Abstract
Acidithiobacillus thiooxidans A01 is widely used in bioleaching processes and commonly thrives in most metal-rich environments. However, interactions between different heavy metals remain obscure. In this study, we elaborated the effect of ferric iron on the growth and gene expression of At. thiooxidans A01 under the stress of nickel. The results showed that 600 mM Ni2+ completely inhibited the growth and sulfur metabolism of At. thiooxidans A01. However, trace amounts of Fe3+ (0.5 mM) facilitated the growth of At. thiooxidans A01 in the presence of 600 mM Ni2+. With the addition of 5 mM Fe3+, the maximum cell density reached 1.84 × 108 cell/mL, and pH value was 0.95. In addition, metal resistance-related and sulfur metabolism genes were significantly up regulated with extra ferric iron. Taking the whole process into account, the promoting effect of Fe3+ addition can be attributed to the following: (1) alleviation of the effects of Ni2+ toxicity and restoring the growth of At. thiooxidans A01, (2) a choice of multiple pathways to export nickel ion and producing precursor of chelators of heavy metals. This can suggest that microorganisms may widely exhibit metabolic activity in iron-rich environments with heavy metals. Our study will facilitate the technique development for the processing of ore bodies with highly challenging ore compositions.
1. Introduction
Biohydrometallurgy is widely used in industrial bioleaching of ores to extract metals such as copper, nickel, gold, uranium. It is done mainly by acidophilic microorganisms [1]. Some of the heavy metals are essential for the metabolic activity of cells at low levels, while they exert toxic effects at concentrations encountered in polluted environments.
Nickel has been identified as a component in a number of enzymes, participating in important metabolic reactions such as ureolysis, hydrogen metabolism, methane biogenesis and acitogenesis [2]. In this way, microorganisms have evolved in the presence of this metal, which is necessary in trace amounts for a variety of metabolic processes but toxic in high concentrations, causing oxidative stress in the cell [3]. The toxicity of nickel is attributed to its replacement of metals in metalloproteins, to its binding to catalytic residues in sulfur dioxygenase, sulfite oxidase and the plasma membrane, and indirectly to its exertion of oxidative stress [4].
Several nickel resistance bacteria have been isolated mainly from heavy metal-contaminated samples [5]. To tackle Ni toxicity, microorganisms have developed diverse mechanisms including extracellular detoxification, intracellular sequestration, modification of cation transport systems and active transport by efflux pumps [6]. Most nickel resistance determinants known to date are efflux pumps isolated from cultivable bacteria. The resistance-nodulation-cell division (RND) protein family was described as a related group of bacterial transport proteins involved in heavy metal resistance [7]. In Gram-negative bacteria, the Cobalt-zinc-cadmium resistance transporter (CzcCBA) (consisting of subunits C, B and A) is a member of the RND system that exports metals including cobalt, zinc and cadmium from the cytoplasm or the periplasm across the outer membrane [8]. The complex achieves heavy metal resistance by active cation efflux driven by a cation-proton antiporter [9,10]. In Cupriavidus metallidurans strain CH34, RND protein is central to the process of metal cation homeostasis that is adapted to high concentrations of Zn2+, Cd2+, Ni2+, Co2+, Cu+ or Ag+ [11].
New nickel resistance systems are still being discovered. A nickel hyperaccumulation mechanism is proposed for a serine O-acetyltransferase [12]. Seven different genes conferring Ni resistance were identified in Acidiphilium sp. PM. Two of them form an operon encoding the ATP-dependent HslVU (ClpQY) protease. Other Ni-resistance determinants include genes involved in lipopolysaccharide biosynthesis and the synthesis of branched amino acids and membrane fatty acid [13,14]. Acidithiobacillus ferrooxidans and Pseudomonas sp. MBR had the ability to reduce Ni2+ to its elementary forms coupled to biomineralization under aerobic conditions [15,16]. It suggests that Ni resistance relies on different molecular mechanisms.
Nickel and other heavy metals are often encountered in an aquatic environment. Gikas observed that Ni2+ and Co2+ acted synergistically at the increasing stimulation and at the intoxication zones, whilst an antagonistic relation was determined at the decreasing stimulation zone [2]. Similar experiments implied that ferrous iron gave significant protection to Acidithiobacillu caldus BC13 against lead and zinc toxicity [17]. Bacillus may widely exhibit catabolic activity in iron-rich environments with heavy metal [2]. It is obvious that interactions between different heavy metals remain obscure, while it should be taken into account in the methodologies used to establish criteria for tolerance levels in the environment.
