Abstract
Bloom filters are widely used in genome assembly, IoT applications and several network applications such as symmetric encryption algorithms, and blockchain applications owing to their advantages of fast querying, despite some false positives in querying the input elements. There are many research works carried out to improve both the insertion and querying speed or reduce the false-positive or reduce the storage requirements separately. However, the optimization of all the aforementioned parameters is quite challenging with the existing reported systems. This work proposes to simultaneously improve the insertion and querying speeds by introducing a Cache-efficient One-Hashing Blocked Bloom filter. The proposed method aims to reduce the number of memory accesses required for querying elements into one by splitting the memory into blocks where the block size is equal to the cache line size of the memory. In the proposed filter, each block has further been split into partitions where the size of each partition is the prime number. For insertion and query, one hash value is required, which yields different values when modulo divided with prime numbers. The speed is accelerated using simple hash functions where the hash function is called only once. The proposed method has been implemented and validated using random strings and symmetric K-mer datasets used in the gene assembly. The simulation results show that the proposed filter outperforms the Standard Bloom Filter in terms of the insertion and querying speed.
1. Introduction
Bloom filters are an approximate membership querying data structure that is widely used in many applications owing to their space efficiency and fast querying behavior. Bloom filters are used in named IP address lookup [1], named data networking [2,3,4,5] genomics [6,7,8] image classification [9,10] and blockchain applications [11] because of their advantages over other data structures. There is a two-fold increase in gene data every year [12] due to the rapid increase in genome studies for preventing diseases in advance and to unravel the properties of the species. Hence, handling a large amount of gene data increases the storage requirement and also the computation speed. To address these problems, the usage of probabilistic data structures is motivated owing to their use of less memory for storing the input data and their efficient querying in less time. There are several probabilistic data structures reported in the literature such as Bloom filter, quotient filter, cuckoo filter, etc. Bloom filter is a probabilistic data structure which stores the index of the input element in the array and executes the query in less time. In the de novo genome assembly applications [13,14,15] the gene sequences are assembled by constructing the de Bruijn graph where K-mers were used in the vertices of the graph. K-mers are the DNA bases of length r partitioned from the sequence of reads in the gene database. There exists symmetry in the genetic sequences generated from the DNA of a genome [16]. The generated K-mers and their reverse complements follow inversion symmetry [17]. The Bloom filters are used to store and query the K-mers in the genome assembly process. It has also been used in other applications such as the search engines [18], networking sites for spotting malicious URLs and most visited sites [19] and in online shopping sites to collect the most or recently viewed products [20]. Bloom filters use hash functions for mapping the input elements into the memory. For a given arbitrary input data, the hash function gives the fixed random output which is the index of the memory. The hash functions are used in symmetric cryptographic applications, networking and block chain applications [21].
The Bloom filter has been optimized in anyone of the following parameters: the hash function used to insert and query the data into the Bloom memory, the time taken for inserting the string and querying for the existence of the string, memory used to store the entire set of elements and the probability of false positives.
The Bloom filter utilizes k hash functions to insert and query the input data and, thus, it takes k memory accesses in the storage. As the size of the Bloom filter grows, the Bloom filter will be stored in the off-chip memory which uses k time’s disk access. As the number of memory access increases, the latency for querying the input increases which in turn deteriorates the system performance. Although the Bloom filter uses O(1) time complexity for querying the input; for hashing each string k times, the hash function time complexity will become significant. The hash function with less collision and fewer memory cycles are preferred for the efficient Bloom filter applications. Many applications use Bloom filter architecture which uses k memory accesses, and the hash functions use more memory cycles and as a result it has more delay and computational cost. To overcome the above issues, in this work we have proposed the following three points:
- A hybrid hash function that uses murmur hash with the single hash (MSH). It computes hash values in very less time compared to the traditional Murmur hash with the double hash function (MDH).
- An improved cache-efficient blocked Bloom filter (CBBF) [19], utilizes single hash technique with fewer hash functions. It also reduces the insertion and querying time of the input elements compared to the standard Bloom filter.
