Abstract
In this paper, inference under dependent competing risk data is considered with multiple causes of failure. We discuss both classical and Bayesian methods for estimating model parameters under the assumption that data are observed under generalized progressive hybrid censoring. The maximum likelihood estimators of model parameters are obtained when occurrences of latent failure follow a bivariate inverted exponentiated Pareto distribution. The associated existence and uniqueness properties of these estimators are established. The asymptotic interval estimators are also constructed. Further, Bayes estimates and highest posterior density intervals are derived using flexible priors. A Monte Carlo sampling algorithm is proposed for posterior computations. The performance of all proposed methods is evaluated through extensive simulations. Moreover, a real-life example is also presented to illustrate the practical applications of our inferential procedures.
Keywords:
bivariate inverted exponentiated Pareto distribution; dependent competing risks; generalized progressive hybrid censoring; maximum likelihood estimation; posterior analysis MSC:
62F10; 62F15
1. Introduction
In many lifetime studies, components or systems frequently fail, owing to more than one reason. Thus, analyzing performance of units under isolated failure causes may not always be practicable, which may sometimes lead to lower accuracy and even misleading results. Therefore, various researchers have explored the analysis of failure patterns of systems/components when multiple causes of failure are available in their life cycle. Such inferences are referred to as competing risk models and associated inferences have received reasonable attention in the literature. In traditional studies, statistical inferences are commonly reported under the assumption that causes of failure are treated as independent. Kundu and Basu [1], Sarhan et al. [2], Ghitany et al. [3], Pascual [4], Xu et al. [5], Wang [6], Mahto et al. [7], and Lodhi et al. [8] discuss the competing risk models by considering exponential, Weibull, Lomax, log–normal, Kumaraswamy, Gompertz, and inverted exponentiated distributions. Crowder [9] also presents many useful applications of competing risk models in life testing experiments. However, due to the complex internal structures of systems, the assumption of independent failure modes may not be reasonable in many practical situations (e.g., see Wang et al. [10], Wang et al. [11], and Ashraf et al. [12]). Thus, it may be realistic to make inferences concerning competing risk models when the causes of failure are dependent. For modeling dependent causes, a two- or higher-dimensional probability distribution is usually taken into consideration when analyzing the lifetimes of the units under investigation. As a popular application, Marshall and Olkin [13] proposed the Marshall–Olkin bivariate exponential distribution approach from the perspective of a shock model in their reliability study. This particular model can be extended to some other approaches, like the Marshall–Olkin bivariate Gompertz distribution and the Marshall–Olkin bivariate Weibull distribution. Due to their simpler statistical structure and attractive distributional properties, these lifetime models appear to be very useful. These models have wide-ranging applications in reliability analyses. We further refer to Kaishev et al. [14], Pena and Gupta [15], Kundu and Gupta [16], Jamalizadeh and Kundu [17], Feizjavadian and Hashemi [18], and Wang et al. [10,11] for useful discussion in this regard.
Often due to time and cost restrictions, it is difficult to conduct lifetime experiments before all testing units have failed. So, information concerning failure times is usually collected in the form of censored data. Here, a test terminates before all the units have failed. In practice, Type-I and Type-II censoring are the most common methods. In such situations, a test is stopped either when a predetermined time is reached or when a predetermined number of failures has been recorded. These schemes do not allow for the removal of live units during experimentation. To overcome such difficulties, progressive censoring schemes are introduced in the literature; here, the removal of units is allowed at various stages of the testing process. Extensive studies have been carried out on traditional and progressive censoring. See, for instance, Panahi and Sayyareh [19], Balakrishnan and Kateri [20], Rastogi and Tripathi [21], and Feroze and Aslam [22] for some recent works. Interested readers may also refer to Balakrishnan and Aggarwala [23] and Lawless [24] for in-depth discussions.
