Improved Recovery and Selectivity of Lanthanide-Ion-Binding Cyclic Peptide Hosts by Changing the Position of Acidic Amino Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Section
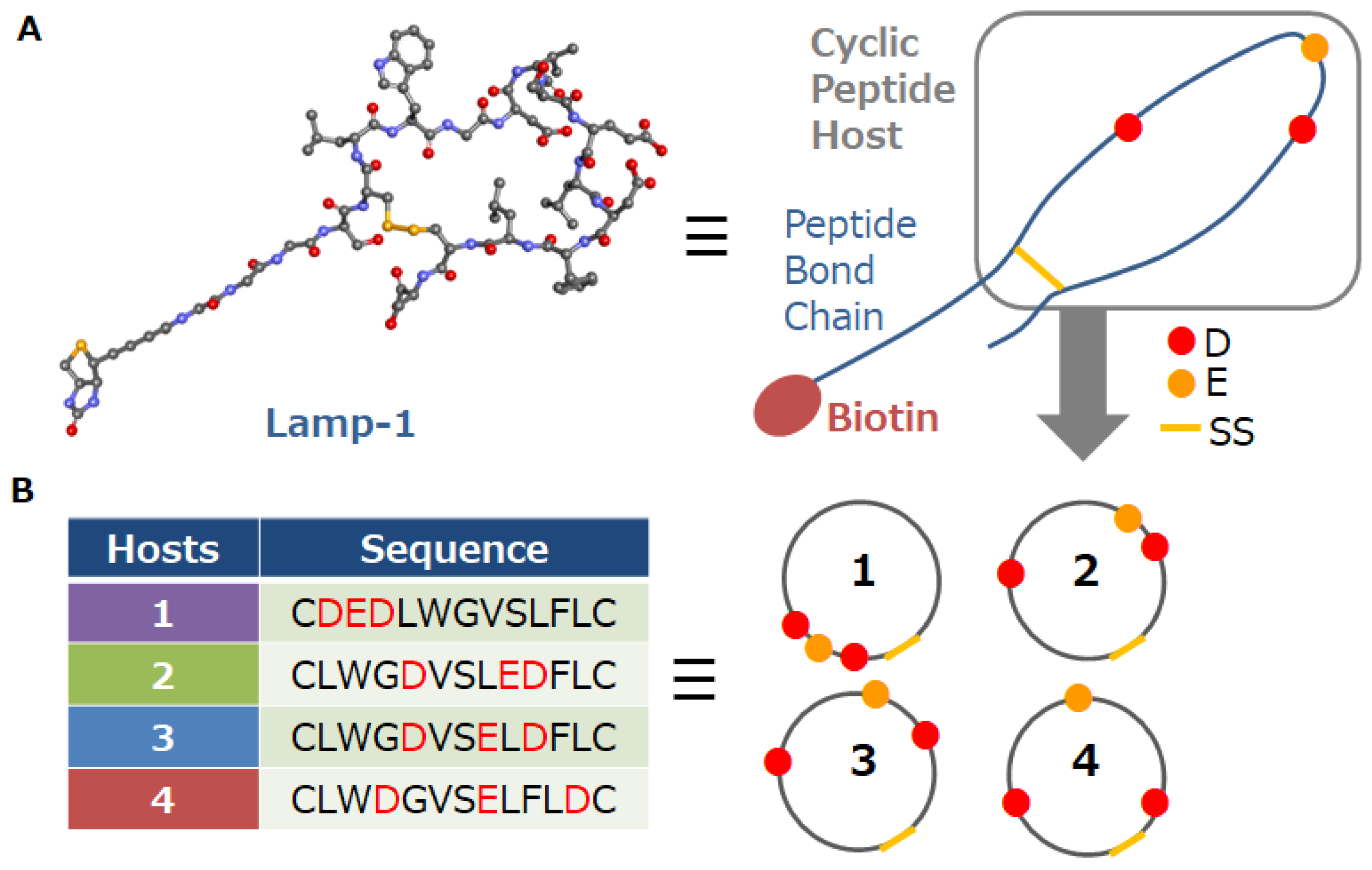
2.1.1. Materials and Instrumentation
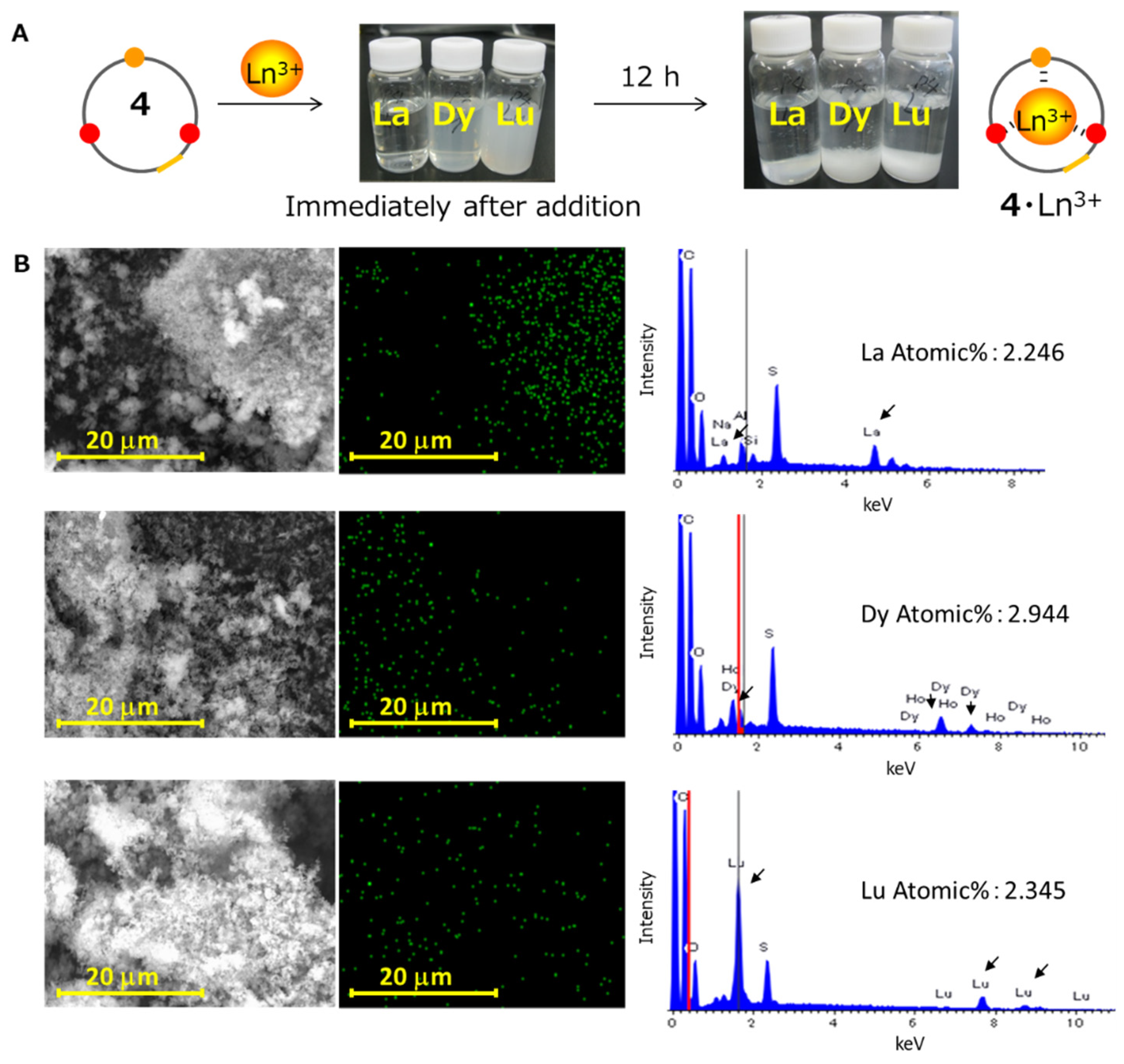
2.1.2. Precipitation of the Peptide Host–Ln Ion Complexes
2.2. Computational Study
2.2.1. Environment, Software, and Methods
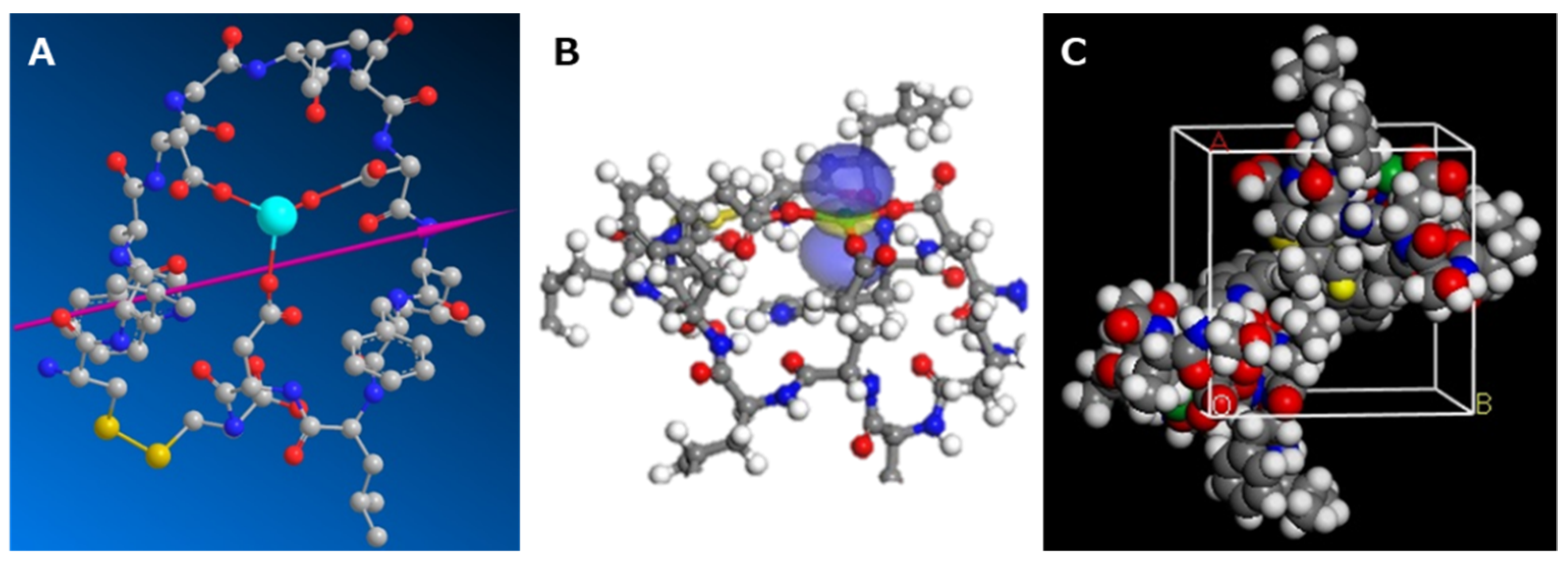
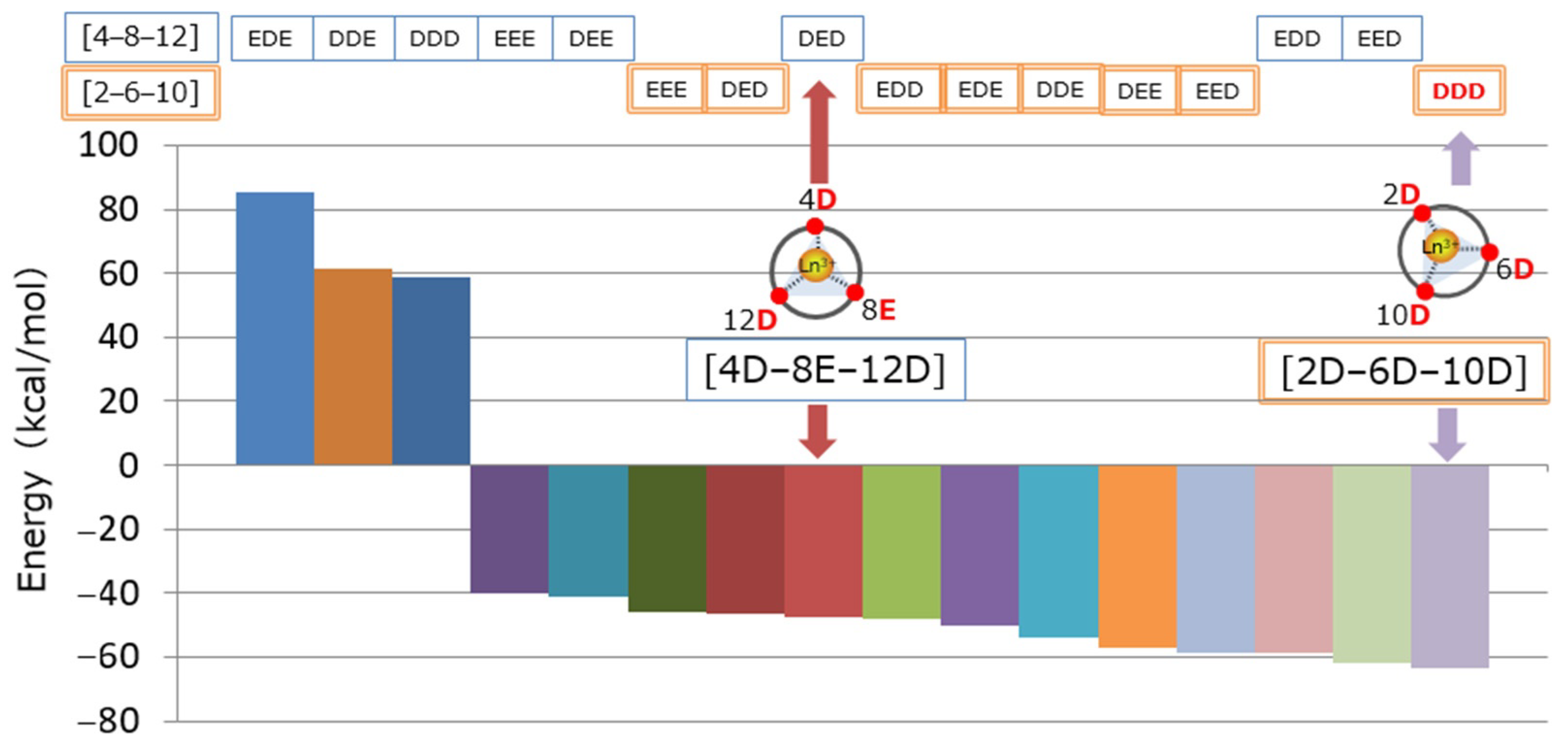
2.2.2. Initial Structure Determination of the Peptide–Ln Complex
2.2.3. Calculating the DM of the Peptide–Ln Complex
2.2.4. Calculating the LUMOs of the Peptide–Ln Complex
2.2.5. Calculating the CE of the Peptide–Ln Complex
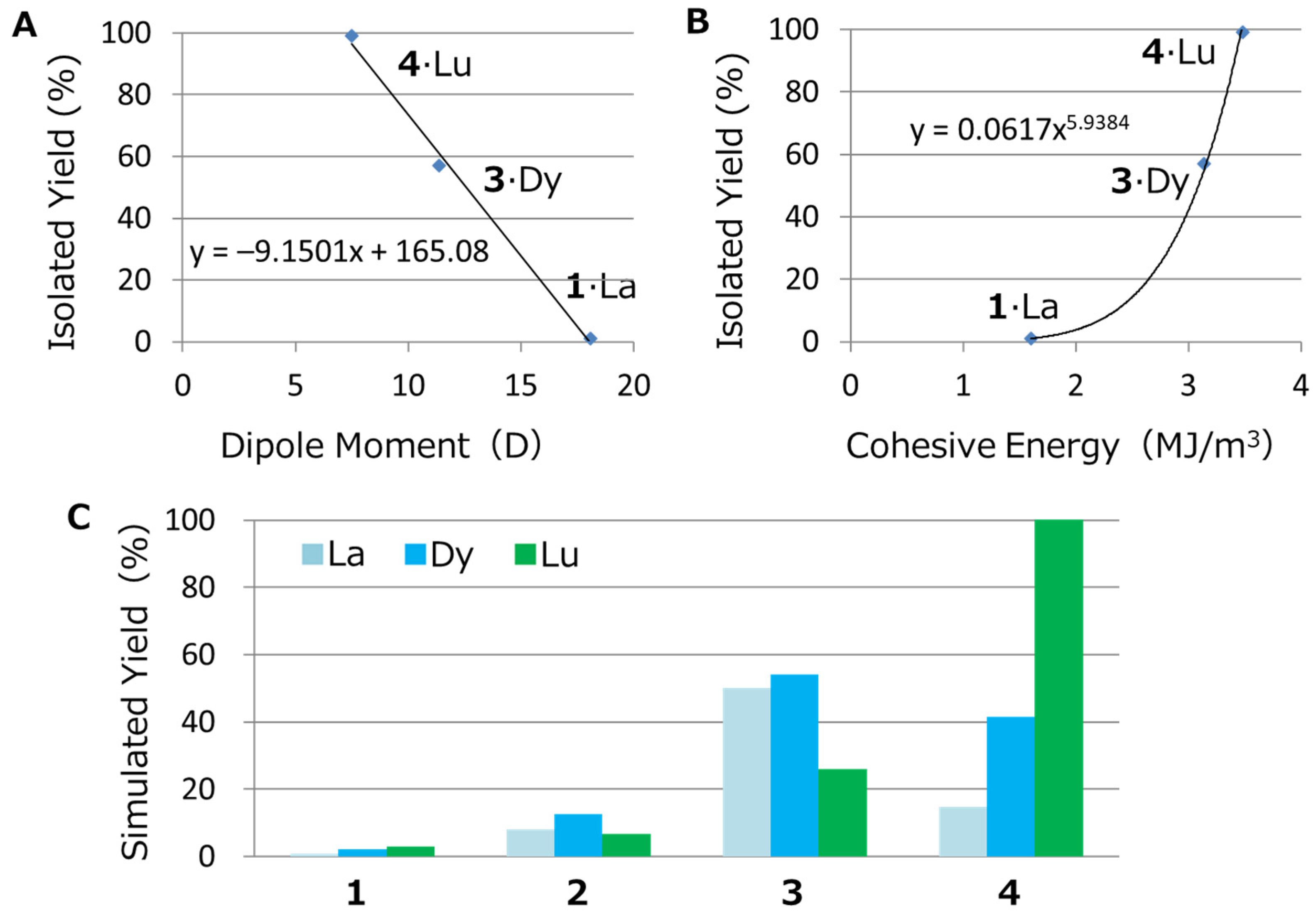
2.2.6. Calculating the Simulated Yield
- YA = (YD + YC)/2,
- YD = −9.1501 (DM value) + 165.08
- YC = 0.0617 (CE value)5.9384
- LUMO = 1 or 0.5
3. Results and Discussion
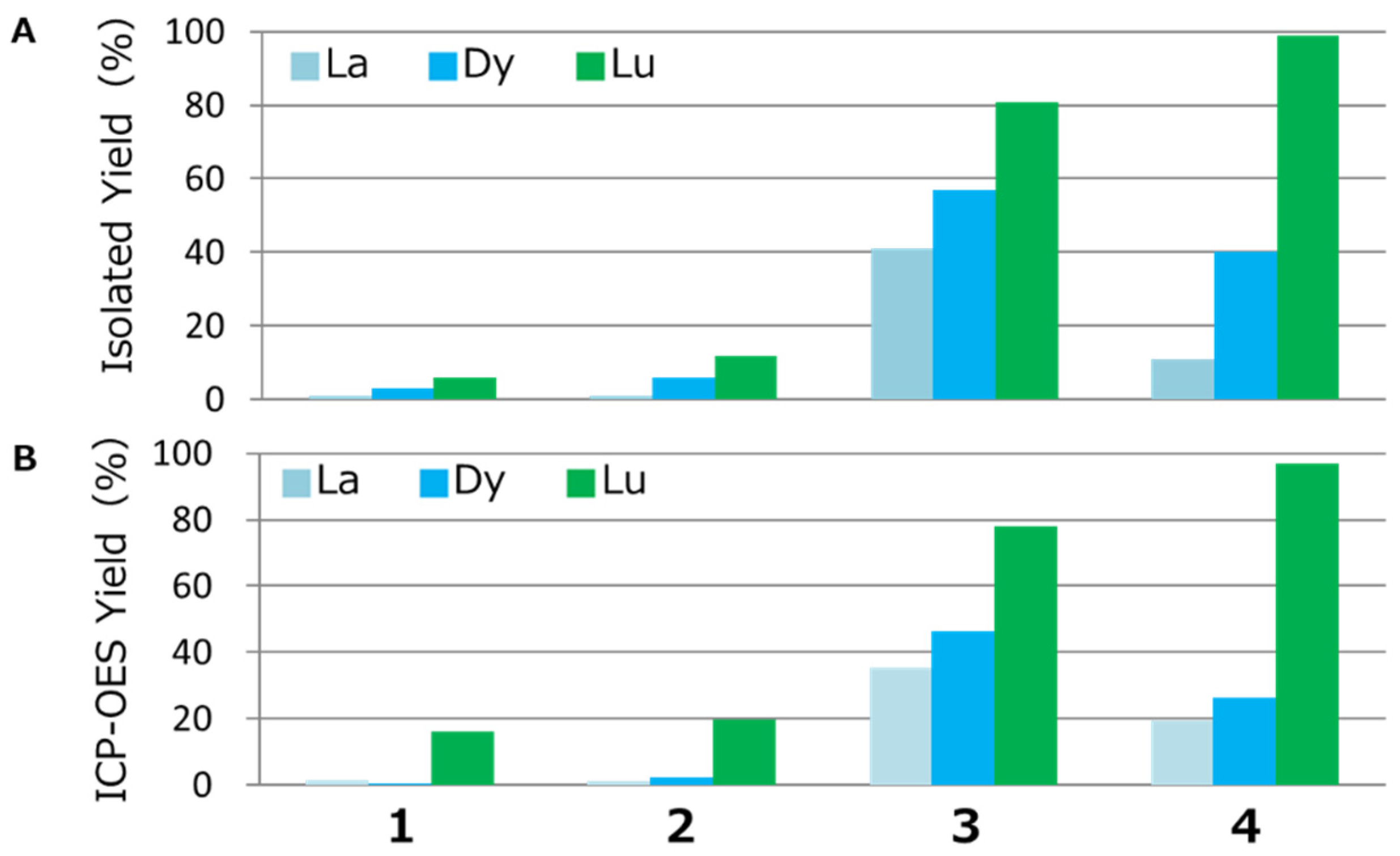
3.1. Lanthanide Ion Recovery
3.2. Prediction of Recovery Performance
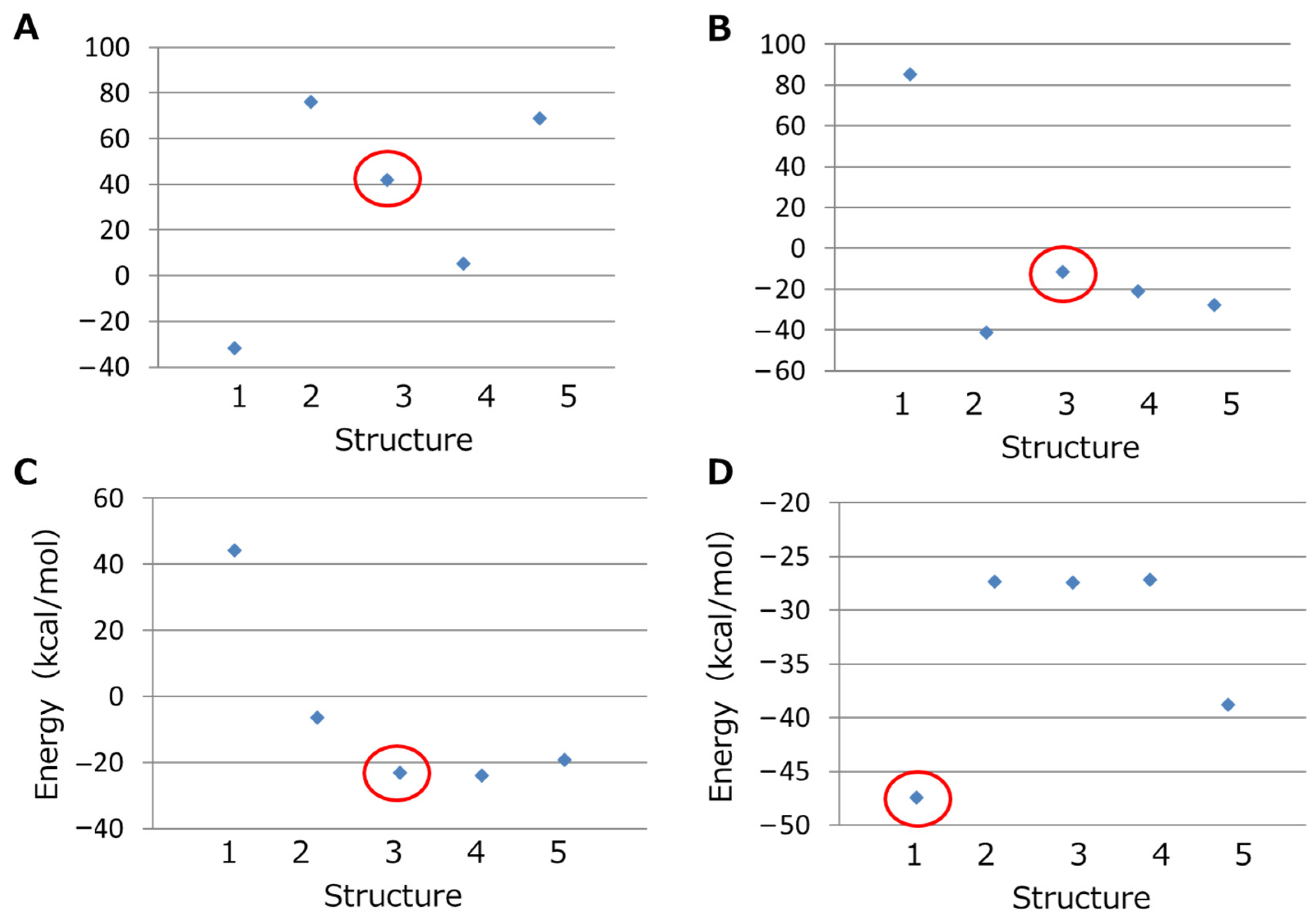
3.2.1. Initial Structure Determination of the Peptide–Ln Complex
3.2.2. Calculating the DM and CE of the Peptide–Ln Complex
3.2.3. Calculation of the Simulated Yield
- YA = (YD + YC)/2,
- YD = –9.1501 (DM value) + 165.08
- YC = 0.0617 (CE value)5.9384
- LUMO = 1 or 0.5
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Binnemans, K.; Jones, P.T.; Blanpain, B.; Van Gerven, T.; Yang, Y.; Walton, A.; Buchert, M. Recycling of rare earths: A critical review. J. Clean. Prod. 2013, 51, 1–22. [Google Scholar] [CrossRef]
