Abstract
Structural aberrations (SA) have been shown to play an essential role in the occurrence and development of cancer. SAs are typically characterized by copy number alteration (CNA) dose and distortion length. Although sequencing techniques and analytical methods have facilitated the identification and cataloging of somatic CNAs, there are no effective methods to quantify SA considering the amplitude, location, and neighborhood of each nucleotide in each fragment. Therefore, a new SA index based on dynamic time warping is proposed. The SA index analysed 22448 samples of 35 types/subtypes of cancers. Most types had significant differences in SA levels ranging between 12p and 20q. This suggests that genes or inter-gene regions may warrant greater attention, as they can be used to distinguish between different types of cancers and become targets for specific treatments. SA indexes were then used to quantify the differences between cancers. Additionally, SA fingerprints were identified for every cancer type. Kidney chromophobe, adrenocortical carcinoma, and ovarian serous cystadenocarcinoma are the three severest types with structural aberrations caused by cancer, while thyroid carcinoma is the least. Our research provides new possibilities for the better utilization of chromosomal instability for further exploiting cancer aneuploidy, thus improving cancer therapy.
1. Introduction
Malignant cells rapidly acquire somatic structural aberrations during proliferation, creating intratumor genetic heterogeneity within the population [1,2]. Structural aberration (SA) refers to the ongoing acquisition of genomic alterations involving either a gain or loss of whole chromosomes [3]. SA is considered a significant type of chromosomal instability (CIN). Another major type of CIN is tumor mutation burden (TMB). Both constitute CIN, which has been proven to underpin much of the intratumoral heterogeneity observed in cancers and drives phenotypic adaptation during tumor evolution [4,5]. Additionally, it has been confirmed that immune features and SA define the most mixed tumor groups [6]. Research on SA may be of great help in the diagnosis of cancers of an unknown primary site (CUP) [6,7], given the fact that even after a complete assessment including immunohistochemistry markers, the identification of the tissue of origin is still a challenge [8,9,10].
Because SAs are typically characterized by both the CNA dosage (corresponding to duplications and deletions) and aberration length (typically measured in base pairs (bp) or kilobase pairs) [11], it is difficult to quantify how severe a SA is in a given region, which makes it difficult to conduct any quantitative analysis in SA-related research. For example, although TMB is only one major form of CIN, it has frequently been used to quantify CIN levels. Regarding TMB, however, the mutation frequency and count is the two most widely used measurements [12,13,14,15]. Unfortunately, the significant mutation genes identified using the mutation count or frequency tend to be long because of their prominence in the sequence length. Moreover, regardless of how many nucleotides are in an insertion or deletion, it is only counted as one mutation without considering its sequence information. Therefore, it is evident that TMB is a highly biased quantifying measure of CIN.
Nonetheless, the high cost of TMB detection limits its use in clinical practice [16,17]. There is a great need for a new method, using only copy number data, for quantifying structural variations in length and amplitude to quantify CIN accurately. Copy number data can be obtained using DNA-based tests, which are more robust when applied to formalin-fixed paraffin-embedded tissues.
Therefore, a new normalized SA index based on dynamic time warping is proposed to quantify global structural variations of somatic CNA profiles for segments. Furthermore, by treating variations in chromosomal sequences as time sequences, the variation length and amplitudes of variation in each nucleotide and the overview “waveform” can be considered.
To demonstrate the usage of our new SA index, 22,448 samples of 35 types/subtypes of cancers downloaded from The Cancer Genome Atlas (TCGA) were analyzed. Their structural aberration fingerprints were identified by analyzing their corresponding SA indexes arm by the arm. The molecular distance was also calculated using genome SA indexes from the perspective of the somatic copy number alteration (SCNA).
By filling the gap in the quantification of global structural variations and mutations, it is possible to use data-driven methods to progress CIN-related research further. Additionally, in combining the SA index with methylations, other features and appropriate machine learning methods, the ultimate goal is to move beyond correlation and classification to achieve new insights into disease mechanisms and treatment targets.
2. Materials and Methods
2.1. Data Preprocessing
TCGA “https://portal.gdc.cancer.gov/projects (accessed on 29 May 2021)” is a landmark cancer genomics program that has molecularly characterized approximately 20,000 primary and non-malignant samples spanning 33 primary cancer types [18,19]. Considering that TCGA is by far the most consistent platform in providing the most cancer types and data testing methods, data from it was used in this study.
The CNA data were measured using the Affymetrix Genome-Wide Human SNP Array 6.0 platform and saved in TCGA “*. grch38.seg.v2.txt” files. In addition, simple Nucleotide Variation (SNV) data of the whole-exome sequencing was measured with the MuTect2 Variant Calling Pipeline and saved in mutation annotation format (MAF) files. These were used in our analysis.
The genes whose CNA data were missing in all samples were removed (132 removed genes out of 24,995 genes, accounting for 0.53%). Moreover, due to our inability to conduct a generalizing analysis on account of the shortage of data on chromosome Y, we removed the genes from it. After removing the samples whose age was unknown, the remaining samples varied from 18 to 90 years.
To ensure that the samples were as typical as possible, all non-primary samples (including recurrent and metastatic tumor samples) were removed before analysis. This meant only primary tumor samples (for LAML, Acute Myeloid Leukemia, and primary blood) were used for analysis. For non-malignant samples, only blood-derived normal (for LAML, solid tissue normal) samples were used. For this study, we separated oesophagal carcinoma (ESCA) into esophageal adenocarcinoma (ESAD) and esophageal squamous cell carcinoma (ESSC), respectively, and cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) into cervical adenocarcinoma (CEAD) and cervical squamous cell carcinoma (CESC), according to the provided clinical information. As a result, there were 22,448 tumor and non-malignant samples of 35 types/subtypes of primary tumors in our study. The data are summarized in Table S1.
