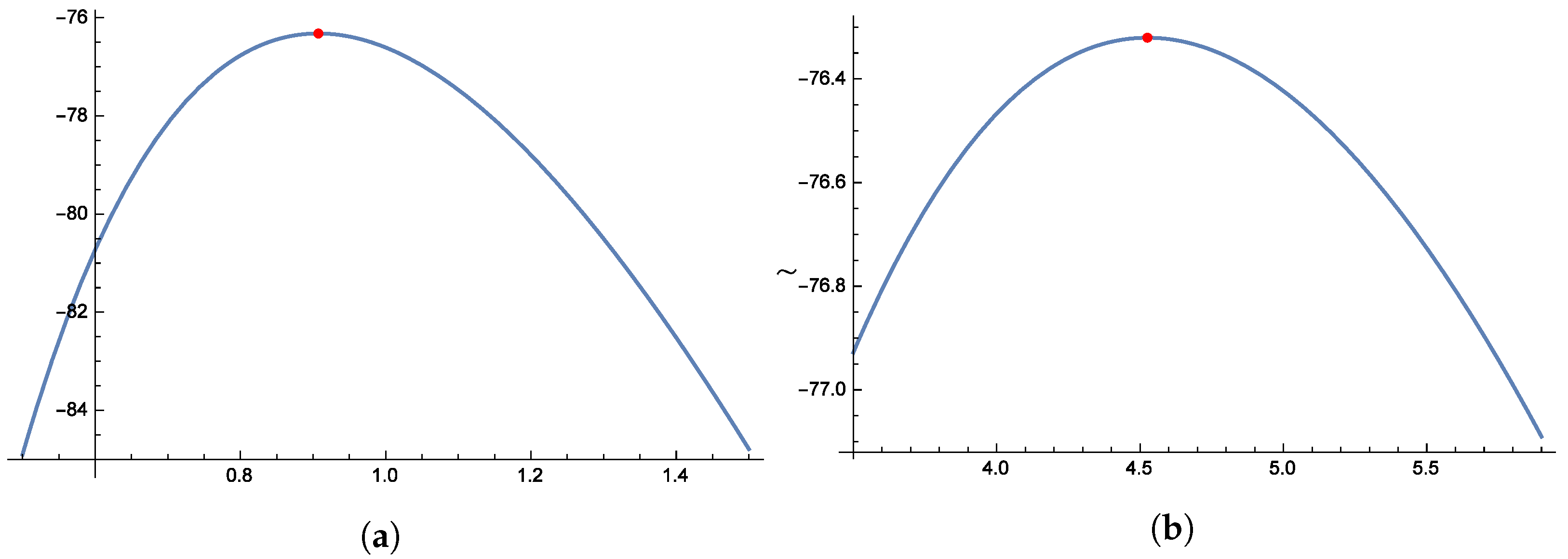1. Introduction
The Gumbel Type-II (G-II) distribution, introduced by the German mathematician Emil Gumbel [
1], serves as an essential model for extreme value analysis, such as floods, earthquakes, and other natural disasters. It is also used in a number of disciplines, such as rainfall studies, hydrology, and life expectancy tables. In comparative longevity tests, Gumbel emphasized its remarkable ability to replicate the anticipated lifespan of products. Moreover, this distribution plays a crucial role in forecasting the likelihood of climatic events and natural catastrophes.
Over the years, numerous researchers have advanced statistical inference techniques for the G-II distribution, such as Mousa et al. [
2], Malinowska and Szynal [
3], Miladinovic and Tsokos [
4], Nadarajah and Kotz [
5], Feroze and Aslam [
6], Abbas et al. [
7], Feroze and Aslam [
8], Reyad and Ahmed [
9], Sindhu et al. [
10], Abbas et al. [
11], and Qiu and Gui [
12]. For
and
, the cumulative distribution function (CDF) of the G-II distribution is given as follows:
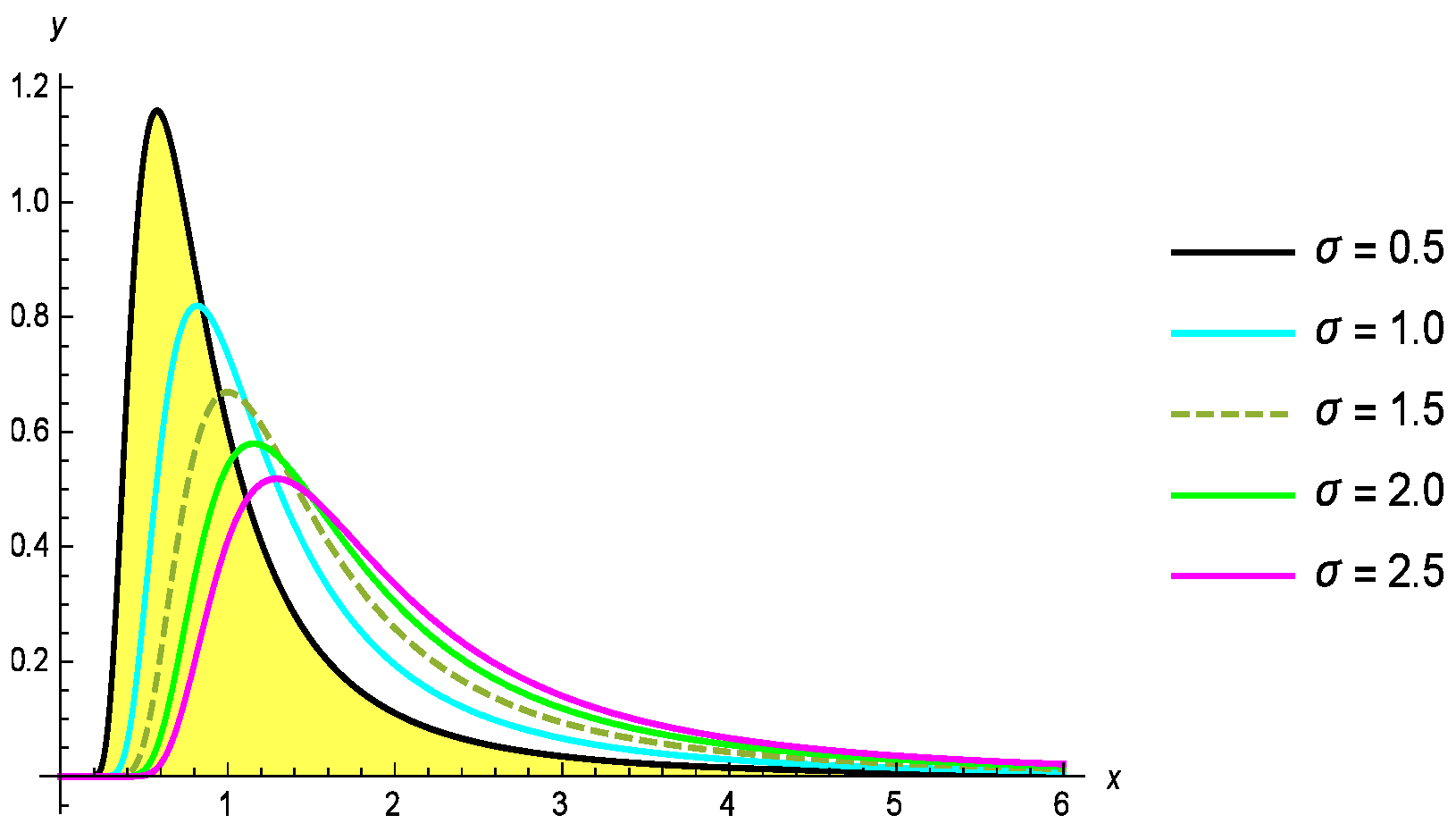
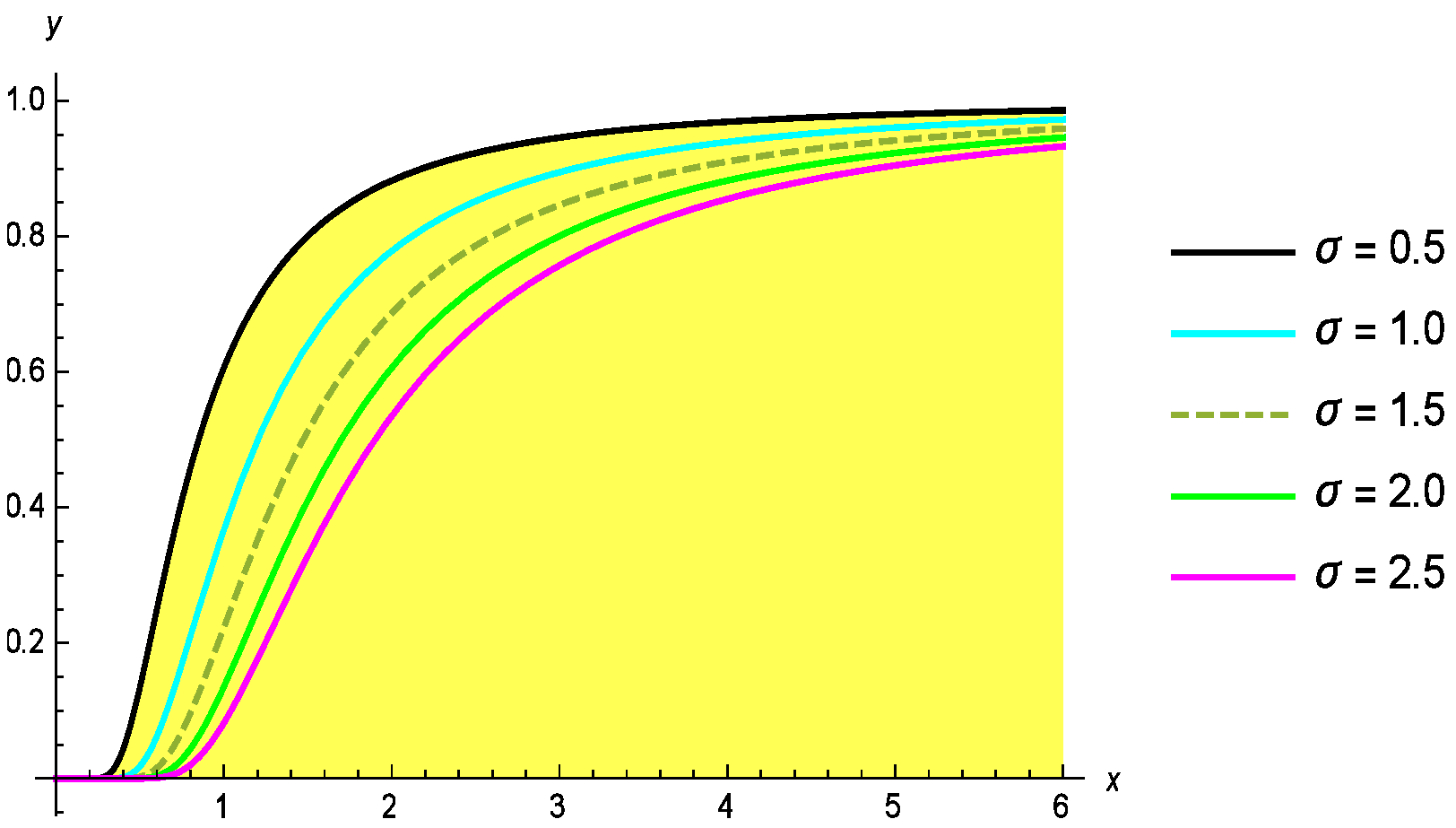
Moreover, the corresponding probability density function (PDF) is provided as follows:
Here, the shape and scale parameters are denoted by
and
, respectively. The probability density function (PDF) and cumulative distribution function (CDF) of the G-II distribution for different values of the parameters
and
are shown in
Figure 1 and
Figure 2.
Censored data is prevalent in various fields, including science, public health, and medicine, making it essential to implement an appropriate censoring scheme. One of the most widely recognized methods is progressive Type-II censoring. In this approach, the number of observed failures, denoted as
m (where
), is predetermined along with a censoring scheme
. At each
i-th failure, a specified number of remaining units are randomly removed from the experiment. This method offers the dual advantage of reducing both cost and time by allowing the removal of surviving units during the trial. Due to these benefits, progressive Type-II censoring has gained significant attention and has been extensively studied in the literature [
13,
14,
15,
16,
17].
Implementing censoring strategies on a single group can present several challenges. Although progressive Type-II censoring allows for the removal of certain data points, obtaining a sufficient number of observations remains costly. Moreover, studying group reliability and interactions, which cannot be adequately captured through experiments involving only one group, is often a key objective. To address these limitations, Rasouli and Balakrishnan [
18] proposed the Joint Progressive Censoring Scheme (Joint PCS). This approach enables the occurrence of failures in two groups simultaneously, effectively halving the time required to collect the same volume of data. Additionally, the Joint PCS facilitates the comparison of failure times between the two groups under identical experimental conditions, enhancing the robustness and applicability of the results.
Initially, groups
A and
B contain
m and
n units, respectively. The Joint PCS method originates from a life-testing experiment involving these two groups. The expected number of failures, denoted as
, is predetermined. The failure points are represented by
. At each failure time,
surviving units from group
A and
units from group
B are randomly removed. Thus, at the