- A cache efficient one hashing blocked Bloom filter (OHBB) that employs only one hash function. The OHBB filter increases the insertion and the querying speed compared to the standard Bloom filter (SBF) and reduces the false positive probability compared to CBBF.
We have used three improvisations to increase the speed and reduce the false positive probability.
First, the prevalently used non-cryptographic hash function with fewer collisions is the Murmur hash function. For computing k hash functions, the two hash functions are used. Instead, by using a single hash function the computation time required to calculate the hash values is reduced with the same collision rate.
Second, the elements are inserted in random order in the Bloom filter at the index given by the hash values. By using the blocked indexing and storing the elements in the single block the traditionally used k memory accesses have been reduced to single memory access. The single hash function is used for hashing the input elements. The querying speed has been further increased by adopting single hash functions.
Third, the block in the blocked Bloom filter has been further partitioned into a number of blocks where the size of each block is a prime number. Here only one hash computation is performed, and the hash value is modulo divided by the size of each block that will result in the random index from the block.
With the above proposed methods, the performance of the Bloom filter and the hash functions were improved in terms of the insertion and query time and there is a significant reduction in the false positive probability. This paper is organized as follows: Section 2 presents the works related to the Bloom filter. The proposed methodologies are described in the Section 3 along with the false positive probability analysis. The experimental evaluations along with the results are discussed in the Section 4 and finally the paper is concluded in the Section 5. The main notations used in the paper are shown in Table 1.

Table 1.
Main Notations.
2. Related Works
2.1. Standard Bloom Filter (SBF)
Bloom filter [22] is a data structure which is probabilistic in nature, used for the approximate membership queries. The Bloom filter data structure is shown in Figure 1. It allows some false positives during the lookup of an element from the set. It stores n elements from the set S, i.e., , in which the elements are hashed and mapped into random bit locations ranging . The element A1 in the set is inserted into the Bloom filter as follows: Initially, the m bit array is initialized to all zeros. The element A1 is hashed by k hash functions {} and each hash function will provide an index to the m bit array. The bit in the index location, is set to 1. For querying the existence of element in the set, the element B being queried is hashed again by k hash functions and the bits in the hashed bit locations are checked for the existence of 1 in all the k locations. There is a possibility that the query results as positive for the element which is not in the set, which is called a false positive.
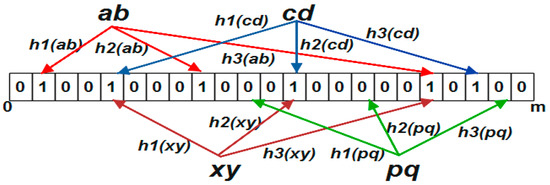
Figure 1.
Standard Bloom filter data structure. The set of elements S1 {ab, cd} were inserted into the Bloom array. The elements in the set S2 {xy, pq} were queried for the existence. All the bits were set to zero in the hashed locations of element “pq”, which means that it is not present in the set. The element “xy” is hashed where all the locations were updated as 1 which falsely identified it as an element of the set S1.
2.2. False Positive Analysis
The probability of a bit in the m bit array to be set as 1 is and the probability of that bit not being set is . Since the element is hashed k times, the probability of the element not set to 1 by any of the hash functions is . The n elements are inserted into the Bloom filter and the bit is still set as zero can be written as . The probability of the bit that is set as 1 can be expressed as . The element that is not in the set is queried in the Bloom filter in k locations where all the k locations are set as one is termed as false positive and it is expressed [23] as
The optimal number of hash functions for the false positive probability FPP with the given memory m and the number of input elements n is given by
There is a trade-off between the memory, accuracy and the processing time. If there is a need for a very small false positive probability, then the size of the memory in turn has to be increased. If the application requires fast processing, the memory size needs to be minimal which results in an increase in false positive probability.