2.2. SA Index Based on the DTW Measure
To quantify how severe the structural aberrations of a certain segment (including focal or local variations) are, here, a SA index based on the dynamic time warping (DTW) measure is proposed. DTW is a widely used method for sequence/curve similarity quantification. DTW is a sophisticated similarity measure that calculates an optimal match between two given sequences (e.g., time series) with certain restrictions. It can be non-trivially transformed. The sequences are “warped” non-linearly in the time dimension to determine a measure of their similarity independent of certain non-linear variations. In chromosomal signals, after representing the time instances by the nucleotide positions and amplitudes by the corresponding CNA levels, the DTW measure is suitable for deriving mountain plots of two CNA curves, denoted as mountain curves [20,21]. This means that the position and order of the CNA points of genes/intergenic regions along chromosome arms can be seen as sequences or curves. Consequently, the DTW measure can be used to quantify the similarity between the mountain curves of any pair of samples, as shown in Equation (1):
Dk symbolizes accumulated distance, and dk is a pairwise distance value. The value of accrued distance Dk(i, j) is determined by a pairwise distance dk(i, j) and the minimum of the previous values of accumulated distances.
For a segment of a given tumor sample, if the mountain curve of the corresponding non-malignant sample is used as the reference, Dk can then be sued to quantify how severe the structural aberration in that segment is compared to its normal status. To put it simply, for sample k, the SA index in a segment can be defined as:
where SAI is normalized to the range <0, 1>. Here, ‘1’ means that the tumor mountain curve is completely different from the corresponding non-malignant mountain curve, which means that the structural aberration varies significantly from the non-malignant profile due to tumour-related or other reasons. In contrast, ‘0’ means that the curves coincide with each other (almost no difference between the tumor and non-malignant CNA profiles).
Note: A mountain curve can be obtained for an individual segment by sorting the CNA values of genes along the nucleotide positions. A mountain curve can be obtained for a group of separate segments by aligning the segments according to their chromosome/genome positions. Each spot represents the median/mean value of the CNAs of all nucleotides in each sample segment or cohort group. A nucleotide, gene, or segment can be treated accordingly as a spot for a mountain curve. This depends on the width covered by the mountain curve and the scale of the granularity of the segment within it. Details on the mountain curve are available in [20,21]. An example is provided in Section 3.1.
2.3. The exoTMB for 35 Different Types of Cancers
TMB is the simple nucleotide mutation counts per million bites [22]. Considering the fact that research has shown that synonymous mutations frequently act as driver mutations in human cancers [23], in our study, the exoTMB (including both nonsynonymous and synonymous mutations) of each tumor sample’s exosome for each type of cancer was calculated. Because only exome mutations are available in MAF files, only the exoTMB of the exome was calculated, as follows:
where exoTMBc represents the nonsynonymous and synonymous mutation counts per million bites in the exon length of the tumor sample for chromosome c, |Nonsynonymous Mutation Counts + Synonymous Mutation Counts| represents the nonsynonymous and synonymous mutation counts in the whole exons of chromosome c, and |Exon Length (MB)| represents the length of all exons in chromosome c, whose unit is a million bites. The average exoTMB values for each arm and the corresponding length of the exons are shown in Table S2. Detailed descriptions of the nonsynonymous and synonymous mutations are available in Supplementary Note S2 [23,24,25,26,27].
3. Results
3.1. SA Index Values of 35 Different Types of Cancers and Their Molecular Distances from Each Other
To calculate a genome-wide SAI for a given type of cancer, it is necessary to create a genome-wide mountain curve for each type of cancer arm-by-arm. Considering the large number of genome-wide genes, a gene was treated as a segmentation. Intergenic regions may be revealed to have essential functions; hence, it is essential to consider these regions. The lengths of intergenic regions are normally several times those of genes. For example, the average length of an intergenic region is 91,008 bps (without counting intergenic regions at the centromere). It is almost three times the average length of a gene, which is 35,328 bps. To obtain more precise genome-wide CNA curves, the following steps were carried out:
- If the length of an intergenic region was longer than 35,328 bps, it was divided into several segments (the number of segments was the rounded-up number of the length of the intergenic region compared with 35,328 bps).
- However, if the length of an intergenic region was shorter than 35,328 bps, it was counted as one segment.
For example, if the length of an intergenic region was 52,992 (1.5 times 35,328), it was divided into two segments of the same length. For each segment, the corresponding CNA is the average value of the CNAs of all nucleotides. In this way, a gene or a segment is represented as a spot, with its starting position as the X-axis value and its average CNA in the cancer cohort as the Y-axis value. Examples of mountain curves of CNAs on Chr3 of all the adenocarcinomas (ADCs) and squamous cell carcinomas (SCCs) are shown in Figure 1 [20,21].
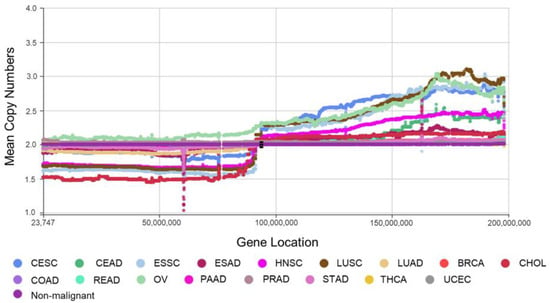
Figure 1.
Mountain curves of ADCs and SCCs for Chr 3. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. The space between the two arms of each chromosome is the location of the corresponding centromere.
Note: When calculating the SA index, our proposed method also took the CNA of the intergenic region into account, which helps one to consider the structural aberration of the whole chromosome fully.