i-th failure, a total of
units are eliminated. Additionally, a second set of random variables,
, is introduced, where each variable takes a value of either 1 or 0. These values indicate the source of the failure as follows:
The censored sample is represented as . Here, represents the total number of failures from group A. Similarly, indicates the number of failures from group B.
The Joint PCS has generated substantial interest among researchers. Several authors have examined Joint PCS and its related inference techniques, contributing to a rich body of literature. Across various applications, numerous scholars have explored a wide range of methodologies and heterogeneous lifetime models. For further reading, the works of [
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31] are recommended.
This manuscript makes significant contributions to the estimation of unknown parameters for the G-II distribution under a joint PCS, which is widely used in reliability analysis and lifetime data modeling. It employs both classical and Bayesian inference methods, utilizing maximum likelihood estimation (MLE) for point and interval estimation in the frequentist framework and Bayesian estimation (BE) via Markov Chain Monte Carlo (MCMC) methods, specifically the Metropolis–Hastings (M–H) algorithm, to handle complex posterior computations. Asymptotic confidence intervals (ACIs) and highest posterior density (HPD) credible intervals (CRIs) are derived to provide a comprehensive uncertainty assessment. The study explores the impact of different loss functions on Bayesian estimators. Through extensive Monte Carlo simulations, it evaluates the efficiency of the proposed estimation techniques by analyzing point estimates, interval estimates, and mean squared errors (MSE), demonstrating that Bayesian methods provide more precise and efficient estimates than MLE, particularly when informative priors are available. Additionally, the practical effectiveness of the proposed methodologies is validated through a real dataset, reinforcing their applicability in reliability analysis. Despite these contributions, the study encounters challenges in handling complex likelihood functions analytically, necessitating computational methods for parameter estimation. The selection of prior distributions in Bayesian inference and the computational burden of MCMC methods also pose difficulties. Moreover, while joint progressive censoring enhances efficiency in data collection, achieving a balance between cost, time, and sufficient observations remains a challenge. Overall, this study advances statistical inference for extreme value distributions by enhancing parameter estimation under joint PCS and offering a robust methodological framework applicable across various scientific and engineering disciplines.
The main goal of this study is to develop and compare estimation methods for the G-II distribution under a joint PCS by employing both classical MLE and Bayesian inference with different loss functions. The study introduces novelty by being the first to provide comprehensive parameter estimation for this distribution using joint progressive censoring, incorporating ACIs and highest posterior density CRIs, and applying squared error and LINEX loss functions in the Bayesian framework. It also uses the M–H MCMC algorithm to handle complex posterior computations and validates the proposed methods with real-world failure data, demonstrating their effectiveness in lifetime and reliability analysis.
