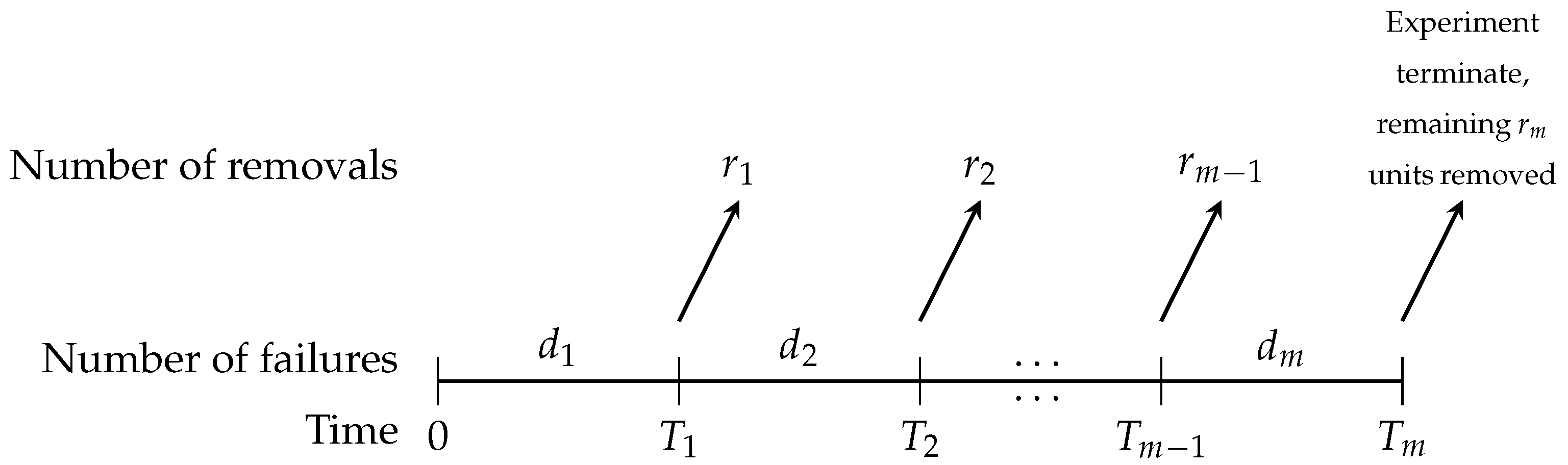1. Introduction
In reliability and survival analysis, selecting an appropriate statistical model is crucial for accurately describing lifetime data. For the data of increasing and decreasing nature of hazards, we have various lifetime distributions existing in the literature like Weibull, exponentiated exponential, gamma, and others. In a recent contribution, Verma et al. [
1] proposed a new two-parameter generalized Kavya–Manoharan exponential (GKME) distribution. This distribution was applied to model the survival times of guinea pigs, life of fatigue fracture of Kevlar 373/epoxy, and time of successive failures of the air conditioning system of jet air-planes. The GKME distribution exhibited a better fit compared to several sound models, including the Weibull, exponentiated exponential, gamma, and the alpha power transformed Weibull distribution. Despite its flexibility and superiority over other models, no further methodological development or inferential work has been carried out for the GKME distribution. Verma et al. [
1] explored several statistical properties of the GKME distribution and conducted parameter estimation within the classical framework using the method of maximum likelihood. However, their analysis was limited to complete data scenarios. In practical applications, particularly in reliability and survival studies, the available data are often subject to censoring. Recognizing this limitation, the present study seeks to bridge this gap by developing a comprehensive inferential framework for the GKME distribution under censored data.