Based on these mountain curves, each arm’s SA index was calculated to quantify the structural aberrations in detail. Then, the chromosome-wide SA index was taken as the summary of the arm-wide SA index values belonging to it. The genome-wide SA index summarised all chromosome-wide SA index values (armSA, chrSA and geSA, respectively). Therefore, the range of armSA was <0, 1>, that of chrSA was <0, 2>, and that of geSA was <0, 41> (as there are no genes on 13p, 14p, 15p, 21p or 22p).
Table S3 lists the arm-wide and genome-wide SA index values of all types of cancers in the TCGA. Compared with the corresponding non-malignant samples, the genome-wide structural variation of THCA (thyroid carcinoma) is the slightest (geSA = 0.07). Furthermore, only Chr14 and Chr22 have minor aberrations among the non-malignant samples, whose armSAs are 0.02 and 0.01, respectively. Figure S1 shows the mountain plot and directional Manhattan plot of Chr22 [28]. It proves that the structural variation pattern of Chr22 is significantly different but shows mild amplitude changes in the tumor samples compared with the corresponding non-malignant samples. Additionally, the armSAs in the p arm and the q arm of each chromosome in GBM (glioblastoma) are almost equal, reflecting the symmetry of structural aberrations in GBM.
On the contrary, the geSAs of KICH (kidney chromophobe, geSA = 8.56), ACC (adrenocortical carcinoma, geSA = 5.67), and OV (ovarian serous cystadenocarcinoma, geSA = 5.06) are the three most severe, which means that they have the most significant changes in their genome-wide structural profiles caused by the tumor. Their corresponding mountain curves are shown in Figure S2.
- Unlike other types of cancers, KICH always shows chromosome-wide deletions or amplifications. Interestingly, no gains or deletions occur in the same chromosome.
- Except for 9p, Chr10, and 14p, ACC also shows arm-wide deletions and amplifications. There are still no gains or deletions that occur in the same chromosome. However, unlike KICH, there are mild fluctuations in these deletions or amplifications.
- Unlike KICH nor ACC, the fluctuation in OV is the greatest among all types of cancers. In addition, there are dramatic fluctuations in almost all the arms except 9p and 21q. Therefore, OV is the most distinguishable due to its dramatic genome-wide ups and downs.
The last row in Table S3 lists the standard deviation (STD) values of the corresponding armSAs across all types of cancers. A larger STD means a comparatively larger difference between the SAs of different types of cancers for the given arm. The STDs on 12p and 20q are the largest, suggesting that the structural aberration profiles of different types of tumors vary significantly at 12p and 20q. Genes or intergenic regions on these arms may be worth greater attention from researchers because they may serve as new biomarkers for tumor origin identification or cancer-specific treatment targets.
On 12p, TGCT (testicular germ cell tumor) shows the greatest variation compared with the corresponding non-malignant samples, whose armSA is 0.55. The second largest arm-wide SA index is 0.30 (the arm-wide SA index of ACC), much smaller than 0.55. This may be considered as the fingerprint of TGCT, aiding in the original identification of CUP. The mountain plots of five cancers possessing the largest SA index values on Chr12 are shown in Figure S3. TGCT has the largest gain on 12p, making it significantly different from the other types of cancers.
Besides 12p, 20q is the second most varied, with the cancers showing great differences compared to the control group and one another. READ (rectum adenocarcinoma), COAD (colon adenocarcinoma), ESAD, UCS (uterine carcinosarcoma), and KICH are the most five varied types of cancers on this arm. Their corresponding mountain plot is shown in Figure S4. The large amplicon between 20q11.1 and 20q11.21 may also be essential in identifying UCS.
Additionally, UVM and UCS are both non-epithelial carcinomas (NEC). Neither UVM nor LIHC has the MYC amplicon, even though it is well-attested in OV [29], BRCA [30,31], and several other types of cancers [12]. Two amplicons are harbored in the area from 8q12.11 to 8q21.12, which may help distinguish UCS from other cancers (Figure S5).
On the contrary, 14q, 6q, 21p, 4p, 9q and 2q are the steadiest arms, with hardly any aberration for all types of cancers. Consequently, these arms provide hardly any helpful information for further structural-aberration-related research on cancer.
3.2. Fingerprints of 35 Different Types of Cancers
Considering the arm-wide variations in all types of cancers, 0.1 (10% variation) was set as the cutoff value. If an armSA ≥ 0.1, the corresponding arm-wide structural aberration is considered significant compared to the super control group (the group of all non-malignant samples of all types of cancers available in the TCGA); if an armSA < 0.1, it is considered to be non-significant. The SA fingerprint of each kind of cancer was identified, and the results are listed in Table 1. Because structural aberrations in these arms can be used to distinguish them from one another, research on the initiation of their chromosome instability, genes/intergenic regions, and so on may be of great importance in deepening our understanding of tumor mechanisms or uncovering new targets.

Table 1.
SA fingerprints of each type of cancer.
Except for the PRAD (prostate adenocarcinoma), PAAD (pancreatic adenocarcinoma), THCA (thyroid carcinoma), and UCEC (uterine corpus endometrial carcinoma) of ADCs and the DLBC (lymphoid neoplasm diffuse large B-cell lymphoma), LAML (acute myeloid leukemia), LGG (brain lower-grade glioma) and THYM (thymoma) of non-epithelial carcinomas (NEC), the other 27 types of cancers have unique SA fingerprints. For example, the SA fingerprints of READ consist of chr7, chr8, 13q, 17p, chr18 and 20q, shown in Figure S6. Therefore, if and only if a sample shows broad variation in the copy numbers for these chromosomes simultaneously, it may be a READ tumor sample.