At. thiooxidans is a conspicuous dweller of acidic metal-rich environments. It is well known for its rapid oxidation of elemental sulfur. It is able to oxidize sulfur and reduced sulfur compounds for energy but not Fe2+. At. thiooxidans A01 is not able to tolerate high levels of Ni2+. The co-culture of At. thiooxidans and iron-oxidizing Leptospirillum ferriphilum showed a stronger sulfur oxidation and ferrous oxidation activity under the stress of heavy metals including Ni2+ and As3+ [18,19]. It indicated that there were synergetic interactions between iron-oxidizing and sulfur-oxidizing bacteria. We suggested that the metabolites of L. ferriphilum may facilitate the growth of At. thiooxidans under the stress of nickel. In this study, we investigated the effect of ferric iron on the growth and sulfur metabolism of At. thiooxidans A01 in the presence of Ni 2+ to clarify the mechanism at transcription level.
2. Materials and Methods
2.1. Bacterial Strain, Medium and Growth Condition
In this study, the bacterial strain was At. thiooxidans A01 (FJ15-4526), which was isolated and identified by our laboratory from Ping Xiang coal mine [20]. It was grown in 9K medium, which contained 3.0 g/L (NH4)2SO4, 0.5 g/L MgSO4·7H2O, 0.5 g/L K2HPO4, 0.1 g/L KCl, 0.01 g/L Ca(NO3)2. The initial pH value of the medium was 1.8 and the medium was autoclaved for 20 min at 121 °C. The energy resource was elemental sulfur (S0, 10 g/L) and it was incubated at 30 °C and 170 rpm.
2.2. Nickel Resistance Experiment with Different Ferric Iron Concentration
To investigate the effect of ferric iron on nickel resistance, this experiment was designed to form a ferric iron concentration gradient during At. thiooxidans A01 incubation with a certain nickel concentration. After exposure to ultraviolet light (10 W, 254 nm) for 30 min, ferric sulfate (Fe2(SO4)3·7H2O; 0, 0.5, 2.5, 5 and 50 mM), nickel sulfate (NiSO4·6H2O; 600 mM) and elemental sulfur (S0; 10 g/L) were added in previously autoclaved 100 mL 9K medium, which was in 250-ML Erlenmeyer flasks. The initial cell density was 1 × 107 cells/mL, the initial pH value of the medium was 1.8, and they were incubated in a shaker incubator set at 30 °C and 170 rpm. Each treatment was performed in triplicate.
The physicochemical parameters including pH and cell density were measured every day. The pH value was measured by pH meter, the density of the microorganism was counted by a Thomas chamber with an optical microscope at 100× magnification.
2.3. Shock Treatment
The shock experiment was conducted to explore the influence of ferric iron or nickel on the relative gene expression. At. thiooxidans A01 was incubated with 10 g/L S0 at 30°C and 170 rpm, and the initial cell density was 1 × 107 cells/mL and the initial pH of 9K medium was also 1.8. We designed three treatments, including 5 mM ferric iron shock, 600 mM nickel sulfate shock, and 5 mM ferric iron, 600 mM nickel sulfate joint shock. The shock treatment was at the 108th hour, the cell density was 1.00 × 108 cells/mL, and the pH was 0.77. The control group was not shock treated. Before utilizing, 9 K medium, S0, ferric sulfate and nickel sulfate were sterilized. After shock treatment, cells were harvested for RNA extraction in the 10th, 30th, 60th and 120th mins, the cell densities were 1.00 × 108, 1.03 × 108, 1.06 × 108, 1.18 × 108 cells/mL in the presence of Fe3+, 1.00 × 108, 7.95 × 107, 8.80 × 107, 4.51 × 107 cells/mL in the presence of Ni2+, 1.00 × 108, 7.96 × 107, 8.80 × 107, 6.00 × 107 cells/mL in the presence of Ni2+ and Fe3+, respectively.
2.4. Cell Collection and RNA Extraction
The process of cell collection was shown in a previous study by Jiang et al. [20]. The cells were filtered out using a filter paper (0.45 μm) to remove the remaining sulfur. The solution was put into centrifuge bottle and centrifuged at 12,000 rpm for 20 min to collect the cells. The total RNA extraction was isolated and purified according to the method described by Wang et al. [8]. About 1 μg purified RNA from each sample was used for cRNA synthesis with the ReverTra Ace Qpcr RT Kit (Toyobo, Osaka, Japan), according to the manufacturer’s protocol. RNA extracts were treated with DNase to remove DNA before cDNA synthesis.
2.5. Primers and Real-Time Polymerase Chain Reaction (PCR)
The genome of At. thiooxidans A01 has been sequenced, completely annotated and deposited at the DDGJ/EMBL/Genbank with the accession number AZMO0000 [21]. Nickel resistance-related genes and growth-related genes were predicted based on the genome sequences. Specific primers used in this study are shown in Table 1. The specificity of primers was checked by conventional polymerase chain reaction (PCR) and sequencing.

Table 1.
Specific primers used for polymerase chain reaction (PCR) and real-time PCR.