Modern products are usually highly reliable. So, it may still be difficult to collect enough failure samples in traditional and progressive censoring. Cho et al. [25] proposed a new censoring method known as the generalized progressive hybrid censoring (GPHC) scheme; here, testing time can be controlled, and an appropriate number of failure times can be observed under this censoring approach. This method can be briefly described as follows: Let a test begin with n units; further, assume that k and m are prefixed integers satisfying . In addition, non-negative integer-valued variables , and a time point, T, are randomly picked in advance, with . Upon the occurrence of the first failure, say , we randomly eliminate number of live units from the test. Similarly, at second failure , we randomly remove number of live units from the test. Finally, at the ith failure, , we randomly remove number of live units from the test. The experiment stops at time , i.e.,
Thus, under this scheme, the recorded observations may occur as one of the following three types:
where d satisfies . The GPHC scheme encompasses various traditional censoring methods, including Type-I, Type-II, progressive, and hybrid censoring schemes. For completeness, we elucidate the impact of the scheme on the inference process and its advantages in analyzing competing risk data. The GPHC scheme integrates the flexibility of progressive censoring which permits the staggered removal of units during the experiment with a predetermined terminal time, thereby ensuring a balance between study duration and data sufficiency. This affects inference in two pivotal ways. Firstly, the GPHC scheme dynamically adjusts the censoring process based on observed failures, thereby enhancing estimation efficiency compared to fixed-time (Type-I) or fixed-failure (Type-II) censoring schemes. For instance, the likelihood function under the GPHC scheme incorporates both time-dependent failure counts and censoring thresholds, thus facilitating more precise recovery of dependency between parameters in the bivariate model. Secondly, while this scheme introduces complexity into the likelihood formulation due to its adaptive censoring rules, the closed-form survival function of MOBIEP distribution simplifies the derivation of maximum likelihood estimators (MLEs) and the computation of posterior distribution under the Bayesian framework. Additionally, it offers distinct advantages in the context of dependent competing risks. Specifically, competing risks data frequently entail dependencies between failure modes and censoring mechanisms. This particular scheme addresses this by explicitly modeling the joint survival function of MOBIEP distribution, thereby ensuring that the applied scheme adapts to the observed failure patterns without introducing bias into the parameter estimates. It allows early termination of testing if a sufficient number of failures occur. The test can further be extended if more failures need to be recorded. Thus, in various situations, the GPHC scheme optimizes resource allocation. This is particularly beneficial in reliability studies, where prolonged testing may be costly or impractical.
In this paper, we investigate inference procedures for a dependent competing risks model under the assumption that failure times are modeled by a Marshall–Olkin-type bivariate distribution. Various estimates of unknown parameters are derived from both classical and Bayesian viewpoints when latent failure causes follow an inverted exponentiated Pareto (IEP) distribution and failure times are observed under the GPHC scheme with partially detected competing risks.
The remainder of the paper is structured as follows. In Section 2, the proposed Marshall–Olkin bivariate inverted exponentiated Pareto (MOBIEP) model is introduced. The associated competing risk data with partially detected failure causes are also described. Section 3 describes results related to the MLEs of unknown parameters. Additionally, the asymptotic confidence intervals (ACIs) are derived based on the Fisher information matrix. Bayesian point estimates and credible intervals for model parameters are discussed in Section 4. Section 5 provides a numerical study. A real data analysis is also considered for illustrative purposes. Finally, concluding remarks are given in Section 6.
2. Data Description Under Marshall–Olkin-Type Bivariate IEP Model
In this section, the Marshall–Olkin-type bivariate IEP model is proposed for modeling dependent competing risks. Furthermore, data on latent failure times are described under the GPHC scheme.
2.1. Marshall–Olkin Bivariate IEP Distribution
Consider a random variable, X, that follows IEP distribution. Then, its cumulative distribution function (CDF) and the probability density function (PDF) are given by
and
where . Furthermore, and are shape parameters. The corresponding survival function (SF) of X can be rewritten as
Hereafter, we use notation to denote the inverted exponentiated Pareto distribution with parameters and . Ghitany et al. [3] presented numerous useful discussions on this model within the context of life test studies. In fact, the authors demonstrated that this particular distribution can appropriately model a variety of physical phenomena. Its hazard rate function may be decreasing or nonmonotone, depending on the values of and . The IEP distribution plays a significant role in life testing and various other practical fields of application, such as communication systems, mortality data analysis, and clinical trials.
Further, the new MOBIEP is constructed as follows. Assume are independent random variables, satisfying . Let quantities and be the random vector . Then, this pair of random variables is said to follow the MOBIEP distribution with parameters , and , denoted as . Hereafter, we use notations , and for , and in subsequent part of the paper.
Theorem 1.
Let ; then, the joint SF of is given by
where .
Proof.
See Appendix A. □
It is noted from Theorem 1 that, when , we have , which indicates that variables and are independent. So, governs the dependency between and .
Corollary 1.
Let ; then, the joint PDF of can be expressed as
where
Proof.
See Appendix B. □
Corollary 2.
Let ; then, the random variable is also IEP distributed with parameters λ and That is,
Proof.