To prove the validity of the cancer fingerprints we identified, we downloaded HNSC data from the GEO database (GSE103322). In addition, we obtained LUSC data from [32], and we then performed an analysis with these data as the control. The comparative results are shown in Table S4. It can be seen that the SA fingerprints obtained with these new data are almost consistent with the original results (only the fingerprint of 7p in LUSC is different, but the difference in the SA index values between the two is very small), which demonstrates the effectiveness of our method and enhances the credibility of SA fingerprints.
In addition to the fingerprints of all types of cancers listed in Table 1, there are some profiles worth mentioning. From the bee swarm plots for the geSAs of four cancer categories (ADC, SCC, NEC, and OEC) shown in Figure 2, it can be seen that the geSAs of the OECs (consisting of six types of cancers) present the greatest difference from each other. The corresponding geSAs range from 0.85 to 8.56 (the gap between the maximum and minimum is 7.71). On the contrary, the SCCs only consist of four cancer types, and their SA index values range from 1.36 to 3.28. From the average geSA of each category of cancer, it can be concluded that the structural aberration patterns of ADCs and OECs fluctuate more violently than those of SCCs and NECs. Additionally, two of the most varied cancers (KICH and ACC) belong to the category of OECs.
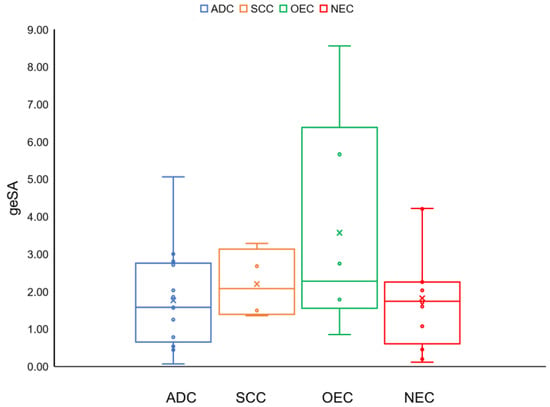
Figure 2.
Bee swarm plots for the geSAs of four cancer categories (adenocarcinoma, squamous cell carcinoma, other epithelial carcinomas, and non-epithelial carcinomas).
According to the armSA results of all the ADCs and SCCs shown in Table S3, we can see the significant difference in the CNA patterns of the ADCs, as one kind, and the SCCs, as another, occur on 3q. Almost all the SCCs have large arm-wide amplifications on 3q; however, the ADCs (except for OV) have only mild amplifications. We can also see that only the SCCs have common aberrations on chr3, 5p and 20q. The other cancer categories do not have any aberration in common.
According to the definition of the SA index, the length of deletion or amplification is more significant than its amplitude. For example, Figure S7 shows mountain curves of 14q and 17p for UCS. Even with the enormous focal amplicons on 14q, its armSA is 0.07. On the contrary, the armSA of 17p is 0.19, and here, there is an arm-wide amplification with a much smaller amplitude. This example shows that the aberration length does play an essential role in the SA index value. Indeed, the SA index value considers both the copy number alteration and aberration length. Considering the fact that, in practice, it is more often the case that false focal amplicons or deletions are observed in a single sample rather than false arm-wide broad alterations being detected due to noise inherent in the data, broad-signature SAs are more practical and ascertainable.
Focal deletions on 9p21.3, where CDKN2A/B is located, are well-attested in GBM, LUAD and LUSC [14]. Figure 3 shows ESSC and LUSC in the SCCs, CHOL, PAAD, LUAD, and STAD in the ADCs, BLCA in the OECs, and GBM, SKCM, and MESO in the NECs all have CDKN2A/B focal deletions. Among them, GBM shows the greatest deletion, followed by SKCM. The relationship between these primary tumors may be worthy of further research. Another two well-known genes, CD274 (the coding gene of PD-L1, programmed death-ligand 1, the main immune checkpoint) and JAK2 (9p24.1) are also located in this area. The amplification of both of these genes has recently been described in pulmonary carcinomas in association with PD-L1 expression [33,34]. However, Figure 3 and Figure S8 show that only a tiny percentage of these samples show copy number amplifications consistent with the published results [35]. For comparison, the bee swarm plot of PDCD1 (the coding gene of PD-1, programmed death receptor 1) is also shown at the bottom of Figure S8.
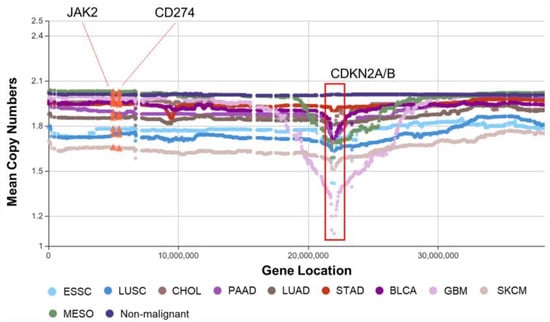
Figure 3.
Mountain plot of 9p with CDKN2A/B focal deletions. Each spot is the mean copy number of genes in the corresponding group. The genes are sorted according to their locations. For example, the mean values of CNV of CD274 (the coding gene of PD-L1) and JAK2 for each type of cancer are highlighted with orange triangles.
COAD and READ are always combined because of the relationship between their tissues of origin. In Table S3, we find that across the whole genome, their SA patterns have the same trend. However, whenever COAD has copy number variations, READ has larger ones, except for Chr2, 3p, 12q, 20p and 21q. According to the deflection plot (shown in Figure S9), across the whole genome (data are not shown), only the SA patterns on 18q and 20q are significantly different from each other. COAD has significantly less loss on 18q and less gain on 20q compared with READ. The gain on 20q of READ and COAD has previously been identified [36,37].
