Temporal Network Link Prediction Based on the Optimized Exponential Smoothing Model and Node Interaction Entropy
Abstract
1. Introduction
- We record the fine-grained interaction information among nodes within the snapshot period and incorporate the concept of information entropy and weak ties to construct the node interaction entropy. This value differentiates the popularity of nodes within the snapshot structure from a more nuanced perspective.
- We combine node interaction entropy and eigenvector centrality to construct an enhanced node similarity that considers the network structure’s distance between nodes, weak ties characteristics, and centrality.
- We normalized the sum of node interaction entropy, and the normalized result will reflect the ratio of the current snapshot’s weak ties during the entire period. With the higher ratio, the node similarity matrices can provide more weights for time series prediction. We combine the smoothing coefficient and the ratio into the exponential smoothing model. This improves the shortcoming of the single reference score of the prediction process.
2. Related Works
2.1. Temporal Network
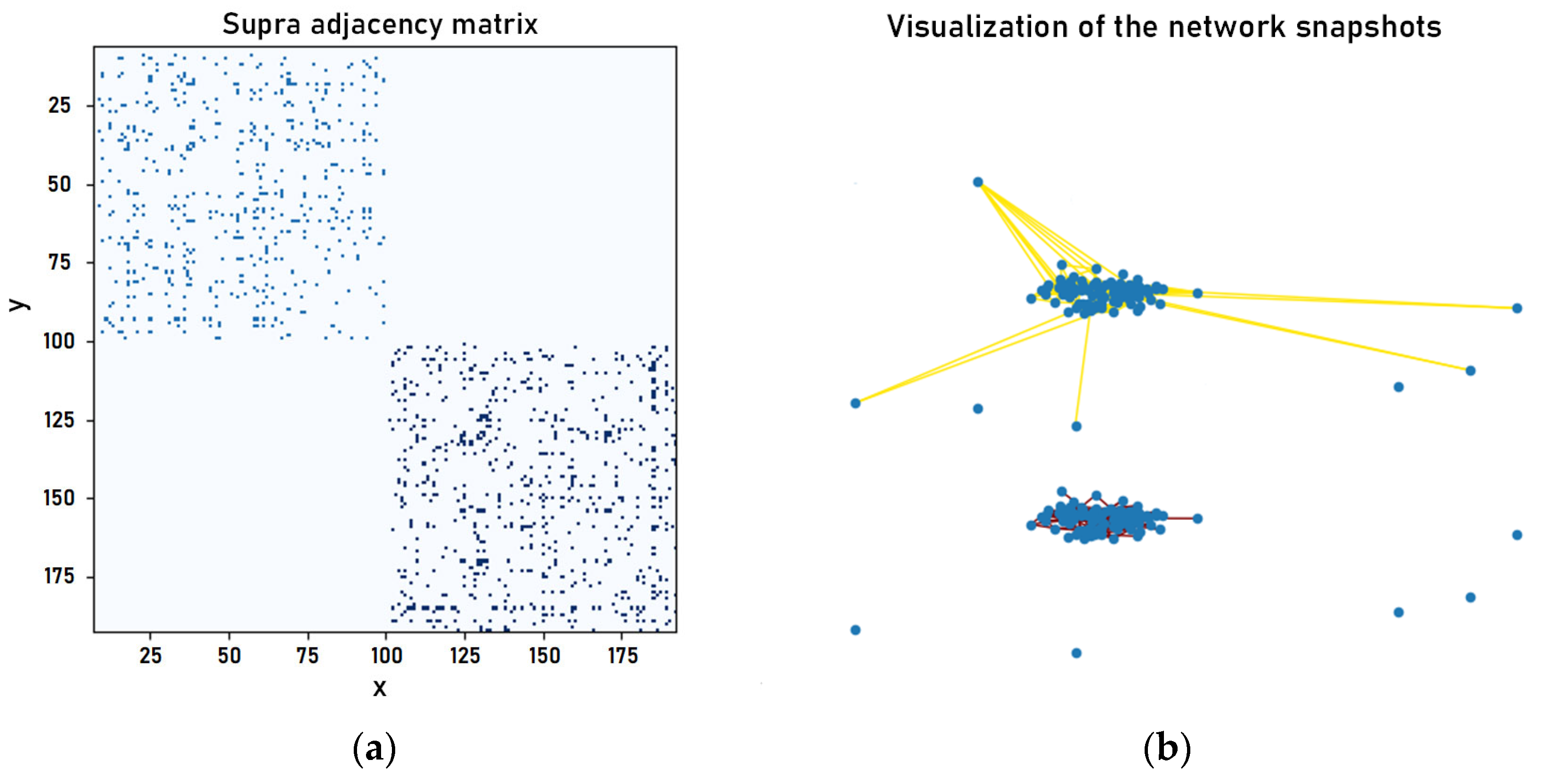
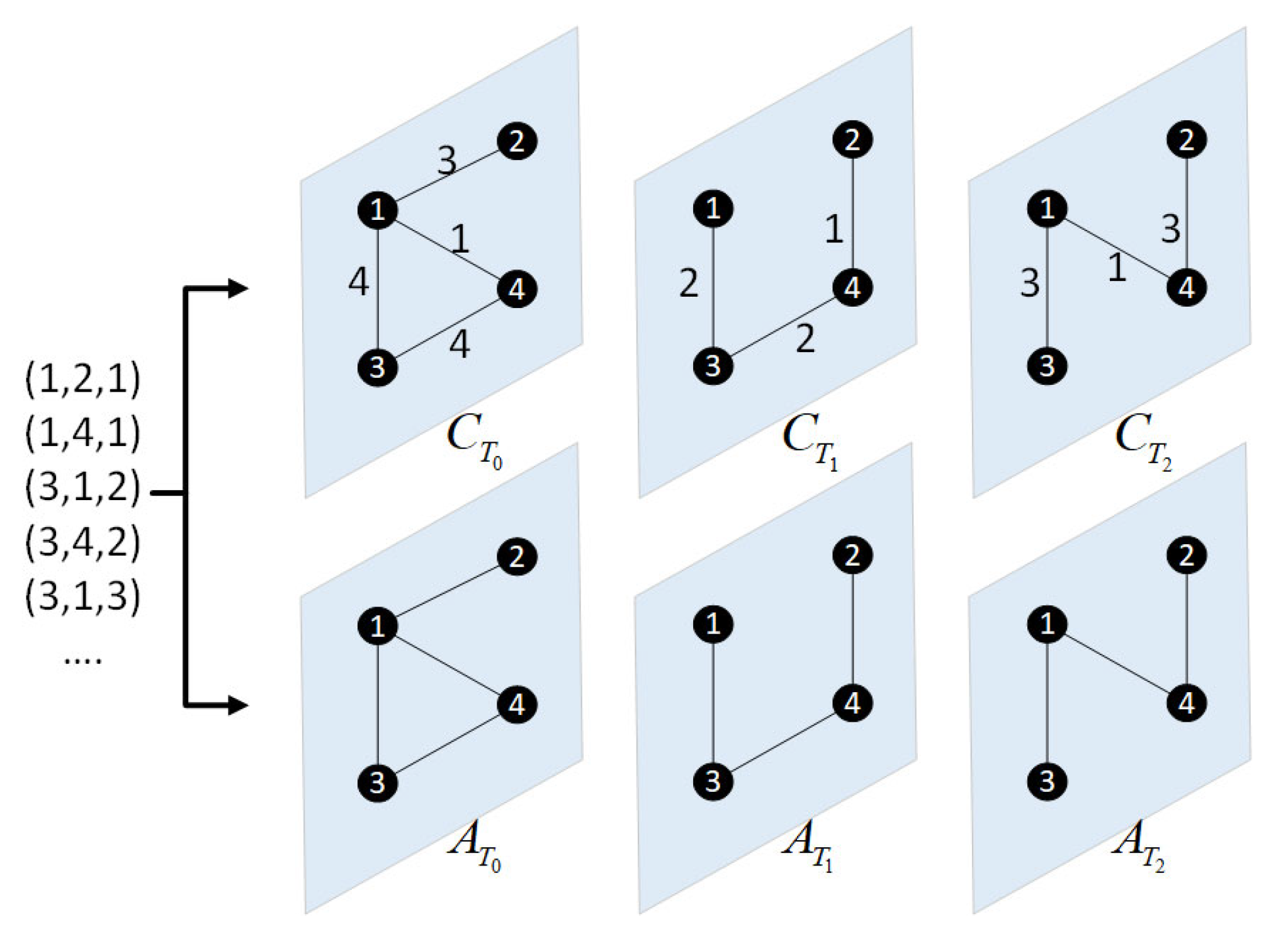
2.2. Construction of Network Snapshots and Multi-Layer Network Model
2.3. Weak Ties Theory
2.4. The Gravity Model
2.5. Information Entropy
2.6. The Eigenvector Centrality of Nodes
2.7. Temporal Network Link Prediction
2.8. The Exponential Smoothing Model in Link Prediction
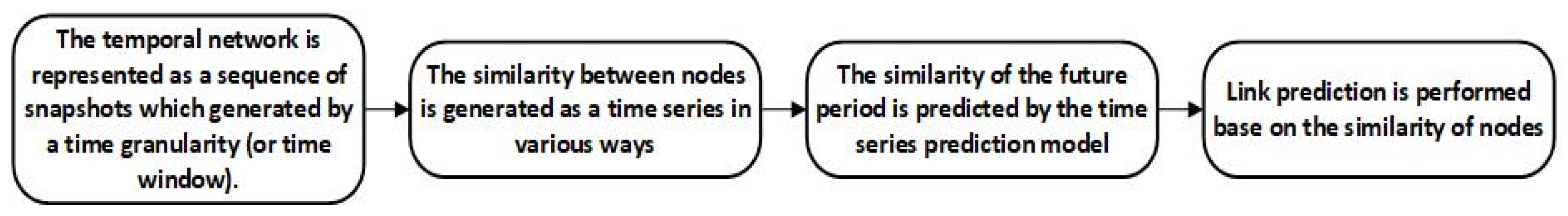
3. Description of the OESMNIE Temporal Network Link Prediction Method
3.1. Establishment of the Node Interaction Entropy
3.2. Establishment of the Improved Node Centrality in Each Snapshot
3.3. Establishment of Node Similarity Matrix by Gravity Model
3.4. Optimization of the Exponential Smoothing Model
3.5. Detailed Explanation of the OESMNIE Method
4. Experiments and Discussion
4.1. Experimental Environment
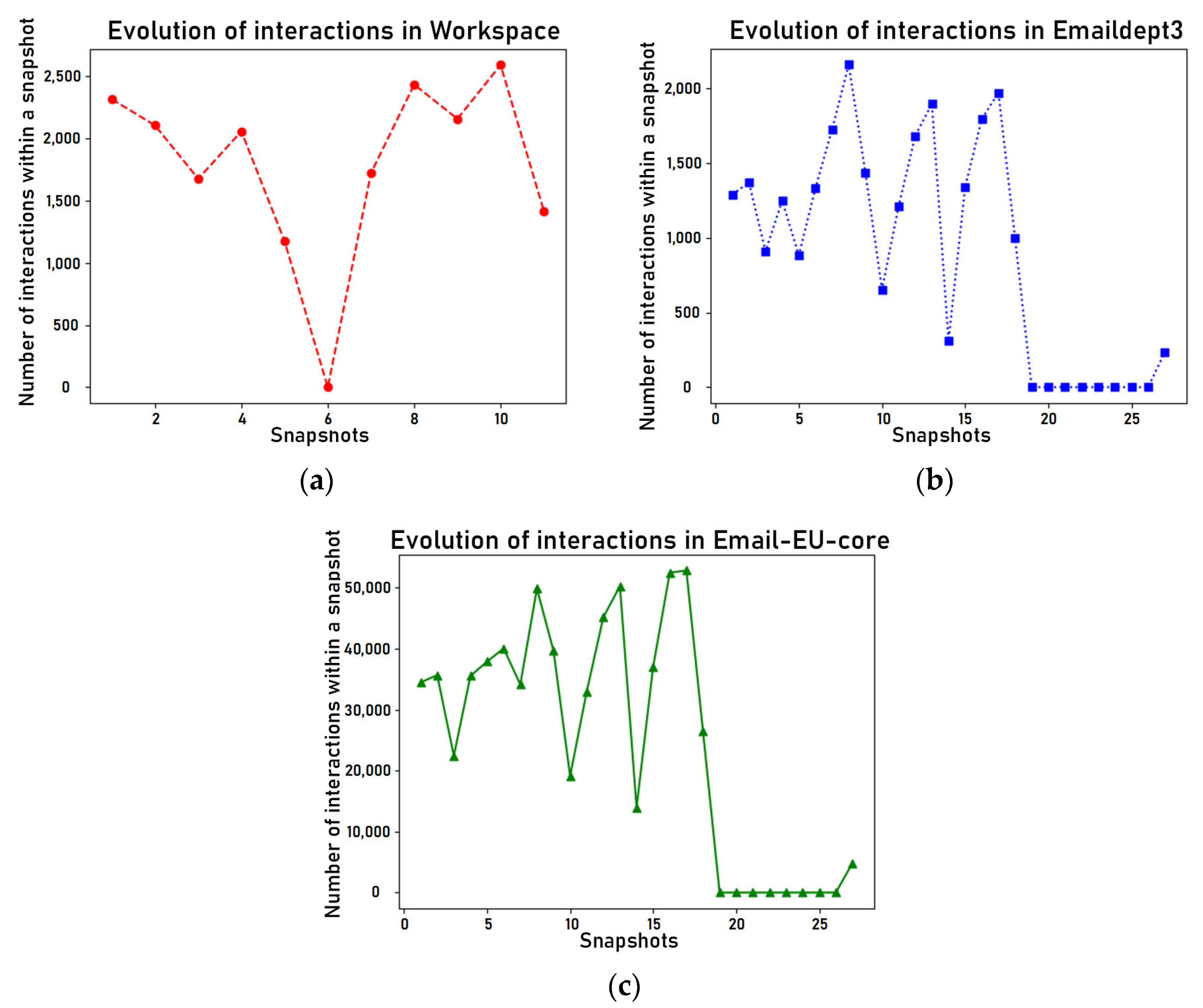
4.2. Data Selection
4.3. Evaluation Method
4.4. Performance Comparison
4.5. Parameters Analysis