- Hatanaka, T.; Matsugami, A.; Nonaka, T.; Takagi, H.; Hayashi, F.; Tani, T.; Ishida, N. Rationally designed mineralization for selective recovery of the rare earth elements. Nat. Commun. 2017, 8, 15670. [Google Scholar] [CrossRef] [Green Version]
- Ishida, N.; Hatanaka, T.; Hosokawa, Y.; Kojima, K.; Iizuka, T.; Teramoto, H.; Sezutsu, H.; Kameda, T. Direct recovery of the rare earth elements using a silk displaying a metal-recognizing peptide. Molecules 2020, 25, 761. [Google Scholar] [CrossRef] [Green Version]
- Bünzli, J.-C.G.; Piguet, C. Lanthanide-containing molecular and supramolecular polymetallic functional assemblies. Chem. Rev. 2002, 102, 1897–1928. [Google Scholar] [CrossRef] [Green Version]
- Kanesato, M.; Yokoyama, T.; Suzuki, M.T. Reaction of tris(2-aminoethyl)amine coordinated to lanthanum(III) and gadolinium(III) with salicylaldehyde. Chem. Lett. 1997, 26, 93–94. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.Y.; Bai, X.P.; Wada, T.; Inoue, Y. Molecular design of crown ethers. 21.(1,2) synthesis of novel double-armed benzo-15-crown-5 lariats and their complexation thermodynamics with light lanthanoid nitrates in acetonitrile. J. Org. Chem. 2000, 65, 7105–7109. [Google Scholar] [CrossRef]
- Bogart, J.A.; Lippincott, C.A.; Carroll, P.J.; Schelter, E.J. An operationally simple method for separating the rare-earth elements neodymium and dysprosium. Angew. Chem. Int. Ed. 2015, 54, 8222–8225. [Google Scholar] [CrossRef]