4. Discussion
As mentioned above, somatic structural aberration is a major form of chromosomal cancer instability observed in most solid tumors and is associated with poor prognosis and drug resistance. However, the mechanism underlying CIN in cancer remains unclear [34]. In part, this may be because there is no effective quantifying method for SA profiles, which hinders the ability of statistical methods, machine learning, and other mathematical methods to assist in research on this mechanism. Moreover, as an important biomarker, TMB plays an essential role in response to immune checkpoint inhibitor therapy for most cancers. Figure 4 shows the Nightingale rose diagrams of 20 types of cancers with the top SPCCs (Spearman’s correlation coefficients) between their SA index values and exoTMBs, both arm-wide and genome-wide. SPCC is used to quantify both linear and non-linear relationships between SA index values and exoTMBs. Additionally, Table S5 lists the SPCCs between genome-wide and arm-wide SA index values and exoTMBs (including synonymous and nonsynonymous) for all 35 types/subtypes of cancers. However, according to these results, it is clear that there is hardly any linear/non-linear relationship between the CNA and exoTMB. Since both a structural aberration and mutation are major parts of CIN, it is not reasonable to use only TMB to evaluate how severe the CIN is for a given sample. This also shows the necessity and importance of quantifying the degree of SA from the global point of view of a given segment (i.e., an arm, a chromosome, or even the whole genome).
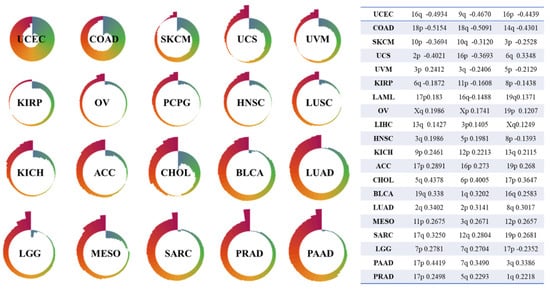
Figure 4.
Nightingale showed SPCCs between SA index values and exoTMBs, both arm-wide and genome-wide, for 20 types of cancers. The table in Figure 4 shows the top 3 SPCCs (the table is sorted by absolute value and shows the arm with the strongest correlation between the SA index value and exoTMB for each cancer).
Although there was not a strong relationship between the exoTMB and SA index values among any of the types of cancers, there were still some interesting points. Table S5 shows that the arm-wide SPCCs of BRCA, LUAD, and PRAD are all positive, which means that a larger SA index value (a much more severe alteration between the tumor and non-malignant SA profiles) might lead to an increased exoTMB. On the contrary, for COAD and UCEC, their arm-wide SPCCs are negative, which indicates that in these two cancers, the tremendous alteration in structure might cause a reduction in exoTMB. More significantly, except for 1q, 5p, and Xp, the SPCCs of the other arms in UCEC show slight negative correlations between the armSA and exoTMB (SPCC < −0.3, p < 0.0001). For BLCA, PAAD and SARC, there are only a few arms with negative SPCCs (BLCA Chr9, PAAD 9p and 16p, and SARC 10p). As for the rest of the cancer types, they all have several arms with significant SPCCs. For example, for DLBC, the SPCC of 6p is 0.5760 (p < 0.0001), and the SPCCs of ESAD for 17q and 19p are 0.3815 and −0.4908 (p < 0.0001), respectively. These features suggest a correspondence between the armSA and arm-wide exoTMB for each cancer; therefore, the SA index abnormality of some typical arms in certain cancers may correspond to a specific exoTMB. As for the correlation between the geSA and whole-exome exoTMB, the SPCCs of PAAD (SPCC = 0.5518, p < 0.0001) and PRAD (SPCC = 0.5881, p < 0.0001) show a high level, which may reveal the potential of geSA to replace exoTMB as a biomarker for immune checkpoint inhibitor therapy. A description of the calculation of SPCC is provided in Supplementary Note S3.
Returning to the SA index, Table S3 lists the arm-wide and genome-wide SA index values of all types of cancers available in the TCGA. Molecular similarities between histologically or anatomically related cancer types provide a basis for focused pan-cancer analyses, such as pan-gastrointestinal, pan-gynecological, pan-kidney, and pan-squamous analyses and those pertaining to stemness features, which, in turn, may inform strategies for future therapeutic development [38]. Our SA index provides a quantification method based on structural aberrations when taking a cancer cohort as a group. The SA index can also quantify the severity of structural aberrations for an individual sample.
More importantly, the molecular fingerprint based on the SA index value, unique to a specific type of cancer, may provide new insight into cancer initiation and development. In turn, it may also inform strategies for future therapeutic development. Therefore, this study represents the largest high-resolution structural aberration profiles generated by a single platform and the first large-scale analysis of absolute copy number data across pan-cancer types. First, we identified the fingerprint SA patterns of these types. Then, we quantified their molecular differences to provide the first comprehensive overview of the molecular factors that distinguish different neoplasms in the TCGA. The corresponding mountain plots of the signature CNAs of READ, ESSC, BLCA, and GBM in different cancer categories are shown in Figures S6 and S10–S12, respectively. Others may easily be created using the web tool provided by us “https://www.clickgenome.org/ (accessed on 10 August 2021)”.
Although structural aberration is only part of CIN or only one aspect of profile genomic variation, methylations, mutations, and mRNA expressions have all been shown to have more direct relationships with cancer mechanisms and treatment targets. Therefore, using only the SA index should not be enough to provide completely satisfactory outcomes. The following examples using the arm-wide SA indexes of the samples as variables still prove that the SA index can extract helpful information for CIN-related research. It is reasonable to assume that with mRNA expressions and other genomic profiles, more breakthroughs in cancer research may be achieved in the near future.
To test whether SA indexes are clinically related to the survival of tumor patients, a multivariate Cox-regression-model-based survival analysis using the arm-wide SA indexes of individual samples of 35 types of cancers was carried out. Then, the samples for each cancer type were divided into high-risk and low-risk groups according to the median of the risk scores across all individuals. The corresponding Kaplan–Meier (KM) survival curves, with log-rank test values, are shown in Figure 5.