The remainder of the paper is structured as follows.
Section 2 develops the model, derives point estimates using the MLE method, and obtains ACIs.
Section 3 presents BEs of the parameters under different loss functions, along with the proposed MCMC algorithm.
Section 4 includes a simulation study to compare the proposed methods.
Section 5 demonstrates the application of the inference procedures using a real-life dataset. Finally,
Section 6 provides concluding remarks.
3. Bayesian Estimation
BE is a cornerstone of modern statistical inference, offering a robust framework for updating beliefs in light of new evidence. At its core, Bayesian methods utilize Bayes’ theorem to combine prior knowledge with observed data, yielding posterior distributions that reflect updated probabilities. This approach is particularly valuable in situations where prior information is available or when dealing with complex models that require flexible parameter estimation. The importance of BE lies in its ability to quantify uncertainty in a coherent and intuitive manner, making it a powerful tool for decision-making and predictive modeling. It is widely applied in diverse fields such as machine learning, epidemiology, finance, and social sciences, where incorporating prior expertise can significantly enhance the accuracy of inferences. Moreover, Bayesian methods excel in handling small datasets, as they leverage prior distributions to compensate for limited data. By providing a probabilistic interpretation of parameters, BE bridges the gap between theoretical statistics and real-world applications, enabling researchers to draw meaningful conclusions while accounting for uncertainty. Its adaptability and interpretability make it an indispensable tool in the statistician’s arsenal.
3.1. Prior Distribution
The prior distribution, which represents our pre-existing beliefs or prior knowledge about a parameter before we see any data, is crucial to the field of Bayesian statistics. This distribution serves as the foundation upon which the posterior distribution is constructed, combining prior knowledge with new evidence through Bayes’ theorem. The importance of the prior distribution lies in its ability to influence the inference process, especially in scenarios with limited data, where it can guide the analysis towards more plausible conclusions. However, the choice of prior must be made judiciously, as an overly restrictive or biased prior can lead to misleading results. Conversely, a well-chosen prior can enhance the accuracy and robustness of statistical inferences, making it an indispensable tool in Bayesian analysis. Whether informative or non-informative, the prior distribution bridges the gap between subjective assumptions and objective data, enabling a coherent framework for updating beliefs in light of new information.
In this section, we present the Bayesian estimates for the unknown parameters, along with their corresponding CRIs, utilizing the Joint PCS as outlined earlier. Our main focus is on the SEL and LINEX loss functions.
The prior distributions for
,
,
, and
will now be established. To ensure that the prior and posterior densities belong to comparable families, it is ideal for the model parameters to be independent. These chosen priors allow the posterior distribution to be handled analytically and computed more efficiently. For the priors of
,
,
, and
, it is appropriate to assume that these four parameters follow independent gamma distributions. The probability density functions for these distributions are given by:
To incorporate prior information about
,
,
, and
, the parameters
and
(where
) are selected.
The joint prior density for
,
,
, and
is derived by combining the prior distributions specified in Equations (
15)–(
18) as follows:
3.2. Posterior Distribution
The posterior distribution is a fundamental concept in Bayesian statistics, representing the updated beliefs about a parameter after observing new data. It combines prior information, expressed through the prior distribution, with the likelihood of the observed data to produce a comprehensive view of the parameter’s possible values. This integration is achieved through Bayes’ theorem, which mathematically formalizes how evidence modifies prior beliefs. The importance of the posterior distribution lies in its ability to provide credible intervals and point estimates that reflect uncertainty in parameter estimation. Additionally, it serves as a basis for making predictions and decisions in various fields, including medicine, finance, and machine learning. By incorporating past knowledge with empirical data, the posterior distribution allows statisticians and researchers to make more informed conclusions and enhance the robustness of their analyses.
The joint posterior density is expressed as:
Equations (
4) and (
19) are combined to derive the joint posterior density function for
,
,
, and
, given by:
It can be challenging to derive explicit formulas for the marginal posterior distributions, as shown in Equation (
20). To address this, the MCMC approach can be used to generate samples from Equation (
20). The conditional posterior density functions for
,
,
, and
are given below:
Equations (
21)–(
24) demonstrate issues related to mathematical tractability. To estimate the unknown parameters, we develop Bayes estimators utilizing the SEL and the LINEX loss functions.
3.3. Loss Functions
Loss functions play a crucial role in statistical estimation, influencing the accuracy and robustness of parameter estimates. Two widely used loss functions in BE are the squared error loss (SEL) and the LINEX loss function. Each has distinct properties and applications depending on the nature of the estimation problem.
3.3.1. SEL Function
The SEL function is defined as:
where
is the estimate of the true parameter
. SEL penalizes deviations symmetrically, giving equal weight to overestimation and underestimation. This function is commonly used due to its mathematical simplicity and optimality under the mean squared error criterion. However, it assumes that over- and under-estimation have identical consequences, which may not always hold in real-world applications.
3.3.2. LINEX Loss Function
The LINEX loss function proposed by Varian [
32] is expressed as:
where
is a constant controlling the asymmetry of the loss function. When
, overestimation is penalized more heavily than underestimation, and when
, the opposite occurs. The LINEX function is particularly useful in applications where the cost of overestimating a parameter is not equal to the cost of underestimating it, such as reliability analysis and survival studies.
3.4. MCMC Method
MCMC is a fundamental tool in statistical estimation, especially in Bayesian inference, where direct sampling from complex probability distributions is often infeasible. MCMC generates dependent samples from a target distribution by constructing a Markov chain that converges to the desired distribution over time. This allows for approximating posterior distributions, estimating model parameters, and solving high-dimensional problems. The M–H algorithm is used when the target distribution is complex and cannot be sampled directly (see Metropolis et al. [
33] and Hastings [
34]). It works by proposing candidate samples and accepting or rejecting them based on an acceptance probability that ensures convergence to the correct distribution. Gibbs sampling is a special case of MCMC used when conditional distributions of parameters are available and easier to sample from directly. It is particularly useful for high-dimensional problems where parameters can be updated sequentially, reducing computational complexity. M–H is preferred when conditional distributions are intractable, while Gibbs sampling is efficient when conditional distributions have closed-form solutions.
M–H algorithm
Step 1: The process begins with the initial values , where K represents the burn-in period.
Step 2: Set .
Step 3: Equations (
21)–(
24) are used to generate
,
,
, and
through the M–H algorithm. The proposed normal distributions for this process are
,
,
, and
.
- (I)
Generate the proposed values , , , and from their corresponding normal distributions.
- (II)
Using the steps listed below, determine the probability of acceptance.
- (III)
Select a uniformly distributed random variable u with values ranging from 0 to 1.
- (IV)
If , accept the proposal and update to ; otherwise, retain as .
- (V)
If , accept the proposal and update to ; otherwise, retain as .
- (VI)
If , accept the proposal and update to ; otherwise, retain as .
- (VII)
If , accept the proposal and update to ; otherwise, retain as .
Step 7: Set j = j + 1.
Step 8: Repeat steps two through seven
times. Thus, under the SE loss function, the estimated posterior means of
, denoted by
, can be calculated as follows:
Finally, determine the Bayesian estimates of
using the LINEX loss function.
4. Real Data Analysis
4.1. Example 1
The dataset represents the oil breakdown times of insulating fluid exposed to various constant elevated test voltages. Originally reported by Nelson [
35], the data were collected under different stress levels. In this study, we focus on the dataset recorded at a stress level of 30 kilovolts (kV), representing normal use conditions, and at 32 kV, representing accelerated conditions. To simplify computations, each observed value in the original dataset has been divided by 10.
We employed various goodness-of-fit tests to assess the model’s performance, including the following:
Anderson–Darling (A–D) test.