Several research works [24] have been carried out on Bloom filters to increase their performance. The drawback of the Bloom filter is its poor cache locality. For inserting an element or for querying an element the entire Bloom filter memory has to be read, which requires k memory accesses. To reduce the memory accesses from k memory accesses to 1, Blocked Bloom filters [25] were developed where the Bloom filters are split into blocks. Each block size is designed based on the cache line size of the system. An element being mapped is first hashed and the hash value is used to select the block. The element is then hashed k times and inserted into the single block. As all the k elements were mapped to the single block, the false positive probability is increased due to the collisions, and it also requires k + 1 hash functions. In Bloom-1 filter [26], the memory m is split into blocks where the block size is same as the word size of the system. Each element is hashed by a hash function; the first l hash bits from the hash function is used to select the word and the rest of the hash bits are used to map the element into the word. By adopting this method, the memory cycles are reduced further by mapping k values to a single word of 64 bits and it uses fewer hash bits. This work has been generalized by mapping each element into two words. The insertion process involves checking the flag bit of the first word and inserting into the second word. Thus, it requires an average of two memory access for the querying an element. The Bloom-1 filter has been further optimized by using Single instruction multiple data (SIMD) where parallel instructions were used to speed up the computation. The Ultrafast Bloom filter [27] used the blocked Bloom filter approach and divided the memory into 256 bits. Each block is further divided into eight blocks of 32 bits each. Each element is hashed and distributed in all the eight blocks in parallel. Thus, the number of hash functions k is fixed to the sub block size which is 8.
3. Methodology
The one-hashing blocked Bloom filter (OHBB) method proposed in this work aimed to reduce the time required to insert and query the elements from the set. This is achieved by reducing the number of memory access for each element by splitting the memory into blocks with the size same as the cache-line size and by using fewer hash functions. The performance of the Bloom filter depends on the two stages. The first stage is the hash computation which also plays an important role in speeding up the process. The second stage is the membership check or insertion stage. At first, stage I is optimized by using the efficient hash function, and the second stage is optimized by blocking the memory which is aligned to the size of the cache line in the system.
3.1. Hash Function
Though the Bloom filter takes O(1) time for the membership check and insertion, for each element k hashes need to be calculated which takes O(k) time for the hash computation. Since the hash function directly impacts the speed of the membership check as well as the insertion, there is a need to find a better hash function that takes less time for hash computation with less collision rate. Since Murmur hash is the non-cryptographic hash function with good avalanche properties among other hash functions [28], in this work, Murmur Hash function is used for the hashing process. The Murmur hash [29] is to be hashed k times for each membership check. When the k value increases, there is an increase in the hash computation cost. To overcome this issue, Less hashing [30] was developed to reduce the hash computation where only two hash functions h1 and h2 are needed to compute k hash values with the same collision rate and less computation time. The hash values are calculated as
where i value ranges from 1 to k, h1 and h2 are two hash functions. The hash value is modulo by the size of the Bloom filter to obtain the index. So far, these double hash functions have been widely used in applications where h1 and h2 are the murmur hash function with different seed values. This hash function involves multiplication and addition operations to calculate k hash values. When the k value increases, the cost of the arithmetic operations also increases.
To reduce the hash computation cost further, another hash function called single hash [31] has been developed. In the single hash, the complex multiplication and the addition operations were replaced by the logical operations. The Murmur hash function results in 64-bit or 128-bit hash values where most of the higher order bits were omitted during the modulo operation. Thus, with the minimum bits resulting from the hash operation and by using minimal operations such as bit shift and Xor operations, the speed of the hash computation has been improved in comparison to the widely used double hash methodology. For example, with a 64-bit hash value 264 values can be generated where all the 64 bits are not needed for addressing the index of the Bloom filter. With nearly half of the bits, the index in the Bloom filter can be addressed, and the remaining bits can be used for hashing another element. The hash function can be expressed as
where i value ranges from 1 to k, h(x) is the murmur hash. When an element e is inserted, the element is hashed by the murmur hash function which results in the hash value h(x). It is then hashed k times to update 1 in the index given by H(x).