4.6. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NMF | Nonnegative matrix factorization |
| NRL | Network representation learning |
| WSNs | Wireless Sensor Networks |
| GR | Gravity |
| ARIMA | Autoregressive Integrated Moving Average model |
| SAM | Supra-Adjacency Matrix |
| AUC | Area Under the receiver operating characteristic Curve |
| CN | Common Neighbors |
| RA | Resource Allocation |
| AA | Adamic–Adar |
| JC | Jaccard |
| PA | Preferential Attachment |
| TBNS | Tensor-Based Bode Similarity |
| OESMNIE | Optimized Exponential Smoothing Model and Node Interaction Entropy |
| DC | Degree centrality |
| CC | Closeness centrality |
| BC | Betweenness centrality |
| EC | Eigenvector centrality |
Variables
| The temporal granularity (or time window) for generating snapshots. | |
| The format of node interaction record in temporal network, (source node, target node, timestamp of the interaction occurrence). | |
| The th network snapshot in chronological order. | |
| Smoothing coefficient. | |
| The adjacency matrix (or network snapshot) in . | |
| The set of fully connected edges among nodes within . | |
| The nodes similarity matrix of snapshot . | |
| The similarity score between node and node in . | |
| The nodes similarity tensor. | |
| The snapshot interaction frequency matrix in . | |
| The neighbor nodes of node in the current snapshot. | |
| The set of nodes in snapshot . | |
| The influence indicator of node . | |
| The sum of interactions between node and node in . | |
| The adjacency status between node and node in . | |
| The eigenvector centrality of node in . | |
| The probabilistic value of the interaction frequency between node and node in . | |
| The node interaction entropy of node in . | |
| The sum of node interaction entropy in snapshot . | |
| The degree feature of node in . | |
| The quality of node in the gravity model. | |
| The shortest distance between node and node on | |
| The reference ratio of node similarity in exponential smoothing model for snapshot | |
| Similarity between node and node in . | |
| The predicted score (or similarity) for link prediction in future snapshot in future. |