3. Results
3.1. Growth of At. thiooxidans A01 under Different Ni2+ Concentrations
The growth characteristics of At. thiooxidans A01 under the influence of Ni2+ in different concentrations from 0 to 600 mM was studied with regard to media pH changes and densities of bacterial cell cultures (Figure 1); 600 mM Ni2+ completely inhibited the growth and sulfur metabolism of At. thiooxidans A01, and longer lag phase and shorter exponential phase were observed with the addition of Ni2+. The maximum cell density was 2.02 × 108 cells/mL in blank, while in Ni2+ varied from 50 to 600 mM; the maximum cell densities were 1.16 × 108, 9.6 × 107, 8 × 107, 4.76 × 107, 2.38 × 107, 1.19 × 107, 1.02 × 107 cells/mL, respectively. The final pH value gradually reduced and varied from 0.32 to 1.86 with the increase of Ni2+, and up to 600 mM Ni2+ no notable decline could be observed. It is clear that the presence of 600 mM Ni2+ completely inhibited the growth and sulfur metabolism of A01.
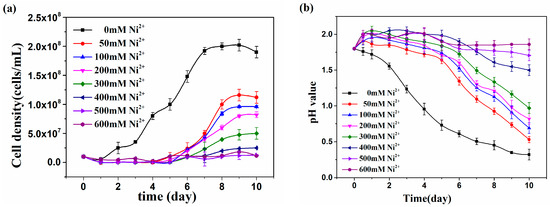
Figure 1.
The cell density (a) and pH value (b) of Acidithiobacillus thiooxidans A01 under different Ni2+ concentrations.
3.2. The effect of Ferric Iron on Growth of At. thiooxidans A01 under Ni2+
Ni2+ and Fe3+ are often encountered in bioleaching systems, so the effect of Fe3+ in different concentrations from 0 to 50 mM was investigated under the influence of 600 mM Ni2+ (Figure 2). This showed that At. thiooxidans A01 could tolerate a high concentration of Ni2+ with the addition of Fe3+. The maximum cell densities increased with the concentration of Fe3+ and were 4.91 × 107, 1.04 × 108, 1.84 × 108 cells/mL respectively, when At. thiooxidans A01 was exposed to 0.5, 2.5, and 5 mM Fe3+. But further increases of Fe3+ showed no further effect on growth. With the addition of 5 mM Fe3+, the pH values decreased from 1.80 to 0.95 quickly after 2 days. All the Eh decreased to 450 mv in 10 days. The results indicated that 5 mM Fe3+ restored the inhibition of Ni2+ on At. thiooxidans A01. There is no doubt that Fe3+ played a vital and positive role in the growth and metabolism of At. thiooxidans A01 under high concentration of Ni2+.
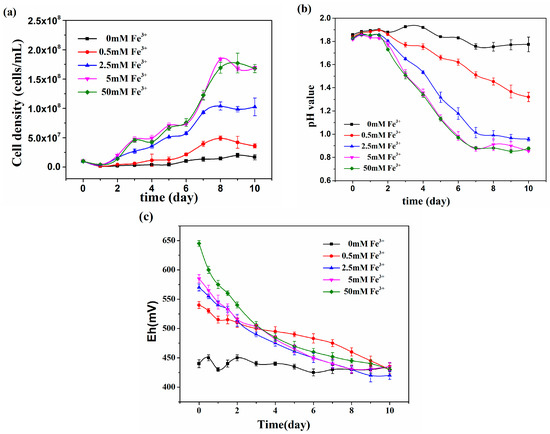
Figure 2.
The effect of ferric iron on the cell density (a), pH value (b) and Eh (c) of At. thiooxidans A01 under the influence of 600 mM Ni2+.
3.3. Growth of At. thiooxidans A01 under Shock Treatment of Individual and Joint Ni2+ and Fe3+
At. thiooxidans A01 at exponential phase (108th hour) was shocked with individual and joint 600 mM Ni2+ and 5 mM Fe3+ (Figure 3). The bacterium fell into recession after Ni2+ shock and cell density decreased from 1.00 × 108 cells/mL to 6.00 × 107 cells/mL in 24 h; however, after joint Ni2+ and Fe3+ shock the cell density restarted increasing until the cell density reached 1.20 × 108 cells/mL after 24 h. The pH value was 0.77 at 108th hour without shock, while in the presence of Ni2+ and in the presence of Ni2+ and Fe3+ the pH value increased from 0.77 to 0.98 in 2 h subsequently down to 0.76 and 0.55 at 240th hour, respectively. It can be seen that the addition of Fe3+ would alleviate the shock of 600mM Ni2+ on At. thiooxidans A01.
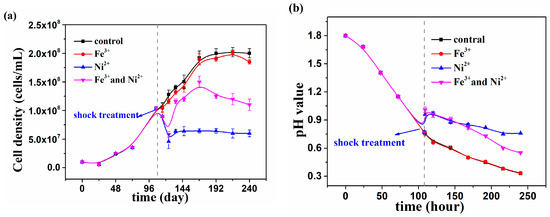
Figure 3.