Cramér–von Mises (C–M) test.
Kuiper test.
Kolmogorov–Smirnov (K–S) test.
Since the
p-values in
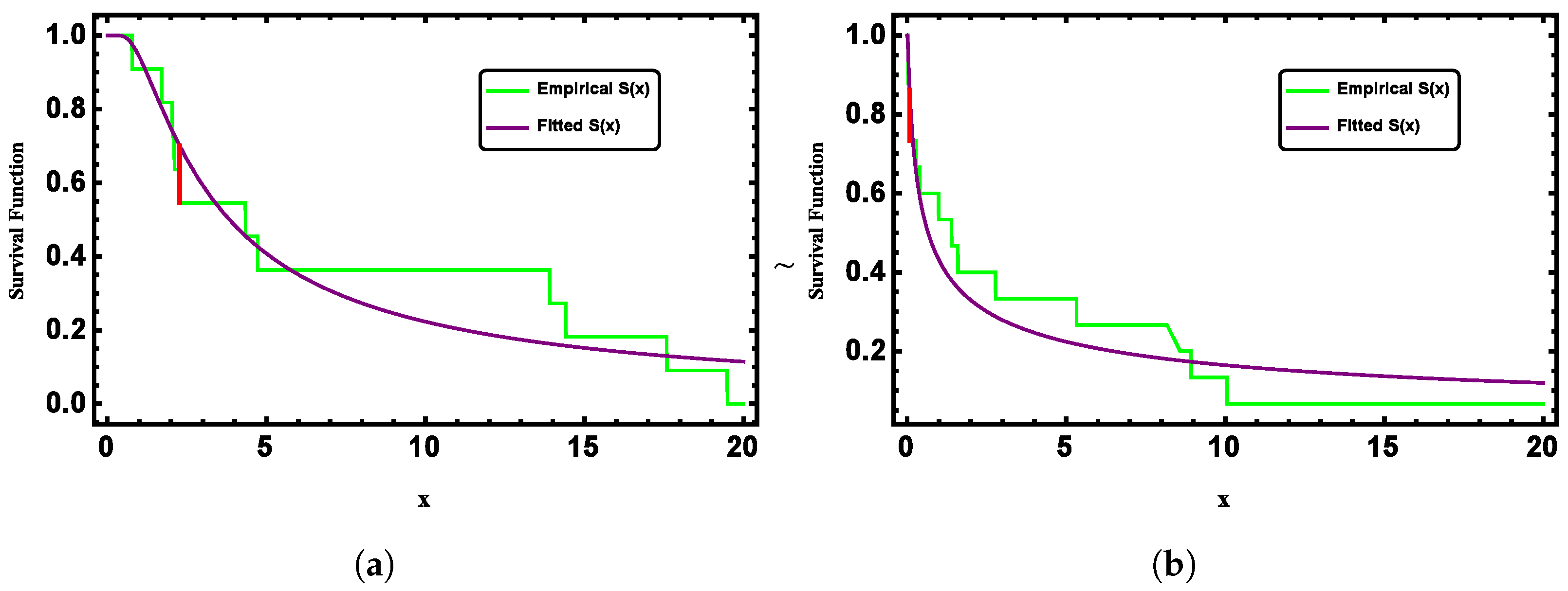
Table 1 are greater than 0.05, the null hypothesis of the distribution cannot be rejected. This indicates that both datasets follow the G-II distribution, demonstrating the applicability of the proposed model to real data. To further illustrate this,
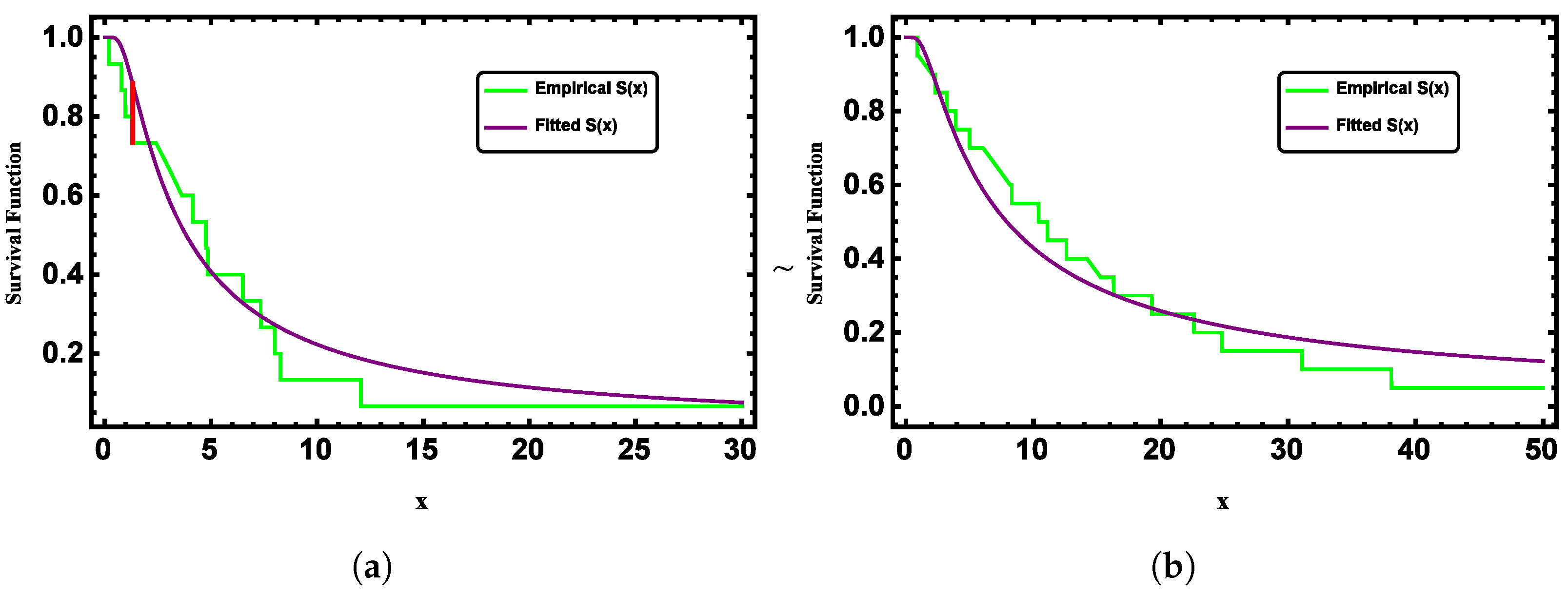
Figure 3 presents the fitted and empirical survival functions of the G-II distribution for both datasets, showing strong agreement between them.