- Laine, S.; Morfin, J.-F.; Galibert, M.; Aucagne, V.; Bonnet, C.S.; Tóth, E. Lanthanide DO3A-complexes bearing peptide substrates: The effect of peptidic side chains on metal coordination and relaxivity. Molecules 2021, 26, 2176. [Google Scholar] [CrossRef]
- Schmidtchen, F.P.; Berger, M. Artificial organic host molecules for anions. Chem. Rev. 1997, 97, 1609–1646. [Google Scholar] [CrossRef]
- Bitta, J.; Kubik, S. Cyclic hexapeptides with free carboxylate groups as new receptors for monosaccharides. Org. Lett. 2001, 3, 2637–2640. [Google Scholar] [CrossRef]
- Hamley, I.W. Small bioactive peptides for biomaterials design and therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitz, M.; Sherawat, M.; Franz, K.J.; Peisach, E.; Allen, K.N.; Imperiali, B. Structural origin of the high affinity of a chemically evolved lanthanide-binding peptide. Angew. Chem. Int. Ed. 2004, 43, 3682–3685. [Google Scholar] [CrossRef] [PubMed]
- Cisnetti, F.; Gateau, C.; Lebrun, C.; Delangle, P. Lanthanide(III) complexes with two hexapeptides incorporating unnatural chelating amino acids: Secondary structure and stability. Chem. Eur. J. 2009, 15, 7456–7469. [Google Scholar] [CrossRef]
- Kotynia, A.; Bielińska, S.; Kamysz, W.; Brasuń, J. The coordination abilities of the multiHis-cyclopeptide with two metal-binding centers—Potentiometric and spectroscopic investigation. Dalton Trans. 2012, 41, 12114–12120. [Google Scholar] [CrossRef] [PubMed]
- Bar, A.K.; Kalita, P.; Singh, M.K.; Rajaraman, G.; Chandrasekhar, V. Low-coordinate mononuclear lanthanide complexes as molecular nanomagnets. Coord. Chem. Rev. 2018, 367, 163–216. [Google Scholar] [CrossRef]