The cell density (a) and pH value (b) of At. thiooxidans A01 under shock treatment of individual and joint Ni2+ and Fe3+.
3.4. Functional Gene Expression of At. thiooxidans A01 by Single or Joint Ni2+ and Fe3+ Shock
The expression of metal resistance-related and sulfur metabolism genes was analyzed at transcriptional level by real-time PCR to investigate the effect of Fe3+ on At. thiooxidans A01 after the shock of Ni2+.
The expression of metal resistance-related genes in the 10th min, 30th min, 60th min, 120th mins after shock treatment is shown in Figure 4. The expression of metal resistance-related genes was more active in the presence of Ni2+ and Fe3+. At first, the expression of genes encoding adenosine triphosphate(ATP)-binding-cassette (ABC) transporter ATP-binding protein (CDB) in the presence of Ni2+ and Fe3+ was threefold higher than that in the presence of Ni2+ and up regulated in the 30th min. The same regulation occurred in the expression of genes encoding CzcAEP protein. During the 30th to 60th min, the expression of most tested genes was consistently upregulated, and the expression in the presence of Ni2+ and Fe3+ was higher than that in the presence of Ni2+. In particular, the expression of genes encoding RND, copper, CDB, and ABC transporter-related proteins CDC1 and CDC2 was more than twofold higher than that in the presence of Ni2+. These data indicated that transporter genes were overrepresented with additional Fe3+. In 120th min, the expression of most genes was down regulated. But in the presence of Ni2+, the expression of genes encoding CzcAEP was four-fold higher than that in the presence of Ni2+ and Fe3+.
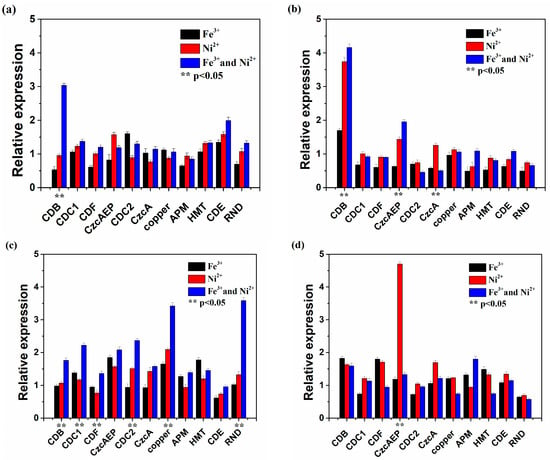
Figure 4.
The expression of metal resistance-related genes in At. thiooxidans A01 in 10th min (a), 30th min (b), 60th min (c), 120th min (d) after single or joint Ni2+ and Fe3+ shock.
The expression of sulfur metabolism-related genes after single or joint Ni2+ and Fe3+ shock was shown in Figure 5. In the 10th min, most genes in the presence of Ni2+ and Fe3+ had higher expression, but from 30 min to 120 min after a single Ni2+ shock, the expression of most investigated genes was higher. It is obvious that Fe3+ would only promote the expression of sulfur metabolism-related genes at the early stage. It is also worth mentioning that the expression of cysA in the presence of Ni2+ and Fe3+ was much higher than that in the presence of Ni2+ or Fe3+, after 30 min the expression of most genes was upregulated in the presence of Ni2+, especially serinO. Compared with the other genes, the expression of soxZ-1, rhd, cysA, cysB and serinO was higher in the presence of Ni2+ and Fe3+ and in the presence of Ni2+ in the 60th min. In the 120th min, the expression of rhd in the presence of Ni2+ was six-fold higher than that in the presence of Ni2+ and Fe3+. The results indicated that these sulfur metabolism-related genes were vital to maintain growth activity under high nickel shock.
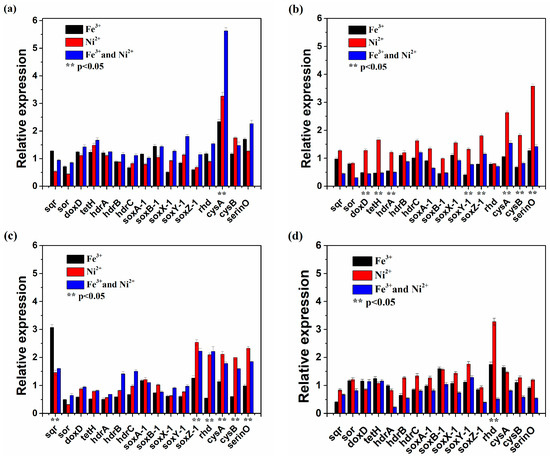
Figure 5.