See Appendix C. □
We illustrate the potential motivation and comparative advantages of this model for modeling dependent competing risks. Firstly, the MOBIEP distribution inherently captures nonlinear dependence structures between latent failure times via its shape and scale parameters. This characteristic is crucial in competing risk scenarios where failure modes may exhibit complex interactions. Secondly, the IEP model is well-suited for modeling extreme-value data or heavy-tailed survival patterns, which are prevalent in reliability and survival analyses. Thirdly, the closed-form survival function facilitates tractable likelihood construction, thereby enhancing computational feasibility for both classical and Bayesian frameworks. Additionally, this model possesses distinct advantages when compared with alternative distributions. We note that copulas allow for flexible dependency modeling; however, they often lack interpretability in competing risk contexts and may necessitate ad hoc assumptions for marginal distributions. The MOBIEP offers a unified parametric framework with direct interpretability of parameters. Similarly, traditional bivariate models such as Weibull/exponential bivariate distributions impose restrictive assumptions (e.g., monotonic hazard rates) and symmetric dependencies, whereas the MOBIEP accommodates asymmetric dependence and nonmonotonic hazards. For ilustration, some plots of the density for the MOBIEP model are provided in Figure 1.
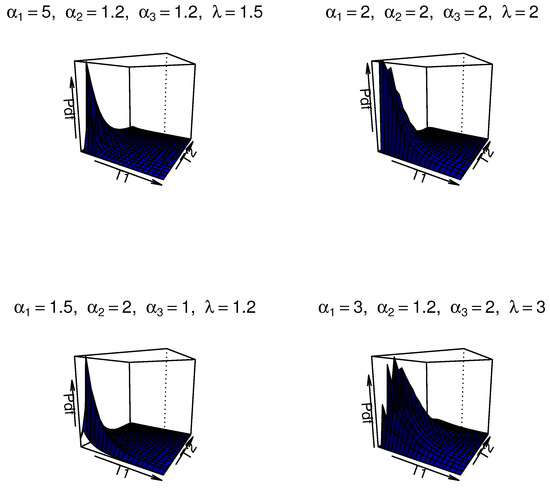
Figure 1.
The surface plots of the joint PDF of and with different values of .
2.2. Data Description
Assume that n units are tested under two dependent causes of failure. The lifetimes of the testing units are examined under two competing risks, and , with . Let be the lifetime of the considered units. Thus, observations under complete sample can be expressed as
with . Here, is the ith failure time of . Further, the notation is the failure indicator, such that
With the GPHC approach, the competing risk data are given by
Here, denotes the specified scheme in advance. Further, T is the monitoring time point and k is a given positive integer. Additionally, d indicates the failure items so that, in Case II, we have .
Remark 1.
In practical situations, identifying failure mechanisms presents a significant challenge; while advanced detection techniques can reveal various causes underlying different physical phenomena, complex internal structures and operating mechanisms often obscure precise identification. However, there are numerous instances where causes of failure cannot be appropriately identified. Therefore, it is observed from (7) that the assumption of partially identified causes of failure in competing risk data is more realistic and suitable for analyzing scenarios where complete knowledge of failure causes is unavailable.
For the sake of simplicity, the competing risk data given in (8) are rewritten as under the GPHC scenario, where is presented as for concision. Further, suppose the dependent causes of failure follow ; then, the likelihood can be expressed as
where in Case I, and in Case II. The function is written as
For cause , the data for the ith unit are completely known in the sense of failure time and related cause of failure. For , the failure time is known; however, the cause of failure may not be detected, satisfying for all . For clarity, a brief explanation about how to establish Equation (9) is given. For data with , the probability contribution of data at is constructed from Equation (7) as
For , data are equivalent to event ; then, the probability contribution can be obtained as
For other cases of with and 3, the probability contributions could be obtained similarly. Then, the joint likelihood function of the GPHC competing risks data (8) can, respectively, be obtained under Cases I, II, and III.
Remark 2.
Under the conditions where and , it should be noted that the probabilities are similar; these are given by and , respectively. Additionally, failure times due to causes one and two are appropriately observed when . In the scenario where , although the failure time has been observed, the specific cause of the product failure remains unknown. Consequently, the cause of failure in this case is masked, and it can be regarded as the failure time of the variable , without a specified cause of failure. Consequently, the corresponding probability contribution is derived from the distribution of . Therefore, the coefficient , which is omitted in the case where , may be considered as an additional factor provided by the cause of failure.