The nature and structure of censoring depend on the experimental protocol and data collection procedures. For example, in many practical scenarios, experiments may be terminated before the failure of all test units due to time constraints, budgetary limitations, or other logistical challenges. This has led to the development of various censoring mechanisms. Among the most frequently used are the classical Type-I and Type-II censoring schemes (see, Sirvanci and Yang [
2]; Basu et al. [
3]; Singh et al. [
4]). Another important consideration in life-testing arises when the exact failure times of units are not directly observable. In many experimental settings, continuous monitoring of all units is infeasible due to limited manpower or technical constraints. Instead, the experiment is structured around periodic inspections, during which only the number of failures occurring within each time interval is recorded. This scenario gives rise to interval censoring, wherein failure times are only known to fall within certain intervals. Interval-censored data are frequently encountered in biomedical studies, especially in longitudinal trials and clinical follow-ups (Finkelstein [
5]). As noted by Jianguo [
6], such data structures are also prevalent in demography, epidemiology, sociology, finance, and engineering disciplines. In their work, Guure et al. [
7] investigated Bayesian inference for survival data subject to interval censoring under the log-logistic model. Later, Sharma et al. [
8] utilized Lindley’s approximation for Bayesian estimation of interval-censored lifetime data based on the Lindley distribution.
However, these approaches generally do not permit the withdrawal of experimental units before the study reaches completion. In real-world applications, maintaining all units under observation until the end is often impractical or even undesirable. Factors such as cost, time, ethical concerns, or logistical limitations may necessitate the withdrawal of units during the course of the experiment. To address this, Balakrishnan and Aggarwala [
9] have developed a progressive censoring scheme that provides a structured yet flexible way to remove subjects partway through the study. Under this approach, the withdrawal of units is planned in advance and carried out systematically, either immediately following certain observed failures or at specified points in time.
Taking this limitation into account, Aggarwala [
10] proposed a hybrid censoring scheme, known as progressive Type-I interval (PITI) censoring, which integrates the features of Type-I interval censoring with those of progressive censoring. This approach has been widely applied, particularly in clinical trial settings. Consider longitudinal study involving
n bladder cancer patients, where the objective is to track the duration of remission. Rather than observing each patient continuously, medical evaluations are conducted at predetermined time points
. At the first check-up (time
), only
individuals attend the visit, implying that
participants have exited the study within the interval
. Of the
patients examined at this stage, the recurrence of cancer is confirmed in
cases. However, the precise time of recurrence for these individuals is not known, but only that the event occurred within the given time interval. Subsequently, at time
, the remaining cohort
is considered, and once again a portion, say
, departs during the interval
. The remaining patients,
, are evaluated, and
cases of recurrence are detected. This process continues through
m successive inspection times, and at the final visit (
), all surviving patients who have not dropped out or shown recurrence are removed from the trial, effectively concluding the experiment. In prior work, Chen and Lio [
11] proposed an approach for parameter estimation under the considered censoring scheme using the generalized exponential distribution. Their method assumes that the proportions
of dropouts at each interval are fixed in advance. More precisely, they set
, where
denotes the number of subjects at risk at the beginning of the
i-th interval. While mathematically tractable, this deterministic assumption about dropouts may not reflect the inherent unpredictability of clinical trials. Patients often withdraw from treatment due to unforeseen events such as death from unrelated causes, side effects, personal preferences, or dissatisfaction with medical care, all of which lie outside the experimenter’s control. Recognizing these practical challenges, several researchers have advocated modeling the number of removals as random variables. For example, Tse et al. [
12], introduced censoring schemes with binomially distributed removals. Ashour and Afify [
13] extended this framework by applying PITI censoring with binomial removal under the assumption that the exact lifetime durations are observable. In the present study, however, we consider a more realistic scenario where exact failure times are unobservable. Instead, only the number of failures occurring within specified intervals is available. We adopt a binomial model for the dropout mechanism, wherein the number of removals at the
i-th stage, denoted
, follows a binomial distribution with parameters that depend on the remaining number of patients and a dropout probability
. Under this more flexible framework, we aim to develop statistical estimators for the unknown shape and scale parameters of the underlying distribution, assuming a progressively Type-I interval censored structure with binomially distributed random removals and unobserved exact failure times. Several studies have extended its use to different lifetime models. For instance, Kaushik et al. [
14,
15] discussed Bayesian and classical estimation procedures under PITI censoring using generalized exponential and Weibull distribution. Lodhi and Tripathi [
16] investigated inference for the truncated normal distribution in the presence of PITI censoring, while Alotaibi et al. [
17] examined Bayesian estimation for the Dagum distribution. More recently, Roy et al. [
18] studied the inverse Gaussian distribution under PITI censoring using the method of moments, maximum likelihood estimation, and Bayesian methods, and also proposed optimal censoring designs. Hasan et al. [
19] considered four strategies for determining inspection times—predefined, equally spaced, optimally spaced, and equal probability—and derived optimal censoring plans by evaluating expected removals or their proportions. Building on these contributions, the present work aims to develop inferential procedures for the GKME distribution under the PITI censoring scheme.