The Hash computation using murmur with single hash (MSH) is shown in Algorithm 1. The Hash1 value gives the index of an element in the Bloom filter. By using the Murmur hash function with the single hash function, only one hash value needs to be calculated with the same false positive probability as compared to the Murmur hash with double hash function (MDH) where two hash values are normally calculated. Moreover, in the double hash function the complex operations such as multiplication and addition were performed as compared to the simple bit shift and the Xor operations.
| Algorithm 1: Hash computation of an element |
| 1: procedure 2: 3: 4: for do 5: 6: end for 7: end procedure |
3.2. Improved Cache Efficient Blocked Bloom Filter (CBBF)
The drawback in the standard Bloom filter is poor cache locality i.e., for an element that needs to be inserted and queried, the element is hashed k times and the index given by the hash functions are random locations in the Bloom array. Hence the entire memory needs to be accessed for a single membership check and insertion. If the Bloom filter memory is large, more cache misses will occur. Since the L1 cache is the fastest memory in the system and the size of the L1 cache is very minimal, the Bloom filter is split into blocks equal to the cache line size. With this approach, the spatial locality of reference (the chance of accessing the elements in the near memory location) is increased, thereby increasing the speed of fetching the data from the memory. The Bloom filter memory is divided to L blocks where each block size W is equal to the size of the L1 cache line. In the processors, the cache line size is 64 bytes. The total memory of the Bloom filter m can be expressed as .
When an element ‘x’ is inserted in the Bloom filter, the element is first hashed by a hash function h(x) to choose a single block in the memory of L blocks. Then, the element is hashed by k hash functions and updates the bit in the k positions in block selected by h(x) as shown in Figure 2.
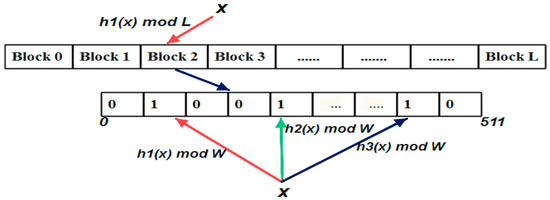
Figure 2.
Cache Blocked Bloom filter data structure. The element x from the set S{x,y,z} is inserted into the Bloom array. The element “x” is hashed by a hash function h1(x) to choose one block from L blocks where each block size is 64 bytes i.e., 512 bits. Then the element is hashed k times and updates the bits in the index positions given by the hash functions h1(x), h2(x), h3(x) to 1.
The insertion procedure is shown in Algorithm 2. The element being inserted is hashed by the Murmur hash with random prime seed and the hash value generated is used to choose the block from L blocks. The same hash value is used to select the index location in the block. Then the hash value is passed to the single hash function to generate k random values. These values are used to select k bits from the block of 512 bits.
| Algorithm 2: Insertion of element into CBBF |
| 1: procedure 2: 3: 4: for do 5: 6: 7: 8: 9: 10: 11: 12: end for 13: end procedure |
For the membership check, the element is hashed by h1(x) to choose the block from L blocks. Then the element is hashed k times h1(x), h2(x), …, hk(x) and checks the bits positions in the index given by the hash functions. If all the bit positions in the block are set as 1, the element is a member of the set. Otherwise, the element is not in the set. The querying process is shown in Algorithm 3.
| Algorithm 3: Membership query of element x in CBBF |
| 1: procedure 2: 3: 4: for do 5: 6: 7: 8: 9: 10: 11: if then 12: 13: else 14: 15: end if 16: for 17: end procedure |
In standard Bloom filter, for positive query k cache misses will occur and for negative queries one to two cache misses occurs on average. In blocked Bloom filter, for each insertion and membership check, only one block of memory with the size equal to the cache line size is accessed. For positive queries, k locations need to be checked and for negative queries a minimum of 1 and maximum k locations are accessed where all the bit locations lie in the single cache line. This reduces the number of cache misses to one and thereby increases the insertion as well as the querying speed of the input elements. Furthermore, instead of k hash functions, the hash value is fixed as five for the lesser false positive probabilities. As a result, the hash computation time is also reduced. Since all the k bits are inserted in one block, there is a little increase in the false positive probability compared to the standard Bloom filter.