Based on Equation (9), let with and . Then, the corresponding likelihood functions and of for the Cases I, II, and III, are further expressed as
3. Classical Inference
In this section, we discuss MLEs of model parameters; subsequently, the asymptotic confidence intervals (ACIs) are obtained.
3.1. Maximum Likelihood Estimation
Due to the different expressions of the likelihood functions under the GPHC scenario, assumptions are made. Accordingly, the likelihood functions and can be rewritten in a single expression. Let be the observed failure number in Cases I, II, and III. Then, we have
The failure times data (Equation (7)) in each case are denoted by . Further, let , and satisfying ,
and
Therefore, the likelihood function of , and , namely is rewritten as
and the associated log–likelihood function is expressed as
The MLEs of , and are presented as follows.
Theorem 2.
Let GPHC competing risk data follow the MOBIEP distribution. Then, for the MLE of , given λ, is obtained as follows
Proof.
See Appendix D. □
Theorem 3.
Let the GPHC competing risk data follow the MOBIEP distribution. Then, for , the MLE of λ uniquely exists as the solution of the following equation
where
Proof.
See Appendix E. □
Since the explicit form of the estimator of does not exist, the required estimate can be obtained using the fixed-point iteration technique. Additionally, the MLEs of , , and can be obtained from Theorem 2, as follows:
3.2. Approximate Confidence Intervals
The observed Fisher information for is given by
where the associated elements are obtained by direct computation. The detailed expressions are omitted here for the sake of concision.
Under mild regularity conditions, the approximate distribution of MLE is such that , where the notation “” means “convergence in distribution”, and represents the inverse of the observed Fisher information matrix, given below as
Now, for some , a confidence interval of is obtained as
where is the percentile of standard normal distribution. Sometimes, these interval estimates may occasionally yield negative lower bounds. In this regard, the delta method can be applied to derive the distribution of , such as . Therefore, the desired interval of is obtained as
where .
4. Bayesian Analysis
In data analysis, the Bayesian approach has gained considerable attention due to its capability to incorporate prior information. This feature is particularly valuable in various fields, such as bio-assay experiments, clinical trials, and competing risk models. This section presents Bayesian estimation using a general flexible prior. Accordingly, estimates of parameters and highest posterior density (HPD) credible intervals are obtained.
4.1. Prior Information
Following the idea of Pena and Gupta [15], a general flexible prior for model parameters is constructed as follows. Assuming a gamma prior for is presented, as follows:
Now, given , has the following prior, with density
Therefore, the prior of , namely is given by
where . The prior (15) is referred to the Gamma–Dirichlet prior, denoted by . This family of priors offers a very flexible approach for Bayesian computation.
In addition, suppose that parameter has an independent gamma prior
Finally, the joint prior distribution of and turns out to be
and, accordingly, the posterior density is obtained as follows:
where .
For arbitrary parametric function , the corresponding Bayes estimator under the squared error loss is obtained as
Clearly, there is no closed form for this Bayes estimator. Therefore, a Monte Carlo sampling method is implemented to compute this estimate.
4.2. Posterior Analysis and Importance Sampling
The posterior density (18) of the parameters , and can be rewritten as
where
and
with notation denting the gamma distribution. So, the estimator of in (19) is rewritten as
We now employ the following sampling technique to compute the point and HPD-credible interval estimates of under the GPHC scheme. The associated computational procedure is given as follows (see Algorithm 1).
| Algorithm 1: Bayesian importance sampling. |
|
5. Simulation Study
In this section, extensive simulation experiments are conducted to assess the performances of the proposed estimates. Additionally, a real-life dataset is discussed to demonstrate practical application.
5.1. Simulation Results
In this part, a simulation analysis is carried out to evaluate the performance of the proposed methods under the GPHC competing risk data. For various sample sizes , censoring schemes (), and monitoring points , the point estimates of parameters , , , and are evaluated using average bias (AB) and mean squared error (MSE) values. The interval estimates are evaluated based on the average length (AL) and the coverage probabilities (CPs).
The GPHC competing risk data are obtained as follows: First, a progressively censored sample for the jth cause is generated by using the algorithm of Balakrishnan and Sandhu [26] based on IEP model with parameters and . These two sets of samples are compared with respect to to obtain the GPHC competing risk data along with the associated causes of failure. Finally, these competing risk data are masked with probability, p, to obtain partially observed GPHC competing risk data. It is noted that a group of iid random data containing either 0 or 1 is generated using Bernoulli trials with probability, p, where the sample size is equal to that of the competing risk data. If a Bernoulli trial yields zero, the cause of failure is considered as unknown. Conversely, if one appears, the cause of failure is considered to be detected.