Let
X be a non-negative continuous random variable that follows the generalized Kavya–Manoharan exponential (GKME) distribution. Adopting the same notations as used in Verma et al. [
1], the cumulative distribution function (CDF) of the GKME distribution is defined as
where
and
are shape and rate parameters, respectively. Henceforth, the GKME distribution with parameters
and
is denoted by GKME
. The probability density function (PDF) and hazard function of the GKME distribution are, respectively,
and
Figure 1 displays the PDF and hazard function of the GKME distribution for various parameter settings. When
, the PDF takes a unimodal form, and the peak of the curve becomes progressively flatter as
increases. In contrast, for
, the PDF resembles an exponential distribution. The behavior of the hazard function also depends on
: it increases monotonically when
, whereas it decreases monotonically when
.
The remainder of this article is organized as follows.
Section 2 provides a detailed description of the PITI scheme with binomial removals and presents the observed likelihood function under this censoring scheme.
Section 3 focuses on the estimation of model parameters based on PITI-censored data using the ML method. Bayesian estimation using informative priors under SELF and GELF is discussed in
Section 4. A Monte Carlo simulation is conducted in
Section 5 to evaluate how various estimators perform when data are subject to PITI censoring. In
Section 6, to illustrate the applicability of the proposed method, a real-world dataset involving survival times of guinea pigs is examined. Finally, conclusions are given in
Section 7.
3. Maximum Likelihood Estimation
For estimating the parameters
and
using ML estimation, the above likelihood function for GKME distribution is written as
The above expression can be decomposed as
where
It can be observed that
does not involve the parameters
and
. Therefore, for the purpose of obtaining the ML estimates of
and
, it suffices to consider only the component
. The associated log-likelihood function is then given by:
Hence, the ML estimators of the parameters
and
are determined by jointly solving the following pair of nonlinear equations:
From the above equations, the probability distributions of the estimators
and
are analytically intractable. Therefore, it is impractical to derive the exact confidence intervals for these parameters. To address this, we utilize the asymptotic normality property of ML estimators to construct
ACIs for
and
. These intervals are derived using the asymptotic variance-covariance matrix associated with their MLEs. The joint asymptotic distribution of
follows a bivariate normal (BN) distribution:
where
represents the observed Fisher information matrix, given by
The components of the Fisher information matrix are given by
where
Based on the observed Fisher information matrix, the two-sided symmetric
ACIs for
and
are obtained as
where
denotes the upper
percentile of the standard normal distribution, and
and
correspond to the estimated variances of
and
, extracted from the principal diagonal of the asymptotic variance–covariance matrix, which is the inverse of the observed Fisher information matrix,
.
4. Bayesian Estimation
In this section, we present the Bayes estimation of the unknown parameters and using the MITI sample. Unlike classical estimation methods that rely only on the likelihood function, the Bayesian paradigm combines prior information about the parameters with the observed data through Bayes’ theorem. This results in the posterior distribution, which serves as the basis for inference. The Bayes estimator of a parameter is then obtained as a functional (such as the mean, median, or mode) of its posterior distribution, depending on the choice of loss function.
The loss function plays a crucial role, as it quantifies the penalty for the difference between the estimated and true values of the parameter. Different loss functions lead to different forms of Bayes estimators. In this study, we consider three widely used loss functions: SELF, LLF, and GELF. The SELF is symmetric and treats overestimation and underestimation equally. On the other hand, the LLF and GELF are asymmetric, making them more realistic for situations where the consequences of overestimation and underestimation are not the same.