False Positive Analysis
The false positive probability is the probability that an element which is not a member of the set is mistakenly identified as the member of the set. To check the presence of element in the set, it is hashed k times and checked in the block. Let x be the number of elements mapped to the same block, the false positive probability is given as
X is a random variable where the members map to the same word, x is a constant ranging from 0 to n elements, k is the number of hash functions and W is the size of the block which is the size of the cache line (512 bits).
The Bloom filter is partitioned into L blocks of 512 bits each. The n elements can be mapped to any one of the L blocks. The probability of the block being chosen from the L blocks follows binomial distribution and it can be represented as
The false positive probability can be written as
The false positive probability of CBBF has been approximated [32] and it is given as
The false positive probability of the cache blocked Bloom filter is increased compared to the standard Bloom filter. There is a trade-off between the insertion andquery speed with the false positive probability, but the difference between both Bloom filter false positive probabilities is very much less.
3.3. One Hashing Blocked Bloom Filter (OHBB)
In the Blocked Bloom filter, the insertion and the querying speed have been accelerated by splitting the memory into blocks where the block size is equal to the cache line size. The number of memory access has been reduced from the k memory access in the standard Bloom filter to one memory access. However, there is an increase in the false positive probability compared to the standard Bloom filter. As the k bits are inserted into the single block of memory there is a probability of collision inside the block.
To overcome the issue of collision, OHBB filter has been proposed in this work. In this approach, the memory size m is split into L blocks where each block is equal to the size of the cache line of the system which is 64 bytes. Thus, the memory access is reduced to one. On the other hand, in cache blocked Bloom filter, k hash functions are needed for the insertion and querying of elements. This has been further reduced by partitioning the block into k blocks where the size of each partition is the unique prime number. The hash function is called only once to choose the block in the memory. The same hash function is used to choose the k where each hash function is modulo divided by the unique prime number as shown in Figure 3. When the hash function is modulo divided by the prime number, the randomness is increased, and the collision rate is decreased.
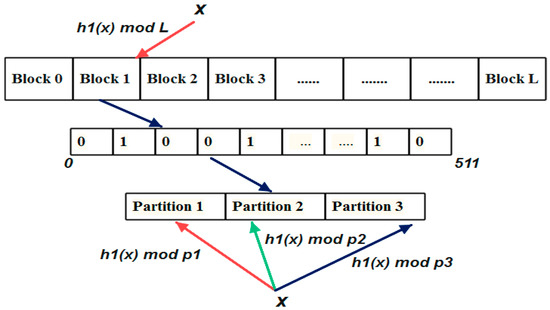
Figure 3.
One hash Blocked Bloom filter data structure. The element x from the set S{x,y,z} is inserted into the Bloom array. The Bloom array is split into blocks of 512 bits. The block is further partitioned into 3 blocks for k = 3. Each partition size is the prime number. The element “x” is hashed by a hash function h1(x) to choose one block from L blocks. Then the hash function is modulo divided by the partition length which gives the random location. The bits at the location were set to 1.
The procedure for choosing the partition length is shown in Algorithm 4. The entire prime numbers were stored in the prime table. Select the nearest prime number equal to m/k as the middle value; Choose the prime numbers before and after the middle value and store k prime values; The sum of all the k prime numbers should be less than or equal to 512; If the value is more or far less than the size of 512, replace the smallest prime number with the next highest prime number; By repeating the procedure the partition length can be chosen nearest to the block size. The sum of the partition length should be near to or equal to the size of the cache line size. If the sum is greater than 512, then two cache lines needs to be accessed for single insertion and query which results in two memory access.