In the simulated illustration, we randomly select parameter (, , , ) as . For the sake of convenience, a traditional Type-II censoring scheme is used with and . Furthermore, for the Bayes estimates, both non-informative priors (NIPs) with hyper-parameters and the informative prior (IP) with hyper-parameters are employed. Various estimates were obtained by considering 3000 replications. The computed results are presented in Table 1, Table 2, Table 3 and Table 4, where the interval estimates are computed with significance level .

Table 1.
ABs and MSEs (within bracket) for OBIEP parameters when .

Table 2.
ABs and MSEs (within bracket) for OBIEP parameters when .

Table 3.
CPs and ALs (within bracket) for OBIEP parameters when .

Table 4.
CPs and ALs (within bracket) for OBIEP parameters when .
The following conclusions regarding point estimates can be drawn from the findings presented in Table 1 and Table 3. The estimated ABs and MSEs of the MLEs decrease with increases in the effective sample size, i.e., n, m, or k, or their combinations. A similar phenomenon is also observed for the Bayes estimates under both proper and improper priors. This findings hold true for estimation of all parameters across different schemes. Additionally, for a fixed sample size, the ABs and MSEs of both MLEs and Bayes estimates decrease with an increase in T or when p decreases. In general, the various estimates of parameters obtained from the Bayesian method under both priors exhibit good behavior compared to their respective MLEs. Furthermore, proper Bayesian estimates are marginally superior to the corresponding improper Bayesian estimates. The interval estimates for all parameters are presented in Table 2 and Table 4 for various schemes. The following remarks are noted for these tables: ALs of both MLE-based and Bayesian intervals decrease with increasing sample sizes, and the CPs of these interval estimates are satisfactory. Moreover, the ALs (CPs) decrease (increase) when T increases or p decreases. The CPs of these intervals are close to significance level. It is observed that lengths of the HPD intervals are smaller than those of the likelihood-based intervals. Additionally, the HPD intervals derived from proper priors show better performances than those based on improper priors.
In summary, the simulation findings indicate that the performances of all the studied point and interval estimates are quite satisfactory under the GPHC competing risk data. The Bayes estimates, in general, are preferable to those estimates derived using the classical technique. Further, when prior information for the unknown parameters is provided, Bayes estimates are a better alternative; otherwise, we use improper prior-based Bayes estimates. Performance of likelihood estimates is also appreciable under given GPHC competing risk data.
5.2. Real Data Example
In this section, a real-life example is presented for application and illustrative purposes. The dataset represents the time, in minutes, taken to kick any goal in the UEFA Champion League for the 2004–2005 and 2005–2006 sessions. Initially, this dataset was analyzed by Meintanis [27]. The data are presented Table 5. Here, the variables X and Y indicate the time of the first kick goal scored by any team and the time of the first goal of any type scored by the home team, respectively. In this illustration, we assume X and Y to be two competing risk variables, corresponding to cause one and cause two, respectively.

Table 5.
Complete competing risks failure data for the proportion time.
Before proceeding, we will verify whether the MOIEP distribution can be utilized to fit these real-life data. For this purpose, we compared their goodness-of-fit with two other distributions, i.e., the inverted exponentiated exponential (IEE) and the inverted exponentiated Rayleigh (IER) models. Additionally, two other models, i.e., the modified half-logistic (MHL) distribution (e.g., Shaheed [28]) and a new mixture of exponential and Weibull (MEW) distribution (e.g., Mohammad [29]) are considered for comparison. We evaluated the marginals and the minimum data to determine whether the IEP distribution provides a satisfactory fit compared to the other distributions. The Kolmogorov–Smirnov distances and p-values (within bracket) for data on X, Y, and are calculated. The estimated results are reported in Table 6. Furthermore, the associated empirical cumulative distributions overlaying the theoretical cumulative distributions are plotted along with the probability–probability (PP) and the quantile–quantile (QQ) plots, as shown in Figure 2, Figure 3 and Figure 4. The estimated results and plots indicate that the IEP distribution fit is quite good compared to those of other models. Therefore, the proposed model can be used for fitting the marginals and minimums of the data for X and Y. Consequently, it may be concluded that the MOBIEP model is suitable for analyzing this football data.

Table 6.
The Kolmogorov–Smirnov (K-S) statistic and the p-values of IER, IEE, and IEP for data on X, Y, and .