References
- Lü, L.; Zhou, T. Link prediction in complex networks: A survey. Phys. A Stat. Mech. Its Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef]
- Abbas, K.; Abbasi, A.; Dong, S.; Niu, L.; Yu, L.; Chen, B.; Cai, S.-M.; Hasan, Q. Application of network link prediction in drug discovery. BMC Bioinform. 2021, 22, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Symeonidis, P.; Iakovidou, N.; Mantas, N.; Manolopoulos, Y. From biological to social networks: Link prediction based on multi-way spectral clustering. Data Knowl. Eng. 2013, 87, 226–242. [Google Scholar] [CrossRef]
- Cannistraci, C.V.; Alanis-Lobato, G.; Ravasi, T. From link-prediction in brain connectomes and protein interactomes to the local-community-paradigm in complex networks. Sci. Rep. 2013, 3, 1613. [Google Scholar] [CrossRef] [PubMed]
- Bocu, R.; Bocu, D.; Iavich, M. Objects Detection Using Sensors Data Fusion in Autonomous Driving Scenarios. Electronics 2021, 10, 2903. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, S.; Zhang, D.; Zhang, X.; Bao, X. Node Importance Identification for Temporal Networks Based on Optimized Supra-Adjacency Matrix. Entropy 2022, 24, 1391. [Google Scholar] [CrossRef]
- Wen, T.; Cheong, K.H. The fractal dimension of complex networks: A review. Inf. Fusion 2021, 73, 87–102. [Google Scholar] [CrossRef]
- Fortunato, S. Community detection in graphs. Phys. Rep. 2010, 486, 75–174. [Google Scholar] [CrossRef]
- Liu, H. Using link prediction to predict network evolution mechanism. Sci. Sin. Phys. Mech. Astron. 2011, 41, 816–823. [Google Scholar]
- Kaya, B. A hotel recommendation system based on customer location: A link prediction approach. Multimed. Tools Appl. 2020, 79, 1745–1758. [Google Scholar] [CrossRef]
- Si, S.; Wang, J.; Yu, C.; Zhao, H. Energy-efficient and fault-tolerant evolution models based on link prediction for large-scale wireless sensor networks. IEEE Access 2018, 6, 73341–73356. [Google Scholar] [CrossRef]
- Poleksic, A. Hyperbolic matrix factorization improves prediction of drug-target associations. Sci. Rep. 2023, 13, 959. [Google Scholar] [CrossRef] [PubMed]