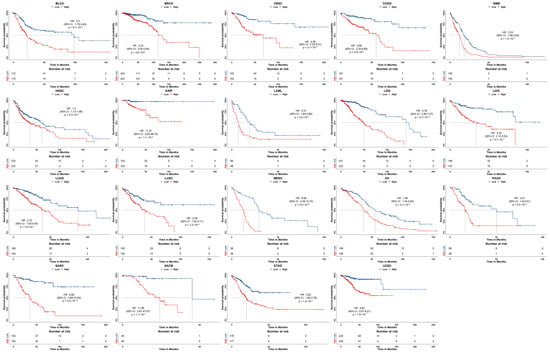
Figure 5.
The KM survival curves of 19 types of cancers with statistical significance (p < 0.05).
It can be seen that 41 arm SA indexes show a high relation to survival risk for 19 out of the 35 types of cancers with a hazard ratio HR > 1, being statistically significant (p < 0.05).
Additionally, to evaluate the diagnosis/classification performance of arm SA indexes, pan-cancer classification tasks were undertaken for the ADC, NEC, OEC, and SCC tumor samples, together with their 41 arm SA indexes, using eXtreme Gradient Boosting (XGBoost) [39]. The sample sizes of CHOL, CEAD, and ESAD in ADC, of UCS, UVM, MESO and SKCM in NEC, and of ESSC in SCC were small, and they were not considered. Of these valid samples, 80% were randomly assigned to the training set, and 20% were assigned to the testing set. The classification confusion matrices and ACC results are shown in Figure 6 and Table 2, respectively. In the model based on the 41-arm SA indexes, most cancer types have a high classification accuracy (≥95%). In particular, for TGCT in NEC, the classification accuracy is up to 100%. Overall, the diagnosis/classification performance of the arm SA indexes is excellent.
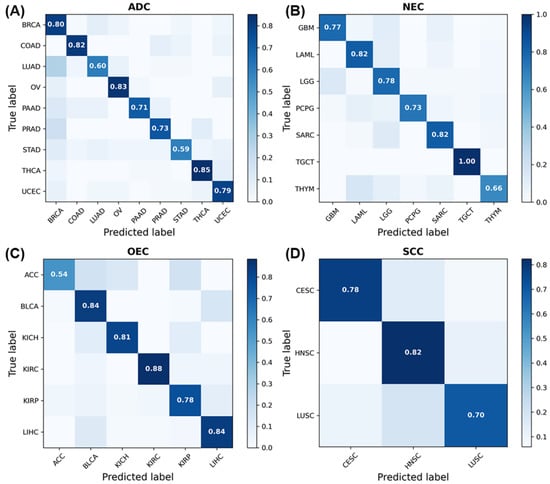
Figure 6.
The confusion matrix results obtained using 41 armSA index values. (A) The confusion matrix of ADC. (B) The confusion matrix of NEC. (C) The confusion matrix of OEC. (D) The confusion matrix of SCC.

Table 2.
The classification ACC results of each cancer type obtained using 41 armSA index values.
5. Conclusions
In this article, we proposed a new SA index based on DTW for quantifying the severity of SAs for given segments considering both the length of the segments and the CNA amplitudes of the genes. By analyzing their corresponding SA indexes arm-by-arm, we identified the structural aberration fingerprints of 35 types/subtypes of cancers. Different cancer types have different fingerprints; thus, SA fingerprints are essential in distinguishing between different cancers. In addition, we considered the clinical role of the SA index. We found a correlation between SA and TMB in some cancers, which may reveal the potential of geSA to replace exoTMB as a biomarker for immune checkpoint inhibitor therapy. Moreover, we confirmed that SA indexes perform well in patient survival analysis and cancer diagnosis/classification.
Supplementary Materials
The following supporting information can be downloaded at: https://doi.org/10.5281/zenodo.7895944. Supplementary Figures. Figure S1. (A) Mountain plot of Chr22 of THCA in the TCGA dataset. Each spot is the mean value of the copy numbers of the genes in the corresponding group. The genes are sorted according to their locations. The space between the two arms of each chromosome is the location of the corresponding centromere. (B) Directional Manhattan plot of Chr22 of THCA in the TCGA dataset. The amplitude of the vertical solid line is -10log10 (p value), where the p value is the significance test for the copy numbers of tumor and non-malignant samples. A positive amplitude means that the mean value of the corresponding gene in the tumor samples is greater than that in non-malignant samples. A negative amplitude means that the mean value of the corresponding gene in the tumor samples is smaller than that in the non-malignant samples. The solid horizontal lines are the cutoff lines according to the Bonferroni correction (8.9 × 10−5). A gap within the individual chromosome data indicates the location of the centrosome. Figure S2. The mountain plots of CNV for the THCA, KICH, OV, ACC, and non-malignant samples in the TCGA dataset. For chromosomes 13, 14, 15, 21, and 22, only genes on the q arm are represented in the microarray. Each spot is the mean copy number of the genes in the corresponding group. The genes are sorted according to their locations. Since the variation in THCA was slight, its curve is almost completely covered by the curve of the non-malignant samples. Figure S3. Mountain plot of Chr12 of TGCT, ACC, KICH, OV, and LUSC in the TCGA dataset. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Figure S4. Mountain plot of Chr20 of READ, COAD, ESAD, USC, and KICH in the TCGA dataset. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. They are the five most varied cancers on 20q. The large amplicon between 20q11.1 and 20q11.21 may also play an important role in identifying UCS. Figure S5. Mountain plot of Chr8 of UCS, KICH, TGCT, and LIHC in the TCGA dataset. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. They are the five most varied cancers on 8q. Two amplicons harbored in the area from 8q12.11 to 8q21.12 may help distinguish UCS from other cancers. Figure S6. Mountain plots of CNV in READ and non-malignant samples in the TCGA dataset. These plots show fingerprints of READ—chr7, chr8, 13q, 17p, chr18, and 20q. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Figure S7. The mountain plots of CNV in UCS and non-malignant samples in the TCGA dataset. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Figure S8. The bee swarm plot of the copy numbers of CD274, JAK2, and PDCD1 in different types of cancers. The numbers are the corresponding percentages of the samples whose copy numbers are above or below the cutoff value (cutoff = 2). Figure S9. (A) and (B) Mountain plots of Chr18 and Chr20 in the COAD and READ tumor and non-malignant samples. Each spot is the mean copy number of each gene in the corresponding group. The genes are sorted according to their locations. The space between the two arms of each chromosome is the location of the corresponding centromere. (C) and (D) The deflection plots of the copy numbers of Chr18 and Chr20 in the COAD and READ tumor and non-malignant samples. The blue color indicates that the deflection (tumor versus non-malignant samples) was greater for READ, whereas the red indicates that the deflection was greater for COAD. The solid horizontal lines are the cutoff lines according to the Bonferroni correction (1.4 × 10−4 and 7.3 × 10−5). A gap within the individual chromosome data indicates the location of the centrosome. Figure S10. The mountain plots of CNV in ESSC and non-malignant samples in the TCGA dataset. These plots show the SA fingerprints of ESSC—chr3, 4p, chr5, 7p, 8q, 9p, 11q, 20q, and chr21. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Figure S11. The mountain plots of CNV in BLCA and non-malignant samples in the TCGA dataset. These plots show the SA fingerprint of BLCA—chr8 and 20q. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Figure S12. The mountain plots of CNV in GBM and non-malignant samples in the TCGA dataset. These plots show the SA fingerprint of GBM—chr7 and chr10. Each spot is the mean value of the copy numbers of genes in the corresponding group. The genes are sorted according to their locations. Supplementary Tables. Table S1. The list of available cancers in the TCGA dataset (.xlsx). Table S2. The arm-wide exoTMB value of each sample (.xlsx). Table S3. The armSA and geSA of 35 types of cancers (.xlsx). Table S4. The armSA and geSA of HNSC and LUSC in control experiments (.xlsx). Table S5. The Spearman correlation coefficient between exoTMB and SA in 35 types of cancers (.xlsx). Supplementary Notes. Note S1: The preprocessing of the SNV data. Note S2: The selection criteria of nonsynonymous mutations and synonymous mutations. Note S3: The calculation of SPCC between the SA index value and exoTMB.
Author Contributions
Y.T. and J.Z. (Junxuan Zhu) downloaded and preprocessed part of the public data and performed the CNA analysis. J.Z. (Junxuan Zhu) and J.Z. (Jinhan Zhang) performed the classification. Q.H., J.Z. (Jinhan Zhang) and L.W. helped with data preprocessing. K.S. supervised the project. K.S. was the major contributor to the design of the project and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the Tianjin Health Science and Technology project (No. TJWJ2021MS013).
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Legacy GDC “https://portal.gdc.cancer.gov/legacy-archive/search/f (accessed on 29 May 2021)” ClickGenome “https://www.clickgenome.org/) (accessed on 10 August 2021)” repositories.
Acknowledgments
In memory of Adi F. Gazdar (who passed away on 29 December 2018), who developed the idea of quantifying copy number alteration profiles. He worked in the Department of Pharmacology, Department of Internal Medicine, and Department of Pathology UT Southwestern Medical Center, Dallas, TX, USA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sansregret, L.; Swanton, C. The role of aneuploidy in cancer evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a028373. [Google Scholar] [CrossRef]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Kuznetsova, A.Y.; Seget, K.; Moeller, G.K.; de Pagter, M.S.; de Roos, J.A.; Dürrbaum, M.; Kuffer, C.; Müller, S.; Zaman, G.J.; Kloosterman, W.P. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 2015, 14, 2810–2820. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Landau, D.A. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a029611. [Google Scholar] [CrossRef] [PubMed]
- Hainsworth, J.D.; Greco, F.A. Cancer of Unknown Primary Site: New Treatment Paradigms in the Era of Precision Medicine. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 20–25. [Google Scholar] [CrossRef]
- Yamane, S.; Katada, C.; Tanabe, S.; Azuma, M.; Ishido, K.; Yano, T.; Wada, T.; Watanabe, A.; Kawanishi, N.; Furue, Y.; et al. Clinical Outcomes in Patients with Cancer of Unknown Primary Site Treated by Gastrointestinal Oncologists. J. Transl. Int. Med. 2017, 5, 58–63. [Google Scholar] [CrossRef]
- Qaseem, A.; Usman, N.; Jayaraj, J.S.; Janapala, R.N.; Kashif, T. Cancer of Unknown Primary: A Review on Clinical Guidelines in the Development and Targeted Management of Patients with the Unknown Primary Site. Cureus 2019, 11, e5552. [Google Scholar] [CrossRef]
- Jones, W.; Allardice, G.; Scott, I.; Oien, K.; Brewster, D.; Morrison, D.S. Cancers of unknown primary diagnosed during hospitalization: A population-based study. BMC Cancer 2017, 17, 85. [Google Scholar] [CrossRef]
- Vibert, J.; Pierron, G.; Benoist, C.; Gruel, N.; Guillemot, D.; Vincent-Salomon, A.; Le Tourneau, C.; Livartowski, A.; Mariani, O.; Baulande, S. Identification of tissue of origin and guided therapeutic applications in cancers of unknown primary using deep learning and RNA sequencing (TransCUPtomics). J. Mol. Diagn. 2021, 23, 1380–1392. [Google Scholar] [CrossRef]