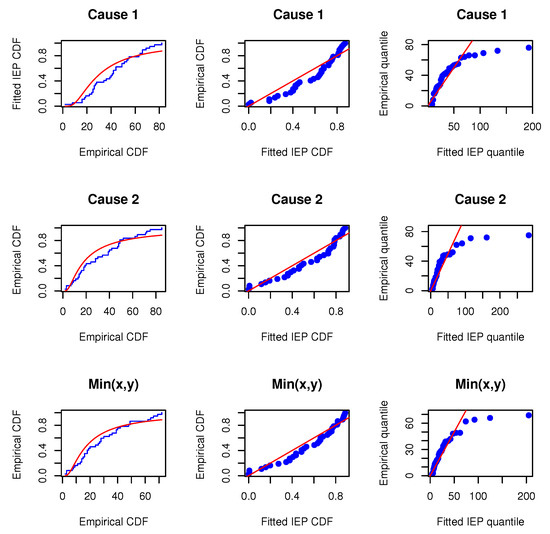
Figure 2.
Empirical distribution and fitted IEP models; PP and QQ plots for UEFA football data.
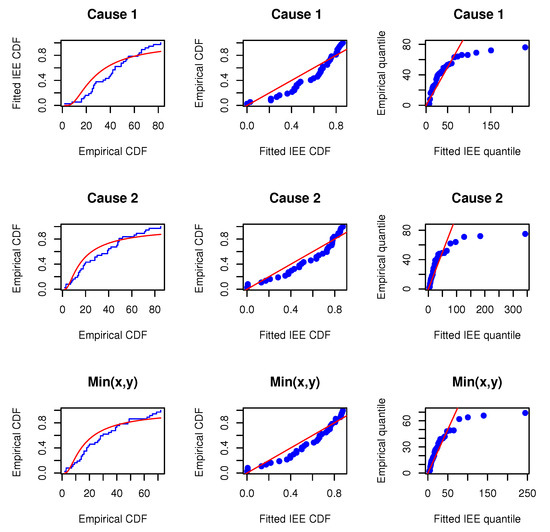
Figure 3.
Empirical distribution and fitted IEE models; PP and QQ plots for UEFA football data.
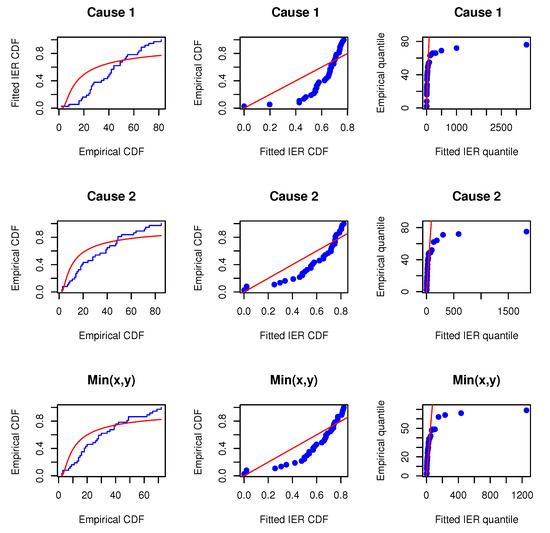
Figure 4.
Empirical distribution and fitted IER models; PP and QQ plots for UEFA football data.
From Table 5, three sets of artificial GPHC competing risk samples are obtained and presented in Table 7. The datasets are generated under the sample size , with the censoring scheme and with various monitoring points, T, and masking numbers, p. Various estimates for , and are listed in Table 8, where the intervals are obtained with a significance level of . In Table 8, the first two rows present point estimates along with their corresponding standard errors. The subsequent two groups of rows provide interval estimates. The associated interval lengths are obtained for asymptotic and Bayesian intervals. Bayes estimates are obtained using improper priors, and all the hyper-parameters are zeros. We observe from Table 8 under datasets I, II, and III that the MLEs and Bayes estimates are quite similar for the different model parameters. Furthermore, the behavior of the credible intervals is observed to be marginally better than that of the MLE-based intervals.

Table 7.
GPHC competing risks for UEFA football data.

Table 8.
Point and interval estimates of MOIEP parameters for the UEFA GPHC competing risks data.