SELF: In keeping with its name, SELF assigns a value equal to the square of the estimation error. Now, if the estimator estimates the true state
by
then the estimation error is to be
, and accordingly, the incurred loss is
From (
17), it is clear that the higher the divergence from the true state, the heavier the penalization. It is needless to mention that the Bayes estimator under the SELF is the posterior mean, i.e.,
.
GELF: The GELF proposed by Calabria and Pulcini [
20] can be written as
The Bayes estimator corresponding to GELF is
. The sign of loss parameter
w signifies the direction of asymmetry, while its size characterizes the intensity of asymmetry.
Prior Distribution and Posterior Analysis
In Bayesian analysis, prior distributions play a central role by incorporating existing knowledge or assumptions about the parameters before observing the data. The choice of an appropriate prior is crucial, as it can influence both the posterior distribution and the resulting inferences. However, there is no universally accepted methodology for selecting “optimal” priors, particularly when dealing with newly developed distributions. For the GKME distribution, determining a joint conjugate prior is analytically challenging due to the presence of two unknown parameters, which complicates the derivation of closed-form posterior distributions. To address this, it is common in Bayesian practice to adopt flexible and computationally convenient families of priors. In this study, we consider independent gamma priors for both parameters. The gamma distribution is a natural choice because it is defined on the positive real line, aligns well with the support of the parameters, and allows straightforward computation within the MCMC framework. This choice strikes a balance between flexibility and tractability, enabling us to obtain reliable posterior estimates while keeping the computational burden manageable. The gamma densities for
and
with hyperparameters
and
are
and
Sometimes, we do not have enough prior information about the phenomenon; in such cases, we use non-informative priors. Here, we have used uniform prior as a non-informative prior. The joint prior density of parameters (
,
) is given by
The joint posterior density of (
,
) based on the likelihood function will be
where
The Bayes estimate of any function
under SELF and GELF can be written as
and
The Bayes estimates of parameters
and
are obtained by substituting
and
in place of general function
. The double integral involved in (
23) and (
24) are analytically intractable. Thus, we compute the Bayes estimates
and
via the MCMC approach. MCMC methods serve as essential tools for Bayesian computation. Notably, the Metropolis–Hastings (M-H) algorithm and Gibbs sampling (Hastings, [
21]; Smith and Roberts [
22]) are among the most commonly employed techniques for generating MCMC samples. For the implementation of the Gibbs sampling algorithm, it is necessary to derive the full conditional posterior densities of
and
, which are given by:
The detailed algorithm to generate posterior samples and to calculate Bayes estimates is given below in Algorithm 1:
| Algorithm 1 MCMC algorithm for Bayesian estimation of and . |
- 1:
Initialize and - 2:
for to M do - 3:
Generate from using M-H:
Generate candidate and . Compute acceptance probability If , set ; else .
- 4:
Generate from using M-H:
Generate candidate and . Compute acceptance probability If , set ; else .
- 5:
end for - 6:
Discard the first samples (burn-in). - 7:
- 8:
Credible Intervals: - 9:
Order the post burn-in samples as and . Then the symmetric credible intervals are The intervals with the shortest length are taken as the HPD intervals.