- Divakaran, A.; Mohan, A. Temporal Link Prediction: A Survey. New Gener. Comput. 2019, 38, 213–258. [Google Scholar] [CrossRef]
- Gao, S.; Denoyer, L.; Gallinari, P. Temporal link prediction by integrating content and structure information. In Proceedings of the 20th ACM International Conference on Information and Knowledge Management, Arlington, VA, USA, 11 November 2011; pp. 1169–1174. [Google Scholar]
- Ouzienko, V.; Guo, Y.; Obradovic, Z. Prediction of attributes and links in temporal social networks. In ECAI 2010; IOS Press: Washington, DC, USA, 2010; pp. 1121–1122. [Google Scholar]
- Zhou, L.; Yang, Y.; Ren, X.; Wu, F.; Zhuang, Y. Dynamic network embedding by modeling triadic closure process. In Proceedings of the AAAI Conference on Artificial intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Huang, X.; Chen, D.; Ren, T. A Feasible Temporal Links Prediction Framework Combining with Improved Gravity Model. Symmetry 2020, 12, 100. [Google Scholar] [CrossRef]
- Yang, X.; Tian, Z.; Cui, H.; Zhang, Z. Link prediction on evolving network using tensor-based node similarity. In Proceedings of the 2012 IEEE 2nd International Conference on Cloud Computing and Intelligence Systems, Hangzhou, China, 30 October–1 November 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 154–158. [Google Scholar]
- Güneş, İ.; Gündüz-Öğüdücü, Ş.; Çataltepe, Z. Link prediction using time series of neighborhood-based node similarity scores. Data Min. Knowl. Discov. 2016, 30, 147–180. [Google Scholar] [CrossRef]
- Lorrain, F.; White, H.C. Structural equivalence of individuals in social networks. J. Math. Sociol. 1971, 1, 49–80. [Google Scholar] [CrossRef]
- Etude, P.J. comparative de la distribution florale dans une portion des Alpes et des Jura. Bull. Soc. Vaud. Sci. Nat 1901, 37, 547. [Google Scholar]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabási, A.-L. Hierarchical organization of modularity in metabolic networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef]
- Molloy, M.; Reed, B. A critical point for random graphs with a given degree sequence. Random Struct. Algorithms 1995, 6, 161–180. [Google Scholar] [CrossRef]