The expression of sulfur metabolism-related genes in At. thiooxidans A01 in 10th min (a), 30th min (b), 60th min (c), 120th min (d) after single or joint Ni2+ and Fe3+ shock.
4. Discussion
At. thiooxidans A01 plays a key role in bioleaching processes and commonly thrives in the Earth’s most metal-rich environments [21]. Previous research indicated that metal ions can affect the heavy metal resistance of bacteria [22,23], so we investigated the effect of Fe3+ on the growth and related genes expression of At. thiooxidans A01 under the stress of Ni2+. At. thiooxidans A01 grows slowly and with a reduced ability to oxidize elemental sulfur in the presence of 300 mM Ni2+ [18], but we found that trace amounts of Fe3+ (0.5 mM) facilitated the growth and sulfur metabolism of At. thiooxidans A01 in the presence of 600 mM Ni2+ and it could not further alleviate the toxicity when Fe3+ was more than 5 mM, indicating that Fe3+ may not weaken the toxicity through the complex with nickel.
Iron participates in a variety of essential biological functions, including metalloregulation, structural stabilization, electron transfer, substrate/cofactor coordination and catalysis [24,25]. Previous studies have shown that growth perturbations induced by metal ion stress can be attributed to disruption of cellular metal ion homeostasis, commonly resulting from protein mismetallation [26,27]. In this study, high concentrations of Ni2+ inhibited the growth and sulfur metabolism of At. thiooxidans A01. Ni2+ could improperly bind to metalloproteins and cause oxidative stress (production of reactive oxygen species, including free radicals and peroxides), which damage major molecular components. The iron-sulfur clusters were the primary intracellular targets of metal ion toxicity. Higher concentrations of metal ion led to total disintegration of the Fe-S cluster in central catabolic and biosynthetic pathways, evidently displacing iron atoms [24,28,29]. Some iron-containing enzymes are inhibited by nickel, with some shown to have nickel replacing the active metal [26]. To avoid such a process, the intracellular concentration of metal is tightly regulated and bacteria require precise homeostatic mechanisms to balance the uptake and storage of different metals. Our results revealed that the external supplementation of ferric iron in the media could correct the effects of Ni2+ toxicity. This can suggest that there were competitive interactions between Fe3+ and Ni2+, which resulting in the reconstruction of the metalloproteins and restore the growth of At. thiooxidans A01 under 600 mM Ni2+. A similar experiment implied that ferrous iron gave significant protection to At. caldus BC13 from lead and zinc toxicity [30].
At. thiooxidans A01 grows and survives by autotrophically utilizing energy derived from the oxidation of elemental sulfur and reduced inorganic sulfur compounds (RISCs). In the presence of Ni2+ and Fe3+, the expression of most sulfur metabolism-related genes was higher compared to the control without Ni2+. Sulfur metabolism-related genes including sox and rhd were upregulated after a shock with high nickel concentration. Sox complex encodes the sulfur oxidation protein, and it can directly catalyze thiosulfate to sulfate [21], and rhd encodes a mitochondrial enzyme that detoxifies cyanide (CN−) by converting it to thiocyanate (SCN−) and yielding to the formation of sulfite [19]. Similar to our results, Zn2+ and Cd2+ increased the expression of genes involved in the sulfur assimilation pathway in At. ferrooxidans [31]. In addition, Cd2+, Zn2+, and Cu2+ exposure increased the expression of a putative high-affinity sulfate transporter gene and root sulfate uptake capacity in maize roots [32]. In the presence of Ni2+ and Fe3+, the expression of most sulfur metabolism-related genes was higher and then downregulated compared to the individual nickel shock. This indicated that At. thiooxidans A01 could respond rapidly to the disturbance. Some bioleaching organisms utilize only Fe3+ as an electron acceptor although most of the sulfite acceptor oxidoreductases were shown to use c-type cytochromes as electron acceptors [33]. Additional Fe3+ is probably involved in sulfur assimilation and uptake in At. thiooxidans A01.
It is worth noting that cysA, cysB and serineO were upregulated in the presence of Ni2+ and Fe3+. Sulfate/thiosulfate import ATP-binding protein (cysA) and transport sulfate as a sulfur source for cysteine biosynthesis [34]. Moreover, serine O-acetyltransferase (serine O) catalyze the reaction: L-serine + acetyl-coenzyme A ⇆ O-acetyl-L-serine + coenzyme A [35], subsequently, the O-acetyl-L-serine can be converted into organic sulfur molecules cysteine [36]. The cysteine is the precursor of well-known chelators of heavy metals such as metallothioneins, glutathione and phytochelatins [37]. Some studies indicate that appropriate amounts of L-cysteine can significantly improve the bioleaching of Ni–Cu sulfide and pyrite [38,39]. Chelation of heavy metals is a ubiquitous detoxification strategy in a wide variety of organisms. Our results indicated that Fe3+ promotes the expression of sulfur assimilation-related genes that have an impact on the survival of At. thiooxidans in high concentrations of nickel by producing precursor of chelators of heavy metals.