- Brucker, A.; Lu, W.; West, R.M.; Yu, Q.-Y.; Hsiao, C.K.; Hsiao, T.-H.; Lin, C.-H.; Magnusson, P.K.; Sullivan, P.F.; Szatkiewicz, J.P. Association test using Copy Number Profile Curves (CONCUR) enhances power in rare copy number variant analysis. PLoS Comput. Biol. 2020, 16, e1007797. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Ju, Y.S.; Haase, K.; Van Loo, P.; Martincorena, I.; Nik-Zainal, S.; Totoki, Y.; Fujimoto, A.; Nakagawa, H.; Shibata, T.; et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016, 354, 618–622. [Google Scholar] [CrossRef]
- Moore, L.; Cagan, A.; Coorens, T.H.H.; Neville, M.D.C.; Sanghvi, R.; Sanders, M.A.; Oliver, T.R.W.; Leongamornlert, D.; Ellis, P.; Noorani, A.; et al. The mutational landscape of human somatic and germline cells. Nature 2021, 597, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Samstein, R.M.; Valero, C.; Chan, T.A.; Morris, L.G.T. Tumor mutational burden as a predictive biomarker for checkpoint inhibitor immunotherapy. Hum. Vaccin. Immunother. 2020, 16, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhang, G.; Zhang, C.; Xue, L.; Yang, Z.; Lu, Z.; Huang, J.; Zang, R.; Che, Y.; Mao, S.; et al. The average copy number variation (CNVA) of chromosome fragments is a potential surrogate for tumor mutational burden in predicting responses to immunotherapy in non-small-cell lung cancer. Clin. Transl. Immunol. 2021, 10, e1231. [Google Scholar] [CrossRef]
- Liu, L.; Bai, X.; Wang, J.; Tang, X.R.; Wu, D.H.; Du, S.S.; Du, X.J.; Zhang, Y.W.; Zhu, H.B.; Fang, Y.; et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy Across Metastatic Cancer. Clin. Cancer Res. 2019, 25, 7413–7423. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e296. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Thu, K.L.; Papari-Zareei, M.; Stastny, V.; Song, K.; Peyton, M.; Martinez, V.D.; Zhang, Y.A.; Castro, I.B.; Varella-Garcia, M.; Liang, H.; et al. A comprehensively characterized cell line panel highly representative of clinical ovarian high-grade serous carcinomas. Oncotarget 2016, 8, 50489–50499. [Google Scholar] [CrossRef]
- Qiu, Z.W.; Bi, J.H.; Gazdar, A.F.; Song, K. Genome-wide copy number variation pattern analysis and a classification signature for non-small cell lung cancer. Genes Chromosomes Cancer 2017, 56, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Supek, F.; Miñana, B.; Valcárcel, J.; Gabaldón, T.; Lehner, B. Synonymous Mutations Frequently Act as Driver Mutations in Human Cancers. Cell 2014, 156, 1324–1335. [Google Scholar] [CrossRef]
- Sharma, Y.; Miladi, M.; Dukare, S.; Boulay, K.; Caudron-Herger, M.; Gross, M.; Backofen, R.; Diederichs, S. A pan-cancer analysis of synonymous mutations. Nat. Commun. 2019, 10, 2569. [Google Scholar] [CrossRef]
- Chu, D.; Wei, L. Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer 2019, 19, 359. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Yu, C.P.; Chang, S.; Xie, L.L.; Wang, S. Synonymous mutations that regulate translation speed might play a non-negligible role in liver cancer development. BMC Cancer 2021, 21, 388. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.-H.; Tong, Y.-F.; Qiu, Z.-W.; Yang, X.-F.; Minna, J.; Gazdar, A.F.; Song, K. ClickGene: An open cloud-based platform for big pan-cancer data genome-wide association study, visualization and exploration. BioData Min. 2019, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Camacho, N.; Van Loo, P.; Edwards, S.; Kay, J.D.; Matthews, L.; Haase, K.; Clark, J.; Dennis, N.; Thomas, S.; Kremeyer, B.; et al. Appraising the relevance of DNA copy number loss and gain in prostate cancer using whole genome DNA sequence data. PLoS Genet. 2017, 13, e1007001. [Google Scholar] [CrossRef]
- Chen, R.C.; Rumble, R.B.; Loblaw, D.A.; Finelli, A.; Ehdaie, B.; Cooperberg, M.R.; Morgan, S.C.; Tyldesley, S.; Haluschak, J.J.; Tan, W.; et al. Active Surveillance for the Management of Localized Prostate Cancer (Cancer Care Ontario Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J. Clin. Oncol. 2016, 34, 2182–2190. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Carter, H.B.; Lepor, A.; Loeb, S. Active surveillance for prostate cancer: Current evidence and contemporary state of practice. Nat. Rev. Urol. 2016, 13, 205–215. [Google Scholar] [CrossRef]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018, 23, 194–212.e6. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef]
- Varadhachary, G.R.; Raber, M.N.; Matamoros, A.; Abbruzzese, J.L. Carcinoma of unknown primary with a colon-cancer profile—Changing paradigm and emerging definitions. Lancet Oncol. 2008, 9, 596–599. [Google Scholar] [CrossRef]
- Condorelli, D.F.; Privitera, A.P.; Barresi, V. Chromosomal Density of Cancer Up-Regulated Genes, Aberrant Enhancer Activity and Cancer Fitness Genes Are Associated with Transcriptional Cis-Effects of Broad Copy Number Gains in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4652. [Google Scholar] [CrossRef]
- Zhang, B.; Yao, K.; Zhou, E.; Zhang, L.; Cheng, C. Chr20q Amplification Defines a Distinct Molecular Subtype of Microsatellite Stable Colorectal Cancer. Cancer Res. 2021, 81, 1977–1987. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).