- Adamic, L.A.; Adar, E. Friends and neighbors on the web. Soc. Netw. 2003, 25, 211–230. [Google Scholar] [CrossRef]
- Zhou, T.; Lü, L.; Zhang, Y.-C. Predicting missing links via local information. Eur. Phys. J. B 2009, 71, 623–630. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Albert, R. Emergence of scaling in random networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Lü, L.; Jin, C.-H.; Zhou, T. Similarity index based on local paths for link prediction of complex networks. Phys. Rev. E 2009, 80, 046122. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lü, L. Link prediction based on local random walk. Europhys. Lett. 2010, 89, 58007. [Google Scholar] [CrossRef]
- Fouss, F.; Pirotte, A.; Renders, J.-M.; Saerens, M. Random-walk computation of similarities between nodes of a graph with application to collaborative recommendation. IEEE Trans. Knowl. Data Eng. 2007, 19, 355–369. [Google Scholar] [CrossRef]
- Klein, D.J.; Randić, M. Resistance distance. J. Math. Chem. 1993, 12, 81–95. [Google Scholar] [CrossRef]
- Liu, M.; Tu, Z.; Su, T.; Wang, X.; Xu, X.; Wang, Z. BehaviorNet: A Fine-grained Behavior-aware Network for Dynamic Link Prediction. ACM Transactions on the web. 2023. [CrossRef]
- Taylor, D.; Myers, S.A.; Clauset, A.; Porter, M.A.; Mucha, P.J. Eigenvector-Based Centrality Measures for Temporal Networks. Multiscale Model. Simul. 2017, 15, 537–574. [Google Scholar] [CrossRef]
- Granovetter, M.S. The strength of weak ties. Am. J. Sociol. 1973, 78, 1360–1380. [Google Scholar] [CrossRef]
- Borgatti, S.P.; Halgin, D.S. On network theory. Organ. Sci. 2011, 22, 1168–1181. [Google Scholar] [CrossRef]
- Lü, L.; Zhou, T. Role of weak ties in link prediction of complex networks. In Proceedings of the 1st ACM International Workshop on Complex Networks Meet Information & Knowledge Management, Hong Kong, China, 2–6 November 2009; pp. 55–58. [Google Scholar]
- Levy, M.; Goldenberg, J. The gravitational law of social interaction. Phys. A Stat. Mech. Its Appl. 2014, 393, 418–426. [Google Scholar] [CrossRef]