Microorganisms have evolved ways to protect themselves from metal overload. A common microbial response to elevated concentrations of toxic metal is to synthesize efflux system, thus reducing the internal concentration of metal [7]. In the presence of Ni2+ and Fe3+, genes encoding a series of RND proteins including CzcAEP and the copper resistance gene afe_1073 showed high expressions. The RND protein family is a huge superfamily involved in exporting superfluous heavy metal including Ni2+ and referred to as CBA efflux systems. CzcAEP is part of the CzcCBA complex (RND protein) that mediates heavy metal resistance [40,41,42]. Iron is an essential metal for cellular functions under high nickel stress. It was speculated that Fe3+ promoted the expression of RND-related proteins driven by the cation-proton antiporter to facilitate nickel transport from periplasm to the outside of membrane. Similarly, extracellular zinc acted in a dose dependent manner to competitively inhibit manganese uptake by Streptococcus pneumonia [43]. Zinc stress induces copper depletion in Acinetobacter baumannii [44].
The ABC transport system is another basic defense against heavy metals [45]. Genes encoding ABC transporter-related proteins (CDB, CDC1, CDC2) were upregulated with additional Fe3+. They were shown to increase resistance to nickel in L. ferriphilum YSK and Escherichia coli strains [41,46]. In addition, extra metal ions including Ni2+, Cu2+ and Mn2+ induced siderophores’ production under an iron-limited condition [47]. Competition experiments between iron and other metals for pyoverdine showed a clear preference for iron [48]. Once iron is chelated by siderophores, it was delivered via various types of transporters including the ABC transport system to sequester this metal [2]. Thus, iron can possibly keep Ni2+ out of cytoplasm by completion. These data indicated that At. thiooxidans A01 might acquire specialized mechanisms to sense, transport and maintain Fe3+ within physiological concentrations and to detoxify non-essential metal Ni2+.
5. Conclusions
Collectively, this work reveals the resistance strategies utilized by At. thiooxidans A01 to survive Ni2+ stress with additional Fe3+, allowing it to thrive in diverse environments. Our results indicated that Fe3+ restored the growth of At. thiooxidans A01 in the presence of Ni2+ by alleviating Ni2+ toxicity, exporting intracellular nickel ions, and producing chelators of heavy metals. However, further metallomics, biochemistry and biophysics analysis such as cellular elemental distribution, enzymatic activity, spectroscopy, thermodynamics, and redox chemistry will be performed to decipher the precise mechanism.
Author Contributions
Conceptualization, Y.L., A.C., Y.X. and X.H.; Methodology, A.C. and Y.X.; Resources, G.Q., X.L., Y.L., H.Y. and H.L.; Data Curation, A.C., Y.L., K.Z. and Y.X.; Writing—Original Draft Preparation, A.C.; Writing—Review and Editing, A.C., Y.X., X.H. and Y.L.; Project Administration, Y.L.
Funding
This research was funded by [National High Technology Research and Development Program of China] grant number [2012AA061502], [National Key Research and Development Program of China] grant number [2016Y FB0101310] and [the National Natural Science Foundation of China] grant number [31570113].
Acknowledgments
We are grateful for constructive comments by three reviewers.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schippers, A.; Hedrich, S.; Vasters, J.; Drobe, M.; Sand, W.; Willscher, S. Biomining: Metal Recovery from Ores with Microorganisms. Geobiotechnol. I Met.-Relat. Issues 2014, 141, 1–47. [Google Scholar]
- Gikas, P. Kinetic responses of activated sludge to individual and joint nickel (Ni(II)) and cobalt (Co(II)): An isobolographic approach. J. Hazard. Mater. 2007, 143, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef]
- Macomber, L.; Hausinger, R.P. Mechanisms of nickel toxicity in microorganisms. Metallomics 2011, 3, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Rouch, D.A.; Lee, B.T.; Morby, A.P. Understanding cellular responses to toxic agents: A model for mechanism-choice in bacterial metal resistance. J. Ind. Microbiol. 1995, 14, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 1999, 51, 730–750. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Wang, W.; Xiao, S.; Chao, J.; Chen, Q.; Qiu, G.; Liu, X. Regulation of CO2 Fixation Gene Expression in Acidithiobacillus ferrooxidans ATCC 23270 by Lix984n Shock. J. Microbiol. Biotechnol. 2008, 18, 1747–1754. [Google Scholar]