| Algorithm 4: Selecting the Partition length (prime numbers) in OHBB filter |
| Input: Prime Table (Prime), mb = 512, k Output: Primes 1: from the prime table and the index as 2: Total ← 0 3: for i = 1 to k do 4: P(i) - round(k/2) + i) 5: end for 6: Total = ∑(P) 7: = Prime(P(k)) //Index of kth prime number in prime table 8: = Prime(P(1)) //Index of 1st prime number in prime table 9: while true do 10: ) then 11: Break 12: else 13: with the next prime number from the prime table 14: Total= ∑(P) 15: end if 16: end while 17: for i = 1 to k do 18: Primes(i) = P(i) 19:end for |
The prime numbers should be the consecutive values or nearby values and also the difference between the partitions should not be too large. If the difference is more, then the distribution of the hash bits in the Bloom array will not be uniform which leads to an increase in the false positive probability. For example, the Bloom filter memory m = 10,000 is divided into 19 blocks. The size of each block is 512 bits. The 512 bits are further partitioned into three partitions for k = 3 where P1 = 151, P2 = 179, and P3 = 181. The sum of all the partitions is less than the size of the block.
The insertion procedure is shown in Algorithm 5. The prime table contains the partition length is loaded initially. The element is being stored into the Bloom filter is hashed by Murmur hash which gives a 64-bit hash value. The modulo operation is performed on the hash value to find the block index and then the same hash value is modulo divided by the k prime numbers which provide the bit index in each partition. The bits in the index provided by the hash function were set to 1.
| Algorithm 5: Insertion of element x into OHBB filter |
| 1: 2: 3: 4: 5: do 6: 7: 8: 9: 10: 11: 12: 13: procedure |
The same hash value is used for finding the block index, partition, and also the bit index because the hash value divided by different prime numbers gives different index values. The membership querying procedure of the proposed approach is explained in Algorithm 6. The element being queried is first hashed by the murmur hash function. The block index, the bit index from each partition is calculated by the same hash function. The bits in the corresponding index positions were checked for 1. If all the bits were set as 1, then the element is the member of the set, otherwise the element is not a member of the set.
| Algorithm 6: Membership query of element in OHBB filter |
| 1: 2: 3: 4: 5: 6: 7: 8: 9: 10: 11: 12: return True 13: else 14: return False 15: end if 16: 17: |
The partition length is decided based on the number of hash functions. Each partition is a unique prime number where the sum of all the partition lengths is equal to the block size. By blocking and partitioning, the number of memory access has been reduced to 1 which directly impacts the insertion and querying speed. By partitioning the block further into the prime number of blocks, the collision rate is reduced compared to the cache blocked Bloom filter. The false positive probability is in between the standard Bloom filter and the cache blocked Bloom filter.
False Positive Analysis
The element which is not in the set being queried in the Bloom filter is mistakenly identified as the element is termed as false positive. The element is hashed and checked in all the partitions for the existence of the element. The probability that the element is present in all the partitions [33] is given below:
The Bloom filter memory is split into L blocks of 512 bits each. The probability of selection of one block out of L blocks follows binomial distribution. The probability that each element can be mapped to any one block with equal probability is given by
where X is a random variable and x is a constant ranging between 0 and n, n is the number of inputs elements.
The probability of the bit in the partition pi being set as 1 is given by 1/pi. The probability that the bit in all the partitions set as 1 by the element x is given as
where k is the number of hash functions and pi is the partitions in each block.
It can be approximated as
The false positive probability of the OHBB filter is the product of the probability of choosing a single block from L blocks ( and the probability of the bits set in the partitions pi from the block selected .
The theoretical false positive probability of OHBB filter and the standard Bloom filter for the different load factor (n/m) and different hash count (k) is given in Table 2 and Table 3.

Table 2.
Theoretical false positive probability of Standard Bloom filter and One hashing blocked Bloom filter where n = 10,000 and k = 3.

Table 3.
Theoretical false positive probability of Standard Bloom filter and One hashing blocked Bloom filter where n = 10,000 and k = 5.
From the above table it is concluded that the false positive probability is increasing with the increase in the load factor. The false positive probability of OHBB is greater than the standard Bloom filter but the difference is very minimal, and it is within the acceptable limits of the applications.