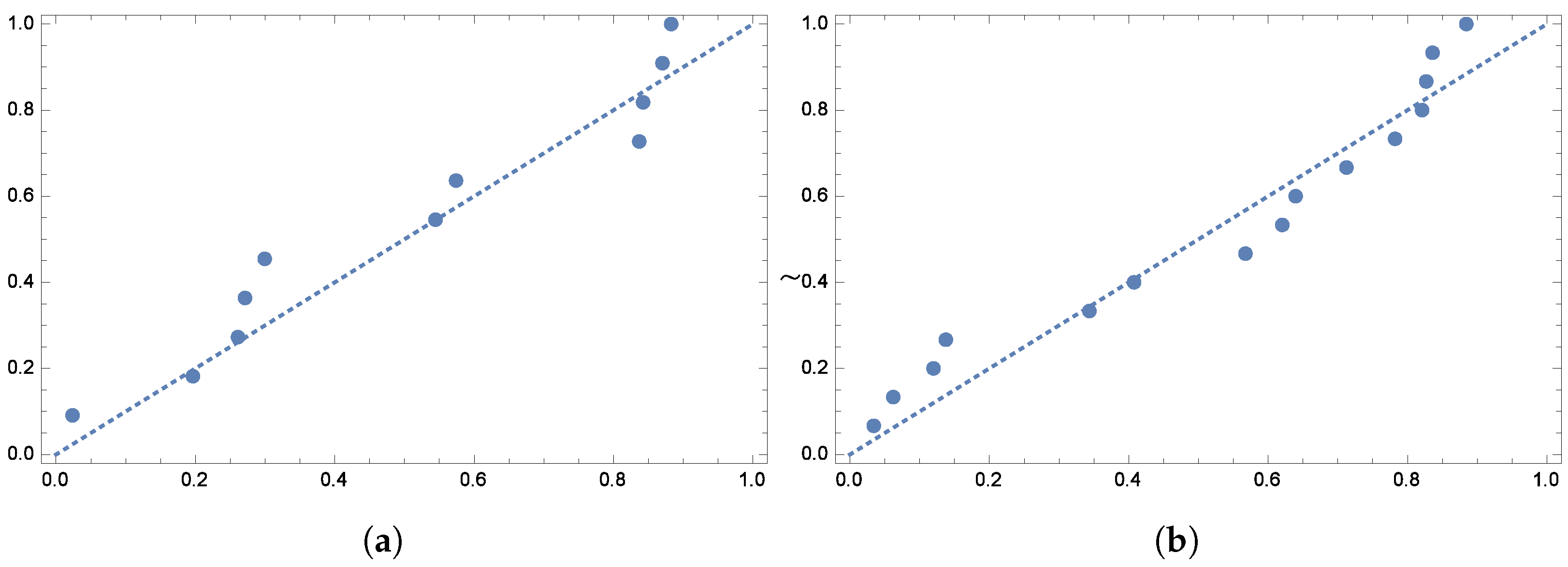
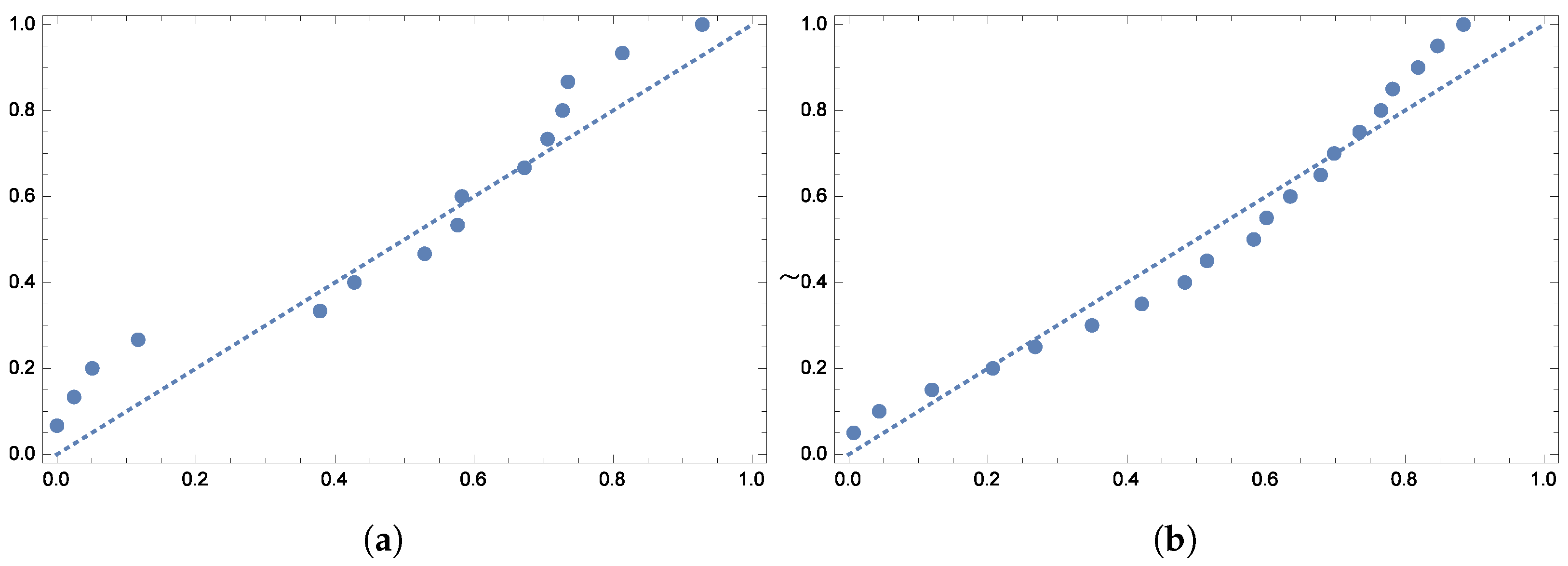
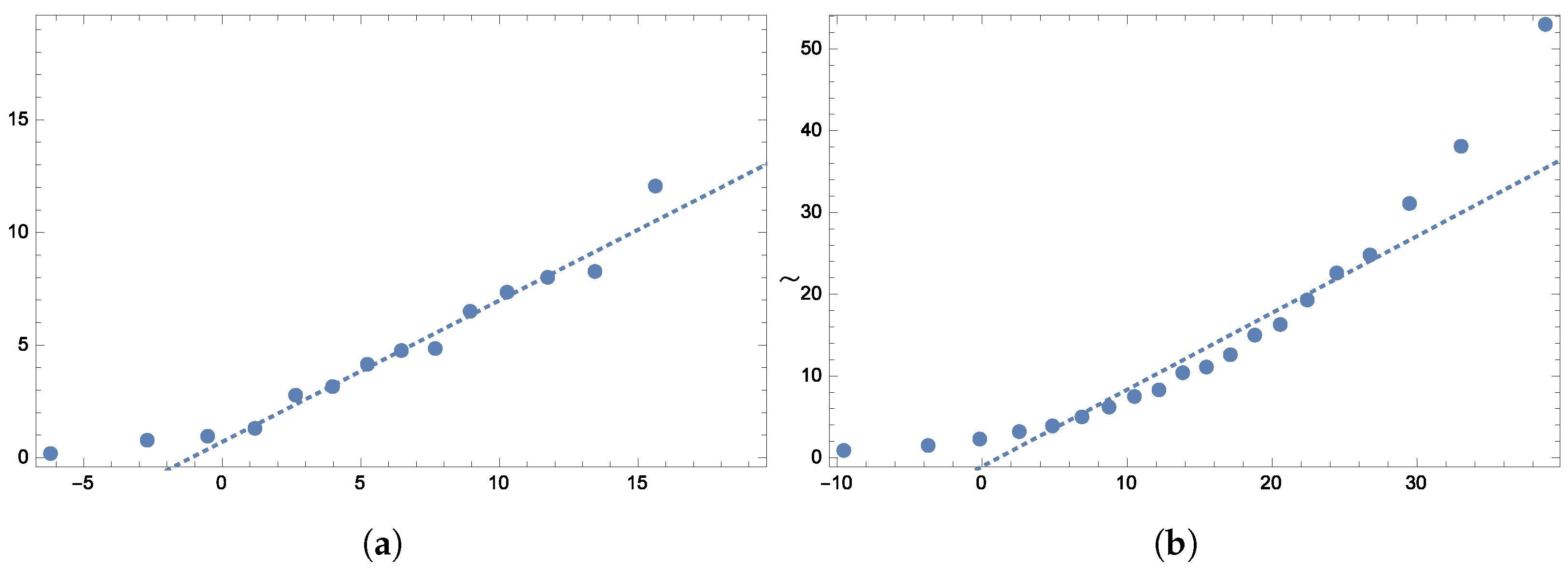
To further validate the model’s fit,
Figure 4 and
Figure 5 display the quantile plots and the observed versus expected probability plots, respectively.