- Gahan, L.R.; Cusack, R.M. Metal complexes of synthetic cyclic peptides. Polyhedron 2018, 153, 1–23. [Google Scholar] [CrossRef]
- Sherrington, D.C.; Taskinen, K.A. Self-assembly in synthetic macromolecular systems via multiple hydrogen bonding interactions. Chem. Soc. Rev. 2001, 30, 83–93. [Google Scholar] [CrossRef]
- Zhao, X.; Pan, F.; Xu, H.; Yaseen, M.; Shan, H.; Hauser, C.A.E.; Zhang, S.; Lu, J.R. Molecular self-assembly and applications of designer peptide amphiphiles. Chem. Soc. Rev. 2010, 39, 3480–3498. [Google Scholar] [CrossRef]
- Hamley, I.W. Peptide nanotubes. Angew. Chem. Int. Ed. 2014, 53, 6866–6881. [Google Scholar] [CrossRef]
- Chan, W.-L.; Xie, C.; Lo, W.-S.; Bünzli, J.-C.G.; Wong, W.-K.; Wong, K.-L. Lanthanide–tetrapyrrole complexes: Synthesis, redox chemistry, photophysical properties, and photonic applications. Chemcal Soc. Rev. 2021, 50, 12189–12257. [Google Scholar] [CrossRef]
- Celine, P.; Marilena, F.; Gadi, R.; Stefania, T. Lanthanide-Based Metal Organic Frameworks: Synthetic Strategies and Catalytic Applications. ACS Catal. 2016, 6, 6063–6072. [Google Scholar]
- Muhammad, S.; Amr, G.; Mika, L.; Maarit, K. Lanthanide-based inorganic-organic hybrid materials for photon-upconversion. J. Mater. Chem. C 2020, 8, 6946–6965. [Google Scholar]
- Ji, F.; Shao, S.; Li, Z.; Wang, S.; Chaudhuri, R.; Guo, Z.; Perkins, N.G.; Sarkar, P.; Xue, M. A cyclic peptide antenna ligand for enhancing terbium luminescence. Analyst 2021, 146, 3474–3481. [Google Scholar] [CrossRef] [PubMed]