4. Results and Discussion
4.1. Experimental Setup
This work is implemented in Intel(R) Xeon(R) processor (E5-2620v4, 2.10 GHz, 8 cores) with 32 GB DDR Memory and 1TB SDD running on Ubuntu 16.04 LTS in HPZ640 machine. The proposed one hashing cache blocked Bloom filter is implemented in C++ in the above platform.
A set of random strings with different numbers were generated for inserting into the Bloom filter and another set of disjoint random strings were generated to check the false positive probability and the membership query speed. These random strings are used as Dataset 1, while Dataset 2 is comprised of a set of K-mers used in genome assembly, gene sequencing, and genome studies. Random K-mers of different sizes were generated and inserted into the Bloom filter and, another set of distinct random K-mers were used for the querying purpose. The two datasets were used to evaluate the performance of the proposed methods as shown in Table 4. All the experiment results were repeated five times and the average result is taken for evaluation.

Table 4.
Datasets used for experimental evaluation.
4.2. Hash Computation
The hashing process is tested with double hash and single hash along with the Murmur hash to find the best hash function in terms of the hashing speed since both the hash functions have similar false positive probability. The hash functions were tested using the two mentioned data sets and their results were shown in Figure 4. There is an increase in the speed of the MSH compared to the MDH. The Bloom filter is used to evaluate the insertion and the membership check speed. Random K-mers were inserted into the Bloom filter and another set of distinct K-mers were queried. The performance of MSH outperforms the other for both random K-mers as well as with the random strings.
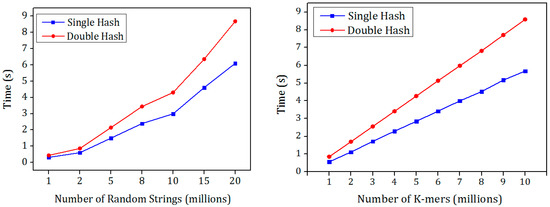
Figure 4.
Comparison of single hash and double hash with random strings and random K-mers as input for hash function k = 7.
The increase in the performance is due to the murmur hash function is called only once while in the double hash the murmur hash is called two times with different seed values. In some applications, the second hash value is generated from the first hash function with few shift operations. Moreover, the arithmetic operations in the double hash involve complex multiplication and addition operations which are called k times. In the single hash the complex operations were replaced by simple shift and Xor operations.
To check the performance of both hash functions for different string length, one million random strings and random K-mers were generated with different lengths. The input files were hashed using single hash and double hash and the time taken for hashing the strings were shown in Figure 5. It is clear from the results that the single hash outperforms the double hash irrespective of the string length. This hash function can be used in applications using input strings with variable lengths.
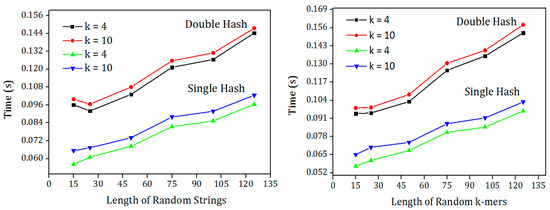
Figure 5.
Comparison of single hash and double hash with one million random strings and random K-mers of different length for hash function k = 4 and k = 7.
4.3. Insertion Speed Check
The CBBF is compared with the SBF in terms of the insertion speed with the two datasets. The standard Bloom filter with the double hash function and single hash function is also implemented to study the behavior of hash functions in the Bloom filter with different datasets. All the versions were implemented for the false positive probability (FPP) of 0.1, 0.01, 0.001, and 0.0001. Different applications use different FPP values as their benchmarks based on their requirement. For some applications, the querying speed is the major area of interest and the FPP has been given least significance, but for some applications the FPP values should be very minimal.
The Bloom filter is designed for different input sizes with different FPR values. The insertion speed in the cache blocked Bloom filter is improved because of the reduction in the memory access from k to 1.