6. Conclusions
In this paper, inferences for the dependent competing risk model are discussed under the GPHC scheme. The Marshal–Olkin bivariate IEP distribution approach is employed to model dependent competing risk failure data. Various inferences for the unknown parameters are derived using classical and Bayesian methods in the presence of partially observed failure causes. The existence and uniqueness of the MLEs for the model parameters are established. Bayes point and interval estimates are computed on based flexible priors. The proposed sampling technique is used for this purpose. Generally, numerical findings have demonstrated that Bayes estimates yield superior results compared to likelihood estimates when the proper prior distribution is applied. Specifically, the intervals obtained from the Bayes methods are shorter in length than those based on the MLE intervals. Although a dependent MOBIEP-generalized, progressively hybrid, censoring-based competing risk model is discussed in this paper, further inferences incorporating the EM algorithm and bootstrap method also appear interesting, these will be discussed in future research.
Author Contributions
Methodology, R.K.; Formal analysis, Y.M.T.; Investigation, R.K.S.; Writing—review & editing, L.W.; Funding acquisition, L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work of Liang Wang was supported by the Yunnan Fundamental Research Projects (No. 202401AT070116), the Yunnan Key Laboratory of Modern Analytical Mathematics and Applications (No. 202302AN360007), the Cross-integration Innovation team of Modern Applied Mathematics and Life Sciences in Yunnan Province, China (202405AS350003), and National Natural Science Foundation of China (12201395).
Data Availability Statement
Data are contained within the article.
Acknowledgments
Authors would like to thank the Editor and reviewers for their valuable comments and suggestions which significantly improved this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
Proof of Theorem 1.
Using the independent properties of , and we have
Therefore, the assertion is completed.
□
Appendix B
Proof of Corollary 1.
Based on Theorem 1, the joint PDF of can be obtained directly from under cases and , respectively. In addition, with the help of the regularity of the IEP model and the direct integral transformation, it is seen that and . Note that
Also, . So, the probability density of for case is given by
Therefore, the assertion is completed. □
Appendix C
Proof of Corollary 2.
From the independence of , and , the SF of can be expressed as
Therefore, the assertion is completed. □
Appendix D
Proof of Theorem 2.
Taking the derivatives of with respect to , , and , respectively, and setting them to zero, it is observed that
For fixed using inequality for and , respectively, with one has
and
As the function of , and , the log–likelihood function in Equation (12) can be rewritten as . Then, using , and the above inequalities, we have
Since , we further have
Equality holds if Therefore, the assertion is completed. □
Appendix E
Proof of Theorem 3.
From Theorem 2, substituting in expression (12) and ignoring the constant terms, the profile log–likelihood of can be expressed as
Subsequently, the required estimate is obtained from the following equation:
which will have an unique solution. The proof follows from Lodhi et al. [8]. In our case, we assume that , where is an increasing function; thus, we have for . Furthermore, we set up for and for for Case I; for , for and for for Case II; and for and for for Case III. Based on these setups, we can easily investigate from Theorem 3.2 in Lodhi et al. [8] that , , and decrease in , which implies that Equation (13) has a unique solution. Therefore, the assertion is completed.
□
References
- Kundu, D.; Basu, S. Analysis of incomplete data in presence of competing risks. J. Stat. Plan. Inference 2000, 87, 221–239. [Google Scholar] [CrossRef]
- Sarhan, A.M.; Hamilton, D.C.; Smith, B. Statistical analysis of competing risks models. Reliab. Eng. Syst. Saf. 2010, 95, 953–962. [Google Scholar] [CrossRef]
- Ghitany, M.E.; Tuan, V.K.; Balakrishnan, N. Likelihood estimation for a general class of inverse exponentiated distributions based on complete and progressively censored data. J. Stat. Comput. Simul. 2014, 84, 96–106. [Google Scholar] [CrossRef]
- Pascual, F. Accelerated life test planning with independent lognormal competing risks. J. Stat. Plan. Inference 2010, 140, 1089–1100. [Google Scholar] [CrossRef]
- Xu, A.; Basu, S.; Tang, Y. A full Bayesian approach for masked data in step-stress accelerated life testing. IEEE Trans. Reliab. 2014, 63, 798–806. [Google Scholar] [CrossRef]