- Singer, H.; Drobot, B.; Zeymer, C.; Steudtner, R.; Daumann, L.J. Americium preferred: Lanmodulin, a natural lanthanide-binding protein favors an actinide over lanthanides. Chem. Sci. 2021, 12, 15581–15587. [Google Scholar] [CrossRef]
- Hatanaka, T.; Kikkawa, N.; Matsugami, A.; Hosokawa, Y.; Hayashi, F.; Ishida, N. The origins of binding specificity of a lanthanide ion binding peptide. Sci. Rep. 2020, 10, 19468. [Google Scholar] [CrossRef]
- Yoneya, M.; Yamaguchi, T.; Sato, S.; Fujita, M. Simulation of metal–ligand self-assembly into spherical complex M6L8. J. Am. Chem. Soc. 2012, 134, 14401–14407. [Google Scholar] [CrossRef]
- Bonnet, C.S.; Fries, P.H.; Crouzy, S.; Sénèque, O.; Cisnetti, F.; Boturyn, D.; Dumy, P.; Delangle, P. A gadolinium-binding cyclodecapeptide with a large high-field relaxivity involving second-sphere water. Chem. Eur. J. 2009, 15, 7083–7093. [Google Scholar] [CrossRef]
- Brichtová, E.; Hudecová, J.; Vršková, N.; Šebestík, J.; Bouř, P.; Wu, T. Binding of lanthanide complexes to histidine-containing peptides probed by raman optical activity spectroscopy. Chem. Eur. J. 2018, 24, 8664–8669. [Google Scholar] [CrossRef]
- Walsh, T.R. Pathways to structure–property relationships of peptide–materials interfaces: Challenges in predicting molecular structures. Acc. Chem. Res. 2017, 50, 1617–1624. [Google Scholar] [CrossRef] [Green Version]
- Carvajal-Diaz, J.A.; Cagin, T. Electrophoretic transport of Na+ and K+ ions within cyclic peptide nanotubes. J. Phys. Chem. B 2016, 120, 7872–7879. [Google Scholar] [CrossRef]