|
5. Simulation Study
This section presents a comprehensive Monte Carlo simulation designed to evaluate the performance of the proposed estimators under the PITI censoring framework. The assessment involves both point and interval estimation criteria, measured using the MSE of the estimators and average width (AW) and coverage probability (CP) of the confidence and credible intervals. For generating PITI censored samples from a given distribution, we adopted Algorithm 2 as suggested by Balakrishnan and Cramer [
23]. Data samples of size
, and 50 were generated from the GKME distribution with two different parameter combinations. To explore the effect of censoring time
, five variations of
and 7 are considered and for censoring intervals,
m, three different values of
m are considered, i.e.,
, and, 10. For observing the behavior of removal scheme, we have considered four random removal schemes, the first scheme considers no withdrawal of units or say, the risk of dropping at all the intermediate stages to be zero i.e., probability or removing the units
p is zero at all the stages before final termination. The second scheme assumes an equal and non-zero probability of removal at each intermediate stage before termination, particularly, we have taken
. The third scheme assigns a higher probability
p of removal at the beginning, which then decreases as the process advances. This situation can be seen in the fitness programs where many participants register initially but drop out early if they lose interest; those who persist beyond early stages are more likely to complete the program. Conversely, in the fourth scheme, probability of removal
p is small in the initial stages and gradually increases in the later stages. This removal scheme can be seen in clinical trials where patients rarely drop out early because they are motivated or monitored closely at the beginning. As the trial progresses, fatigue, side effects, hospital services, or long follow-up may cause higher dropout rates in later stages. In particular, removal schemes are taken as
;
,
, and
, where
and [.] denotes the greatest integer number. Under each configuration, ML estimates and Bayes estimates are computed alongside their MSEs. Confidence intervals for
level of significance were derived using ACI methods, are their corresponding AW and CP are also reported. Bayes estimates of the parameters
and
are calculated for informative prior, under SELF and GELF. The weight parameters used in GELF were
and
. For IP, we propose the use of independent gamma priors for
and
. The hyper-parameters of prior distribution are selected using the method of moments as outlined in Singh et al. [
4] and Yadav et al. [
24]. For NIP, we have used uniform prior. The MSEs of Bayes estimators is calculated along with AW and CP of
highest posterior density credible intervals (HDIs) under both prior settings are also calculated. All analyses are conducted using R software (
https://www.r-project.org/) along with relevant additional packages and computational tools required for estimation, simulation, and plotting.
Results from the simulation are summarized in
Table 1,
Table 2,
Table 3 and
Table 4. From these tables, the following conclusions can be drawn:
- 1.
Table 1 represents the average estimates and MSEs for various combinations of
for fixed
and
for a fixed removal scheme. It is observed that the MSE of
decreases as
increases, suggesting that higher scale parameter values lead to more precise estimation of
. Conversely, the MSE of
increases as
increases, indicating that shape parameter variation introduces more estimation variability. For
, the MSE increases with both
and
, implying that higher parameter values make shape estimation relatively more difficult.
- 2.
Bayes estimators under GELF for loss parameter exhibit the lowest MSE across all the parameter combinations. This indicates that Bayesian inference with asymmetric loss functions can effectively reduce estimation error.
- 3.
Table 2 represents the average estimate and MSE for varying
n, removal schemes,
R and keeping
,
,
and
fixed. From
Table 2, the MSE of the estimators
and
is the least for removal scheme 1 followed by 4, 3 and 2 for a fixed
n. And the MSE of estimators decreases as the sample size increases.
- 4.
Table 3 represents the ACI and HDI with their AW and CP for varying
n,
R and fixed
,
. The AW of HDI is lower than ACI for both
and
, meaning Bayesian HDIs provide narrower intervals, reflecting greater precision. CP associated with HDI is higher than that of ACI, showing that HDI not only gives narrower intervals but also maintains better coverage.
- 5.
AW corresponding to removal scheme is lowest, followed by the AW corresponding to removal schemes , and in most of the cases. CP is also highest for removal scheme 2 in most cases.
- 6.
Table 4 represents the average estimates and their MSEs (in parentheses) for varying
m,
for
,
and
. MSE decreases as censoring time
increases, because extending the study period allows more failures to be observed, leading to improved estimation. Conversely, MSE increases as the number of intervals
m increases, because more intervals mean more chances of incomplete information due to multiple censoring points.
- 7.
The Bayes estimators can be seen to have a smaller MSEs than the classical estimators for all the considered cases.
| Algorithm 2 Simulation of Progressive Interval Type-I Censored Sample |
- 1:
Specify the value of ; model parameters . - 2:
- 3:
Set , , . - 4:
- 5:
Set . If , exit the algorithm. - 6:
- 7:
Generate . - 8:
Update . - 9:
if
then - 10:
- 11:
Generate . - 12:
- 13:
else - 14:
Set . - 15:
- 16:
end if - 17:
Update and go to Step 3.
|