- Wahid-Ul-Ashraf, A.; Budka, M.; Musial, K. How to predict social relationships—Physics-inspired approach to link prediction. Phys. A Stat. Mech. Its Appl. 2019, 523, 1110–1129. [Google Scholar] [CrossRef]
- Xu, Z.; Pu, C.; Yang, J. Link prediction based on path entropy. Phys. A Stat. Mech. Its Appl. 2016, 456, 294–301. [Google Scholar] [CrossRef]
- Xu, H.; Luo, R.; Winnink, J.; Wang, C.; Elahi, E. A methodology for identifying breakthrough topics using structural entropy. Inf. Process. Manag. 2022, 59, 102862. [Google Scholar] [CrossRef]
- Yuyu, M.; Jing, G. Link prediction algorithm based on node structure similarity measured by relative entropy. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2021; p. 012078. [Google Scholar]
- Baltakiene, M.; Baltakys, K.; Cardamone, D.; Parisi, F.; Radicioni, T.; Torricelli, M.; de Jeude, J.; Saracco, F. Maximum entropy approach to link prediction in bipartite networks. arXiv 2018, arXiv:1805.04307. [Google Scholar]
- Bonacich, P. Factoring and Weighing Approaches to Clique Identification. J. Math. Sociol. 1971, 92, 1170–1182. [Google Scholar]
- Huang, Z.; Lin, D.K. The time-series link prediction problem with applications in communication surveillance. Inf. J. Comput. 2009, 21, 286–303. [Google Scholar] [CrossRef]
- Erkol, Ş.; Mazzilli, D.; Radicchi, F. Influence maximization on temporal networks. Phys. Rev. E 2020, 102, 042307. [Google Scholar] [CrossRef]
- Génois, M.; Vestergaard, C.L.; Fournet, J.; Panisson, A.; Bonmarin, I.; Barrat, A. Data on face-to-face contacts in an office building suggest a low-cost vaccination strategy based on community linkers. Netw. Sci. 2015, 3, 326–347. [Google Scholar] [CrossRef]
- Paranjape, A.; Benson, A.R.; Leskovec, J. Motifs in temporal networks. In Proceedings of the Tenth ACM International Conference on Web Search and Data Mining, Cambridge, UK, 6–10 February 2017; pp. 601–610. [Google Scholar]












| Indicator | Topology | Definition 1 | Complexity 2 |
|---|---|---|---|
| CN [20] | Local | ||
| Jaccard [21] | Local | ||
| HPI [22] | Local | ||
| HPD [23] | Local | ||
| AA [24] | Local | ||
| RA [25] | Local | ||
| PA [26] | Local | ||