- Kim, E.H.; Nies, D.H.; McEvoy, M.M.; Rensing, C. Switch or Funnel: How RND-Type Transport Systems Control Periplasmic Metal Homeostasis. J. Bacteriol. 2011, 193, 2381–2387. [Google Scholar] [CrossRef]
- Rensing, C.; Pribyl, T.; Nies, D.H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 1997, 179, 6871–6879. [Google Scholar] [CrossRef]
- Nies, D.H. RND Efflux Pumps for Metal Cations. In Microbial Efflux Pumps: Current Research; Yu, E.W., Zhang, Q., Brown, M.H., Eds.; Caister Academic Press: Wymondham, UK, 2013; pp. 79–121. [Google Scholar]
- Mirete, S.; de Figueras, C.G.; Gonzalez-Pastor, J.E. Novel nickel resistance genes from the rhizosphere metagenome of plants adapted to acid mine drainage. Appl. Environ. Microbiol. 2007, 73, 6001–6011. [Google Scholar] [CrossRef] [PubMed]
- San Martin-Uriz, P.; Mirete, S.; Alcolea, P.J.; Gomez, M.J.; Amils, R.; Gonzalez-Pastor, J.E. Nickel-Resistance Determinants in Acidiphilium sp PM Identified by Genome-Wide Functional Screening. PLoS ONE 2014, 9, e95041. [Google Scholar] [CrossRef] [PubMed]
- Mykytczuk, N.C.S.; Trevors, J.T.; Ferroni, G.D.; Leduc, L.G. Cytoplasmic membrane response to copper and nickel in Acidithiobacillus ferrooxidans. Microbiol. Res. 2011, 166, 186–206. [Google Scholar] [CrossRef] [PubMed]
- Zhan, G.; Li, D.; Zhang, L. Aerobic bioreduction of nickel(II) to elemental nickel with concomitant biomineralization. Appl. Microbiol. Biotechnol. 2012, 96, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, K.B.; Grail, B.M.; du Plessis, C.A.; Johnson, D.B. Reductive dissolution of ferric iron minerals: A new approach for bio-processing nickel laterites. Miner. Eng. 2011, 24, 620–624. [Google Scholar] [CrossRef]
- Aston, J.E. Effects of Organic Acids and Heavy Metals on the Biomining Bacterium: Acidithiobacillus Caldus Strain BC13. Ph.D. Thesis, Montana State University (MSU), Bozeman, MT, USA, 2010. [Google Scholar]
- Xu, Y.; Yin, H.; Jiang, H.; Liang, Y.; Guo, X.; Ma, L.; Xiao, Y.; Liu, X. Comparative study of nickel resistance of pure culture and co-culture of Acidithiobacillus thiooxidans and Leptospirillum ferriphilum. Arch. Microbiol. 2013, 195, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, K.L.; Scharf, D.H.; Litomska, A.; Hertweck, C. Enzymatic Carbon-Sulfur Bond Formation in Natural Product Biosynthesis. Chem. Rev. 2017, 117, 5521–5577. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Liang, Y.; Yin, H.; Xiao, Y.; Guo, X.; Xu, Y.; Hu, Q.; Liu, H.; Liu, X. Effects of Arsenite Resistance on the Growth and Functional Gene Expression of Leptospirillum ferriphilum and Acidithiobacillus thiooxidans in Pure Culture and Coculture. BioMed Res. Int. 2015. [Google Scholar] [CrossRef]
- Yin, H.; Zhang, X.; Li, X.; He, Z.; Liang, Y.; Guo, X.; Hu, Q.; Xiao, Y.; Cong, J.; Ma, L.; et al. Whole-genome sequencing reveals novel insights into sulfur oxidation in the extremophile Acidithiobacillus thiooxidans. BMC Microbiol. 2014, 14, 179. [Google Scholar] [CrossRef]
- Gikas, P. Single and combined effects of nickel (Ni(II)) and cobalt (Co(II)) ions on activated sludge and on other aerobic microorganisms: A review. J. Hazard. Mater. 2008, 159, 187–203. [Google Scholar] [CrossRef]
- Sugio, T.; Taha, T.M.; Kanao, T.; Takeuchi, F. Increase in Fe2+-producing activity during growth of Acidithiobacillus ferrooxidans ATCC23270 on sulfur. Biosci. Biotechnol. Biochem. 2007, 71, 2663–2669. [Google Scholar] [CrossRef] [PubMed]
- Macomber, L.; Imlay, J.A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Melissa, H.; Braud, A. New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, Y.; Outten, F.W. Fur and the Novel Regulator YqjI Control Transcription of the Ferric Reductase Gene yqjH in Escherichia coli. J. Bacteriol. 2011, 193, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Begg, S.L.; Eijkelkamp, B.A.; Luo, Z.; Counage, R.M.; Morey, J.R.; Maher, M.J.; Ong, C.L.; Mcewan, A.G.; Kobe, B.; Omara, M.L.; et al. Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 2015, 6, 6418. [Google Scholar] [CrossRef] [PubMed]