- Webster, A.M.; Peacock, A.F.A. De novo designed coiled coils as scaffolds for lanthanides, including novel imaging agents with a twist. Chem. Commun. 2021, 57, 6851–6862. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Descoteaux, M.L.; Lin, Y.-S. Structure prediction of cyclic peptides by molecular dynamics + machine learning. Chem. Sci. 2021, 12, 14927–14936. [Google Scholar] [CrossRef] [PubMed]
- Damjanovic, J.; Miao, J.; Huang, H.; Lin, Y.-S. Elucidating solution structures of cyclic peptides using molecular dynamics simulations. Chem. Rev. 2021, 121, 2292–2324. [Google Scholar] [CrossRef] [PubMed]
- Špadina, M.; Bohinc, K. Multiscale modeling of solvent extraction and the choice of reference state: Mesoscopic modeling as a bridge between nanoscale and chemical engineering. Curr. Opin. Colloid Interface Sci. 2020, 46, 94–113. [Google Scholar] [CrossRef]
- Hosseinzadeh, P.; Bhardwaj, G.; Mulligan, V.K.; Shortridge, M.D.; Craven, T.W.; Pardo-Avila, F.; Rettie, S.A.; Kim, D.E.; Silva, D.-A.; Ibrahim, Y.M.; et al. Comprehensive computational design of ordered peptide macrocycles. Science 2017, 358, 1461–1466. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, Y.; Suenaga, A.; Sato, Y.; Kosuda, S.; Taiji, M. All-atom molecular dynamics analysis of multi-peptide systems reproduces peptide solubility in line with experimental observations. Sci. Rep. 2016, 6, 19479. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, S.; Yanagida, S. Density functional theory-based molecular modeling: Verification of decisive roles of van der waals aggregation of triiodide ions for effective electron transfer in wet-type N3-dye-sensitized solar cells. Energies 2020, 13, 3027. [Google Scholar] [CrossRef]