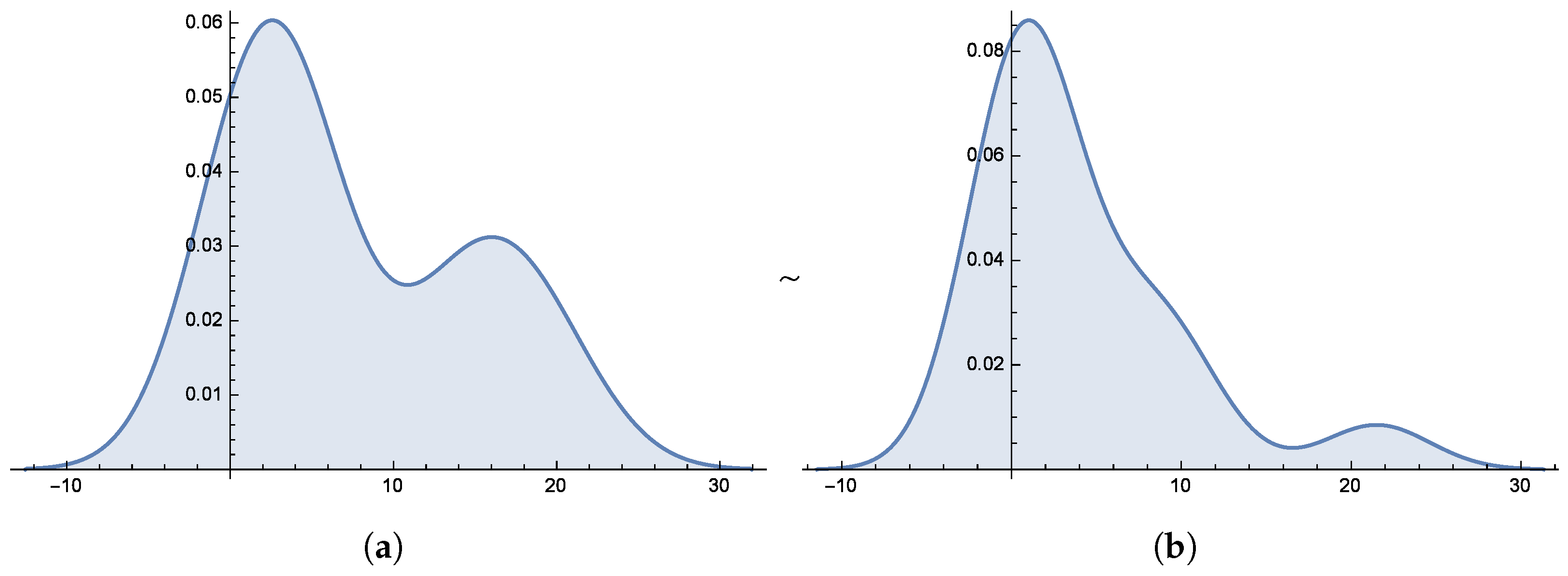
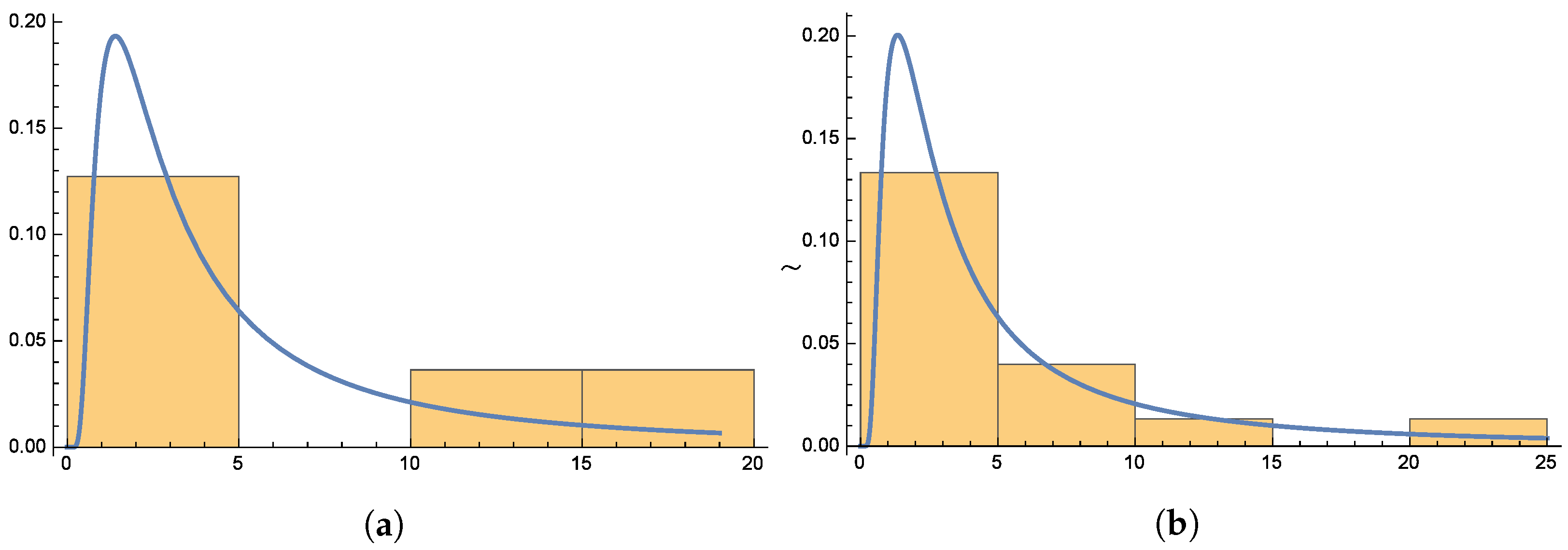
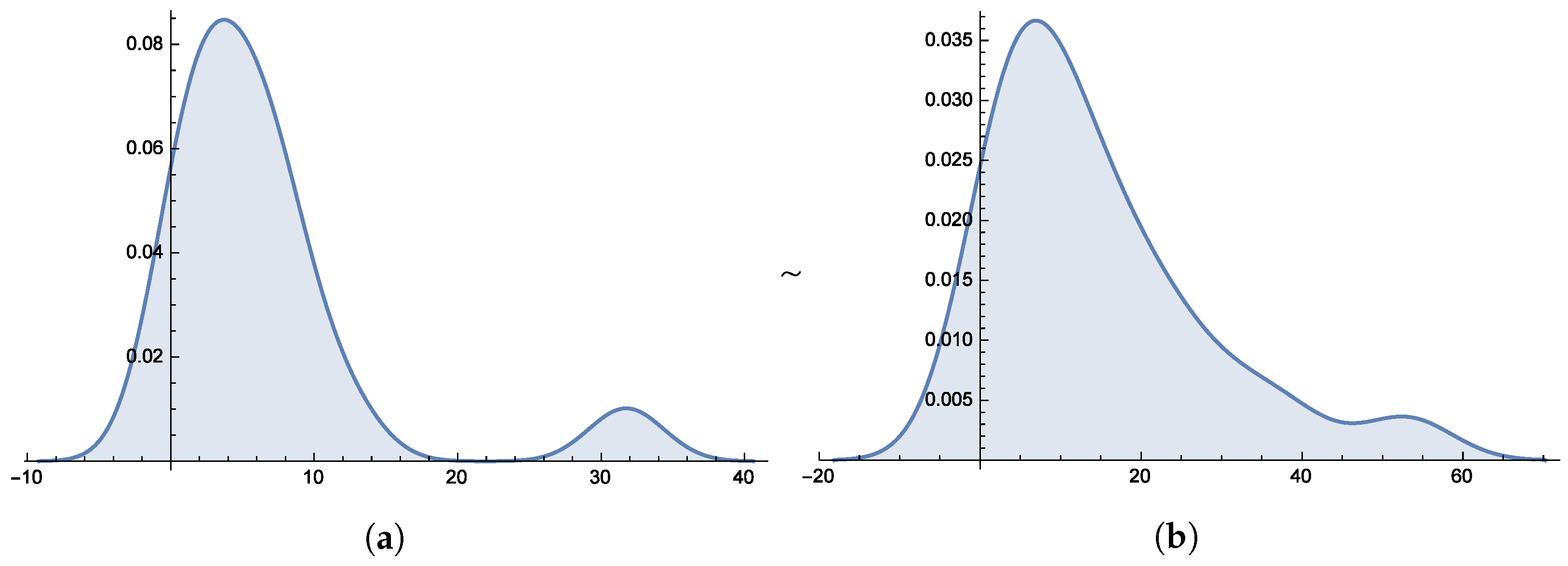
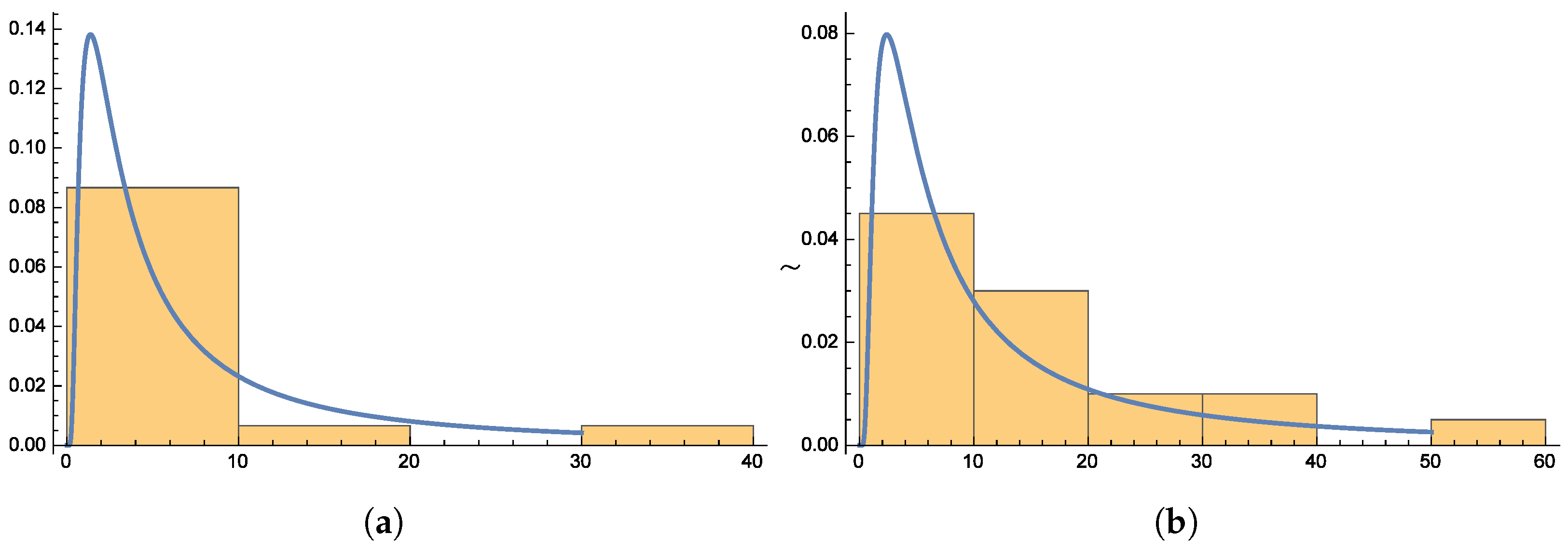
Figure 6 and
Figure 7 present the smooth histogram and box-and-whisker chart for Dataset-I and Dataset-II.
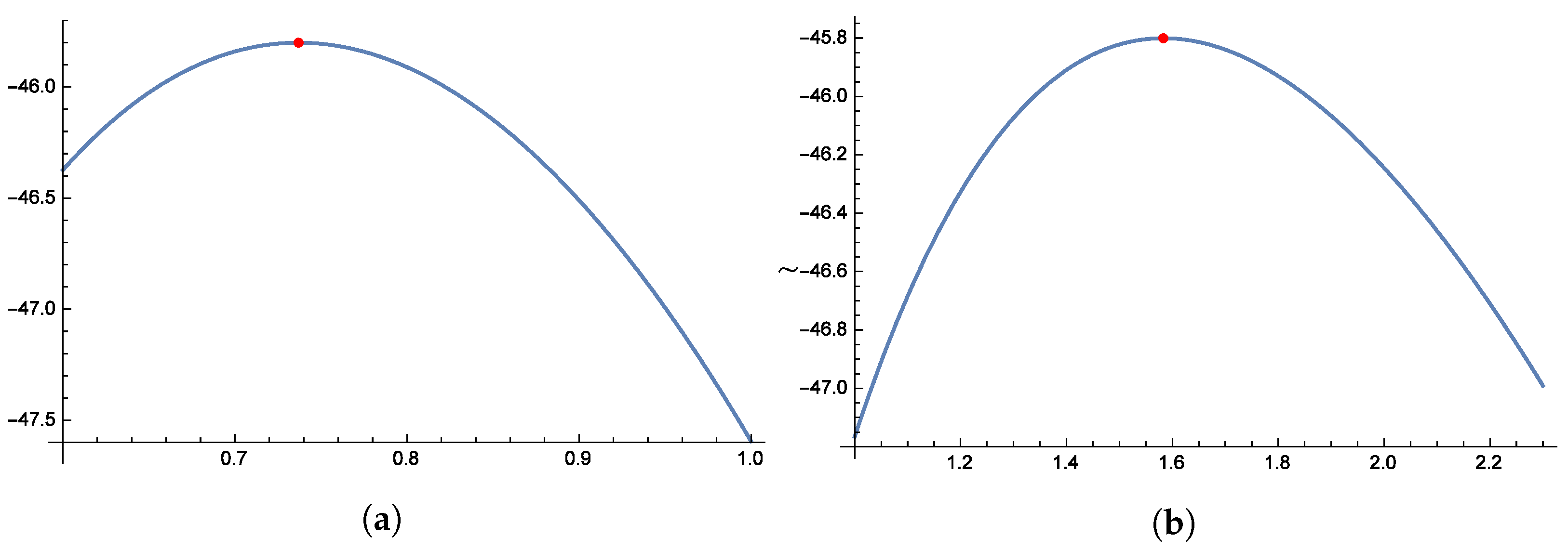
Figure 8 and
Figure 9 illustrate the unimodal behavior of the profile log-likelihood function, which peaks within specific parameter ranges: 1.0 to 1.2 for
, 2.5 to 3.5 for
, 0.4 to 0.6 for
, and 0.5 to 0.7 for
. These distinct peaks confirm the existence of unique MLEs for
,
,
, and
. Finally,
Figure 10 presents the histograms for both datasets, providing further evidence of the distribution’s suitability.
The censoring method outlined below was used to generate a Joint PCS sample based on the previously provided datasets. To perform Joint PCS with
, the first sample was set to
and the second to
. The censoring vectors are defined as follows:
The datasets created are as follows:
To determine estimates for
,
,
, and
, we used the MLE approach, depending on the type of data used in this study.
Table 2 displays the point estimates. While
Table 3 and
Table 4 give the 95% ACIs for
,
,
, and
, We ran 10,000 iterations using the MCMC method for BE, excluding the first 2000 iterations as "burn-in" to guarantee convergence. For the previous distributions, the hyperparameters
and
that we have selected are
, which is near zero. The corresponding point estimates are shown in
Table 2. Bayesian estimates were constructed for
,
,
, and
using both the SEL and LINEX loss functions. The 95% CRIs for
,
,
, and
are also included in
Table 3 and
Table 4.
4.2. Example 2
This section presents a real dataset to illustrate the practical use of the proposed methods. The dataset was originally obtained from Zimmer et al. [
36] and includes the following:
Data III Failure times (in hours) for 15 devices: 0.19, 0.78, 0.96, 1.31, 2.78, 3.16, 4.15, 4.76, 4.85, 6.5, 7.35, 8.01, 8.27, 12.06, 31.75.