| GR [17] | Local | ||
| LP [27] | Semi-Local | ||
| LRW [28] | Semi-Local | ||
| RWR [28] | Semi-Local | ||
| Cos+ [29] | Global | ||
| ACT [30] | Global |
| Data Set 1 | N | C | Span |
|---|---|---|---|
| Workspace | 92 | 9287 | 6/24–7/3, 2013 |
| Emaildept3 | 89 | 12,216 | 802 days |
| Email-EU-core | 986 | 332,334 | 803 days |
| Data Set | T | |
|---|---|---|
| Workspace | 1 day | 10 |
| Workspace | 2 days | 5 |
| Emaildept3 | 7 days | 115 |
| Emaildept3 | 30 days | 27 |
| Emaildept3 | 120 days | 7 |
| Email-EU-core | 7 days | 115 |
| Email-EU-core | 30 days | 27 |
| Email-EU-core | 120 days | 7 |
| Data Set | OESMNIE | GR | CN | AA | JC | PA | RA | |
|---|---|---|---|---|---|---|---|---|
| Workspace | 1 | 0.8772 | 0.7229 | 0.7219 | 0.7254 | 0.7010 | 0.7348 | 0.7249 |
| Workspace | 2 | 0.8928 | 0.7300 | 0.7213 | 0.7284 | 0.7661 | 0.6730 | 0.7264 |
| Email-EU-core | 7 | 0.8856 | 0.7898 | 0.7457 | 0.7327 | 0.7620 | 0.8717 | 0.7787 |
| Email-EU-core | 30 | 0.9118 | 0.8676 | 0.8683 | 0.8438 | 0.8654 | 0.874 | 0.8741 |
| Email-EU-core | 120 | 0.9148 | 0.9063 | 0.9287 | 0.8932 | 0.9235 | 0.8837 | 0.9353 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Zhang, S.; Mao, H.; Liu, R.; Xiong, X. Temporal Network Link Prediction Based on the Optimized Exponential Smoothing Model and Node Interaction Entropy. Symmetry 2023, 15, 1182. https://doi.org/10.3390/sym15061182
Tian S, Zhang S, Mao H, Liu R, Xiong X. Temporal Network Link Prediction Based on the Optimized Exponential Smoothing Model and Node Interaction Entropy. Symmetry. 2023; 15(6):1182. https://doi.org/10.3390/sym15061182
Chicago/Turabian StyleTian, Songyuan, Sheng Zhang, Hongmei Mao, Rui Liu, and Xiaowu Xiong. 2023. "Temporal Network Link Prediction Based on the Optimized Exponential Smoothing Model and Node Interaction Entropy" Symmetry 15, no. 6: 1182. https://doi.org/10.3390/sym15061182
APA StyleTian, S., Zhang, S., Mao, H., Liu, R., & Xiong, X. (2023). Temporal Network Link Prediction Based on the Optimized Exponential Smoothing Model and Node Interaction Entropy. Symmetry, 15(6), 1182. https://doi.org/10.3390/sym15061182