- Cram, D.J. Preorganization—from solvents to spherands. Angew. Chem. Int. Ed. 1986, 25, 1039–1057. [Google Scholar] [CrossRef]
- Yanagida, S. Density functional theory-based molecular modeling: Interaction analysis of PEG with metal ion. Wako Inform. World 2011, 25, 8–9. [Google Scholar]
- Ohyoshi, E. Spectrophotometric determination of formation constants of 1:1 complexes of lanthanides with 4-(2-pyridylazo) resorcinol (par). Talanta 1984, 31, 1129–1132. [Google Scholar] [CrossRef]
- Tsakanika, L.V.; Ochsenkühn-Petropoulou, M.T.; Mendrinos, L.N. Investigation of the separation of scandium and rare earth elements from red mud by use of reversed-phase HPLC. Anal. Bioanal. Chem. 2004, 379, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Itoh, J. Novel analytical applications of porphyrin to HPLC post-column flow injection system for determination of the lanthanides. Talanta 2006, 69, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.P. MOPAC: MOPAC2016, version 16.093W; Stewart Computational Chemistry: Colorado Springs, CO, USA, 2007; Available online: http://OpenMOPAC.net (accessed on 1 May 2016).
- Dassault Systèmes. BIOVIA Materials Studio, version 7.0/8.0; Dassault Systèmes: San Diego, CA, USA, 2018. [Google Scholar]