Data IV First failure times (in months) for 20 electronic cards: 0.9, 1.5, 2.3, 3.2, 3.9, 5.0, 6.2, 7.5, 8.3, 10.4, 11.1, 12.6, 15.0, 16.3, 19.3, 22.6, 24.8, 31.5, 38.1, 53.0.
We applied several goodness-of-fit tests to evaluate the performance of the model, including the following
A–D test.
C–M test.
Kuiper test.
K–S test.
As the
p-values in
Table 5 exceed 0.05, the null hypothesis cannot be rejected. This suggests that both datasets follow the G-II distribution, confirming the model’s applicability to real-world data.
Figure 11 further supports this by comparing the fitted and empirical survival functions for both datasets, which show a strong alignment.
To further verify the model’s fit,
Figure 12 and
Figure 13 show the quantile plots and the observed versus expected probability plots.
Figure 14 and
Figure 15 display the smooth histograms and box-and-whisker plots for Dataset-III and Dataset-IV.
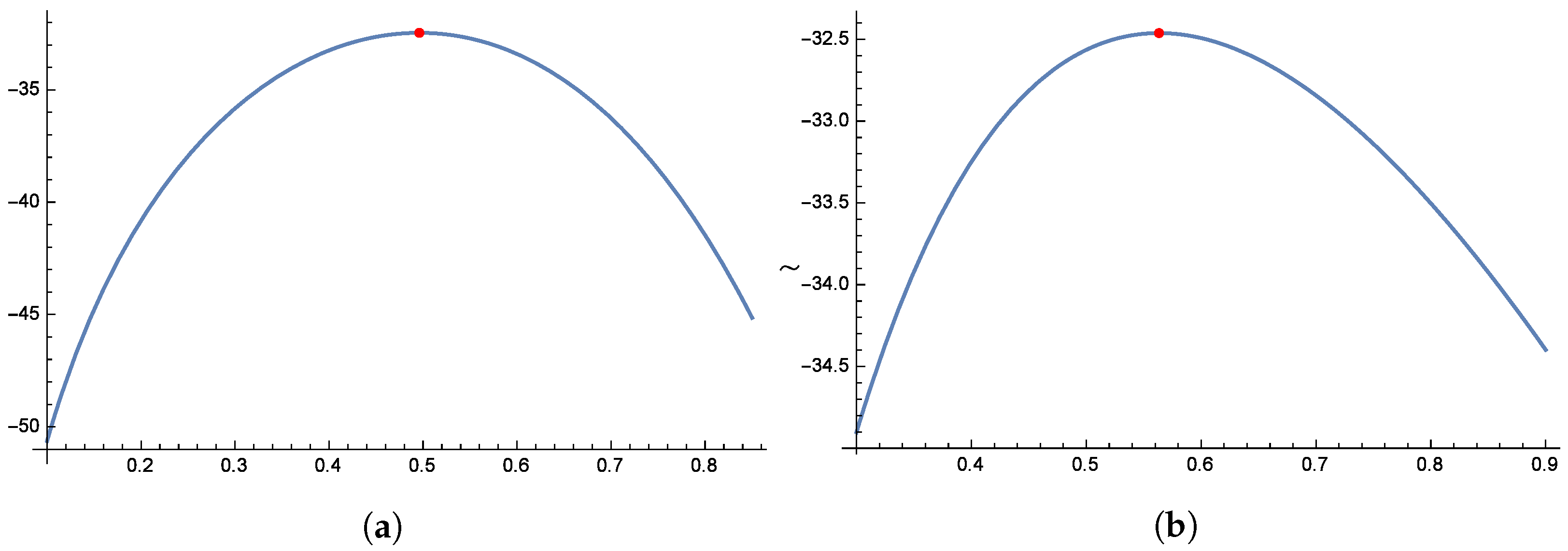
Figure 16 and
Figure 17 illustrate the unimodal shape of the profile log-likelihood functions, with peaks observed in the following parameter ranges: 0.7–0.8 for
, 1.4–1.8 for
, 0.8–1.2 for
, and 4.0–5.0 for
. These distinct peaks confirm the presence of unique maximum likelihood estimates for
,
,
, and
. Finally,
Figure 18 presents the histograms of both datasets, offering additional evidence of the distribution’s suitability.
The censoring scheme described below was used to generate a joint PCS sample based on the previously provided datasets. To implement the joint PCS with
, the first sample size was set to
and the second to
. The censoring vectors are specified as follows:
The generated datasets are as follows:
Estimates for
,
,
, and
were obtained using the MLE method, depending on the data type used in this study.
Table 6 presents the corresponding point estimates, while
Table 7 and
Table 8 provide the 95% ACIs for these parameters. For the Bayesian estimation, we performed 12,000 MCMC iterations, discarding the first 3000 as burn-in to ensure convergence. The hyperparameters
and
were set to
, close to zero, for the prior distributions. The results are summarized in
Table 6. Bayesian estimates for
,
,
, and
were obtained under both the SEL and LINEX loss functions.
Table 7 and
Table 8 also include the 95% CRIs for these parameters.
To further validate the applicability of the proposed model, we conducted an in-depth analysis using the newly added datasets. The evaluation included multiple goodness-of-fit tests, such as the A–D, C–M, K–S, and Kuiper tests. In addition, graphical diagnostics were used, including fitted versus empirical survival functions, quantile plots, and probability plots. The analysis results confirmed that the proposed Gumbel Type-II model under joint progressive censoring provides improved fitting accuracy and narrower CRIs when compared with classical estimation methods. These findings support the practical advantages of the proposed approach in modeling lifetime and reliability data.
5. Simulation Study
This section evaluates the performance of the proposed estimation methods under Joint PCS using Monte Carlo simulations. The simulations were conducted using Mathematica version 10. The comparison focuses on point estimators for lifetime parameters based on the MSE:
A lower MSE indicates improved estimation accuracy. All results are based on 1000 replications. Additionally, average confidence lengths (ACLs) are examined, where a smaller ACL suggests better interval estimation performance. We selected two distinct censoring schemes, detailed below, for different failure counts () and sample sizes (; ).
The parameter values for both datasets are
,
,
, and
. We computed the MLEs and 95% confidence intervals for
,
,
, and
. The mean values of the MLEs and their corresponding confidence interval lengths were determined after 1000 repetitions. The results are presented in
Table 9,
Table 10,
Table 11 and
Table 12.
Additionally, within the BE framework under SEL and LINEX loss functions, we employed informative gamma priors for , , , and . The hyperparameters were set as and for . The values and represented overestimation and underestimation, respectively. Where
adds a constant positive bias, modeling overestimation.
adds a constant negative bias, modeling underestimation.
These shifts test how the estimator performs when the data is systematically too high or too low.
Using the MCMC approach, we obtained Bayesian estimates with 95% CRIs for
,
,
, and
based on 1000 simulations with 21,000 samples. The initial 5000 iterations were discarded as a "burn-in" phase to ensure convergence. The performance of the obtained estimators for
,
,
, and
was evaluated using MSE. After 1000 repetitions, we computed the mean values of the MLEs and their respective lengths. The results are presented in
Table 9,
Table 10,
Table 11 and
Table 12.
Here are the key findings from the manuscript:
BE methods generally outperform classical MLE, particularly in terms of MSE and interval estimation precision.
Joint PCS is shown to be an effective strategy for balancing cost and time while maintaining estimation accuracy.
Bayesian estimators provide shorter CRIs compared with ACIs, making them more precise in uncertainty quantification.
The LINEX loss function results in more efficient Bayesian estimators compared with the SEL function.
The proposed methods effectively model real-world failure data, validating their use in reliability analysis.
Extensive Monte Carlo simulations confirm that Bayesian estimates exhibit lower MSE and narrower CRIs, especially when informative priors are used.
The M–H algorithm successfully approximates complex posterior distributions, ensuring efficient parameter estimation.