- Fisher, B.; Yarmolinsky, D.; Abdel-Ghany, S.; Pilon, M.; Pilon-Smits, E.A.; Sagi, M.; Van Hoewyk, D. Superoxide generated from the glutathione-mediated reduction of selenite damages the iron-sulfur cluster of chloroplastic ferredoxin. Plant Physiol. Biochem. 2016, 106, 228–235. [Google Scholar] [CrossRef]
- Xu, F.F.; Imlay, J.A. Silver(I), Mercury(II), Cadmium(II), and Zinc(II) Target Exposed Enzymic Iron-Sulfur Clusters when They Toxify Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 3614–3621. [Google Scholar] [CrossRef]
- Aston, J.E.; Peyton, B.M.; Lee, B.D.; Apel, W.A. effects of ferrous sulfate, inoculum history, and anionic from on lead, zinc, and copper toxicity to acidithiobacillus caldus strain BC13. Environ. Toxicol. Chem. 2010, 29, 2669–2675. [Google Scholar] [CrossRef]
- Zheng, C.; Chen, M.; Tao, Z.; Zhang, L.; Zhang, X.F.; Wang, J.-Y.; Liu, J. Differential expression of sulfur assimilation pathway genes in Acidithiobacillus ferrooxidans under Cd2+ stress: Evidence from transcriptional, enzymatic, and metabolic profiles. Extremophiles 2015, 19, 429–436. [Google Scholar] [CrossRef]
- Nocito, F.F.; Lancilli, C.; Crema, B.; Fourcroy, P.; Davidian, J.-C.; Sacchi, G.A. Heavy metal stress and sulfate uptake in maize roots. Plant Physiol. 2006, 141, 1138–1148. [Google Scholar] [CrossRef]
- Sugio, T.; Fujii, M.; Takeuchi, F.; Negishi, A.; Maeda, T.; Kamimura, K. Volatilization of mercury by an iron oxidation enzyme system in a highly mercury-resistant Acidithiobacillus ferrooxidans strain MON-1. Biosci. Biotechnol. Biochem. 2003, 67, 1537–1544. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.T.; Moutran, A.; Fessel, M.; Balan, A. The sulfur/sulfonates transport systems in Xanthomonas citri pv. citri. Bmc Genom. 2015, 16, 525. [Google Scholar] [CrossRef] [PubMed]
- Denk, D.; Bock, A. L-cysteine biosynthesis in Escherichia coli: Nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J. Gen. Microbiol. 1987, 133, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Suzuki, K.; Ohtsu, I. Current understanding of sulfur assimilation metabolism to biosynthesize L-cysteine and recent progress of its fermentative overproduction in microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 8203–8211. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S. Evolution and function of phytochelatin synthases. J. Plant Physiol. 2006, 163, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Zhong, H.; Hu, Y.H.; Zhao, J.C.; He, Z.G.; Gu, G.H. Bioleaching of a low-grade nickel–copper sulfide by mixture of four thermophiles. Bioresour. Technol. 2014, 153, 300–306. [Google Scholar] [CrossRef]
- Rojas, C.J.A.; Tributsch, H. Bioleaching of pyrite accelerated by cysteine. Process Biochem. 2000, 35, 815–824. [Google Scholar] [CrossRef]
- Goldberg, M.; Pribyl, T.; Juhnke, S.; Nies, D.H. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 1999, 274, 26065–26070. [Google Scholar] [CrossRef]
- Diels, L.; Dong, Q.; Vanderlelie, D.; Baeyens, W.; Mergeay, M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J. Ind. Microbiol. 1995, 14, 142–153. [Google Scholar] [CrossRef]
- Intorne, A.C.; Deoliveira, W.V.V.; Pereira, L.D.; Desouza, G.A. Essential role of the czc determinant for cadmium, cobalt and zinc resistance in Gluconacetobacter diazotrophicus PAI 5. Int. Microbiol. 2012, 15, 69–78. [Google Scholar]
- Eijkelkamp, B.A.; Morey, J.R.; Ween, M.P. Extracellular Zinc Competitively Inhibits Manganese Uptake and Compromises Oxidative Stress Management in Streptococcus pneumoniae. PLoS ONE 2014, 9, 89427. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Pederick, V.G.; Elbourne, L.D.H.; Ian, T.P.; James, C.P.; Christopher, A.M.; Bart, A.E. Zinc stress induces copper depletion in Acinetobacter baumannii. BMC Microbiol. 2017, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.; Hunke, S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 1998, 22, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mikolay, A.; Nies, D.H. The ABC-transporter AtmA is involved in nickel and cobalt resistance of Cupriavidus metallidurans strain CH34. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2009, 96, 183–191. [Google Scholar] [CrossRef]
- Braud, A.; Geoffroy, V.; Hoegy, F.; Mislin, G.L.; Schalk, I.J. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ. Microbiol. Rep. 2010, 2, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).