- Wang, L. Inference of progressively censored competing risks data from Kumaraswamy distributions. J. Comput. Appl. Math. 2018, 343, 719–736. [Google Scholar] [CrossRef]
- Mahto, A.K.; Lodhi, C.; Tripathi, Y.M.; Wang, L. Inference for partially observed competing risks model for Kumaraswamy distribution under generalized progressive hybrid censoring. J. Appl. Stat. 2021, 49, 2064–2092. [Google Scholar] [CrossRef]
- Lodhi, C.; Tripathi, Y.M.; Wang, L. Inference for a general family of inverted exponentiated distributions with partially observed competing risks under generalized progressive hybrid censoring. J. Stat. Comput. Simul. 2021, 91, 2503–2526. [Google Scholar] [CrossRef]
- Crowder, M.J. Classical Competing Risks; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Wang, L.; Li, M.; Tripathi, Y.M. Inference for dependent competing risks from bivariate Kumaraswamy distribution under generalized progressive hybrid censoring. Commun. Stat. Simul. Comput. 2014, 51, 3100–3123. [Google Scholar] [CrossRef]
- Wang, L.; Tripathi, Y.M.; Dey, S.; Shi, Y. Inference for dependence competing risks with partially observed failure causes from bivariate Gompertz distribution under generalized progressive hybrid censoring. Qual. Reliab. Eng. Int. 2021, 37, 1150–1172. [Google Scholar] [CrossRef]
- Ashraf, R.; Saeed, N.; Saleem, K.; Aslam, M. Optimal design of multiple dependent state repetitive group sampling plan for Inverse Power Lomax distribution. Hacet. J. Math. Stat. 2022, 51, 525–542. [Google Scholar] [CrossRef]
- Marshall, A.W.; Olkin, I. A multivariate exponential distribution. J. Am. Stat. Assoc. 1967, 62, 30–44. [Google Scholar] [CrossRef]
- Kaishev, V.K.; Dimitrova, D.S.; Haberman, S. Modelling the joint distribution of competing risks survival times using copula functions. Insur. Math. Econ. 2007, 41, 339–361. [Google Scholar] [CrossRef]
- Pena, E.A.; Gupta, A.K. Bayes estimation for the Marshall-Olkin exponential distribution. J. R. Stat. Soc. Ser. B 1990, 52, 379–389. [Google Scholar] [CrossRef]
- Kundu, D.; Gupta, A.K. Bayes estimation for the Marshall-Olkin bivariate Weibull distribution. Comput. Stat. Data Anal. 2013, 57, 271–281. [Google Scholar] [CrossRef]
- Jamalizadeh, A.; Kundu, D. Weighted Marshall-Olkin bivariate exponential distribution. Statistics 2013, 47, 917–928. [Google Scholar] [CrossRef]
- Feizjavadian, S.H.; Hashemi, R. Analysis of dependent competing risks in the presence of progressive hybrid censoring using Marshall-Olkin bivariate Weibull distribution. Comput. Stat. Data Anal. 2024, 82, 19–34. [Google Scholar] [CrossRef]
- Panahi, H.; Sayyareh, A. Parameter estimation and prediction of order statistics for the Burr Type XII distribution with Type-II censoring. J. Appl. Stat. 2014, 41, 215–232. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Kateri, M. On the maximum likelihood estimation of parameters of Weibull distribution based on complete and censored data. Stat. Prob. Lett. 2008, 78, 2971–2975. [Google Scholar] [CrossRef]
- Rastogi, M.K.; Tripathi, Y.M. Estimating the parameters of a Burr distribution under progressive Type-II censoring. Stat. Methodol. 2012, 9, 381–391. [Google Scholar] [CrossRef]
- Feroze, N.; Aslam, M. Comparison of improved class of priors for the analysis of the Burr type VII model under doubly censored samples. Hacet. J. Math. Stat. 2022, 50, 1509–1533. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Aggarwala, R. Progressive Censoring: Theory, Methods, and Applications; Birkhauser: Bostan, Iran, 2000. [Google Scholar]
- Lawless, J.L. Statistical Models and Methods for Life Time Data, 2nd ed.; Wiley: Hoboken, NJ, USA, 2003; Volume 63. [Google Scholar]
- Cho, Y.; Sun, H.; Lee, K. Exact likelihood inference for an exponential parameter under generalized progressive hybrid censoring scheme. Stat. Methodol. 2015, 23, 18–34. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Sandhu, R.A. A simple simulational algorithm for generating progressive Type-II censored samples. Am. Stat. 1995, 49, 229–230. [Google Scholar] [CrossRef]
- Meintanis, S.G. Test of fit for Marshall-Olkin distributions with applications. J. Stat. Plan. Inference 2007, 137, 3954–3963. [Google Scholar] [CrossRef]
- Shaheed, G. A new two-parameter modified half-logistic distribution: Properties and applications. Stat. Opt. Inf. Comp. 2022, 10, 589–605. [Google Scholar] [CrossRef]
- Mohammad, G.S. A new mixture of exponential and Weibull distributions: Properties, estimation and relibilty modelling. São Paulo J. Math. Sci. 2024, 18, 438–458. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).