- Dassault Systèmes. BIOVIA Modules Tutorials Materials Studio 2017; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Mohamed, M.E. Synthesis and spectroscopic characterization of l-aspartic acid complexes with metals of the lanthanides family. Int. Lett. Chem. Phys. Astron. 2013, 11, 91–115. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Siu, K.W.M.; Hopkinson, A.C. Generation of [La(peptide)]3+ complexes in the gas phase: Determination of the number of binding sites provided by dipeptide, tripeptide, and tetrapeptide ligands. J. Phys. Chem. A 2007, 111, 11562–11571. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosokawa, Y.; Oshima, A.; Hatanaka, T.; Ishida, N. Improved Recovery and Selectivity of Lanthanide-Ion-Binding Cyclic Peptide Hosts by Changing the Position of Acidic Amino Acids. Minerals 2022, 12, 148. https://doi.org/10.3390/min12020148
Hosokawa Y, Oshima A, Hatanaka T, Ishida N. Improved Recovery and Selectivity of Lanthanide-Ion-Binding Cyclic Peptide Hosts by Changing the Position of Acidic Amino Acids. Minerals. 2022; 12(2):148. https://doi.org/10.3390/min12020148
Chicago/Turabian StyleHosokawa, Yoichi, Ayako Oshima, Takaaki Hatanaka, and Nobuhiro Ishida. 2022. "Improved Recovery and Selectivity of Lanthanide-Ion-Binding Cyclic Peptide Hosts by Changing the Position of Acidic Amino Acids" Minerals 12, no. 2: 148. https://doi.org/10.3390/min12020148
APA StyleHosokawa, Y., Oshima, A., Hatanaka, T., & Ishida, N. (2022). Improved Recovery and Selectivity of Lanthanide-Ion-Binding Cyclic Peptide Hosts by Changing the Position of Acidic Amino Acids. Minerals, 12(2), 148. https://doi.org/10.3390/min12020148