The insertion speed is increased for the Bloom with Single hash because the double hash uses addition and the multiplication in the hash function, which is called k times, whereas the single hash uses the simple bitwise shift and the Xor operations. Because of the improvement in the hash operations as well as the reduction in the cache misses due to blocking of the memory aligned to the size of the cache line, there is a reduction in the insertion time compared to the existing SBF with Double hash approach.
The results shown in Figure 6 and Figure 7 prove that the hashing speed of the single hash is increased compared to the double hash. For the dataset 2, only the SBF with MSH is compared with the CBBF. The random K-mers of length r = 27 were generated and inserted into the Bloom filter with different false positive probabilities. In the genome assembly, the K-mers were generated and stored in the Bloom filter, then a set of K-mers were queried for the existence. Figure 7 shows that the performance of CBBF outperformed the SBF.
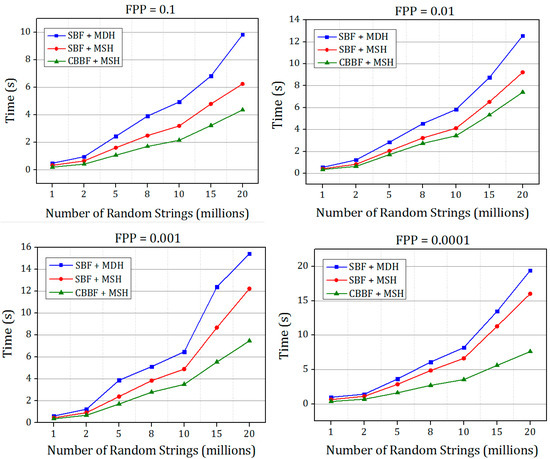
Figure 6.
The comparison of insertion time of SBF + MDH and SBF + MSH and CBBF + MSH for different FPP for Dataset 1.
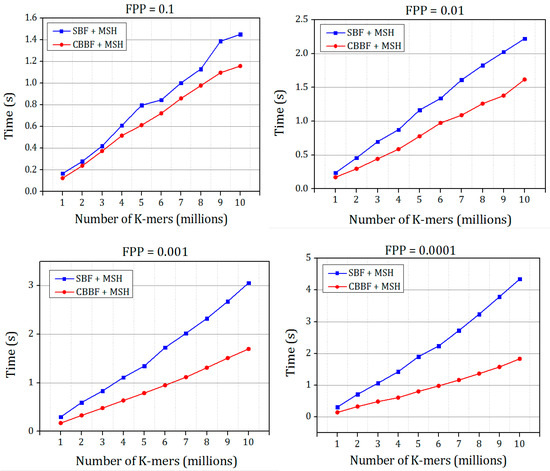
Figure 7.
The comparison of insertion time of Bloom filter with Murmur hash + Single hash and Cache Blocked Bloom filter with Murmur hash + Single Hash for different false positive probabilities for K-mers (Dataset 2).
There is a good improvement in the speed when the value of the FPP is decreased. For FPP = 0.0001 the difference between the SBF and CBBF is substantial due to the number of hash functions used. For FPP = 0.0001, fourteen hash values were used in the SBF while the cache blocked filter uses only five hash functions. When the hash functions are more, the entire memory should be accessed to update k bits in the memory while in the blocked Bloom filter only one memory block is accessed. For lesser false positive probability, the performance of the CBBF is better in terms of the insertion speed. Both of the Bloom filters used the same amount of memory.
The comparison between standard Bloom filter (SBF), cache blocked Bloom filter (CBBF) and one hashing Bloom filter (OHBB) is simulated for Dataset 1 and Dataset 2 to measure the performance of insertion and querying speed with different false positive probabilities.
The time taken for inserting the set of random strings into the Bloom filter for FPP = 0.1, 0.01, 0.001, 0.0001 is shown in Figure 8. In CBBF, the membership and the querying speed increases albeit there is an increase in the FPP compared to SBF due to the collisions in the block because all the k bits were mapped to the same block randomly. In OHBB, the collisions in the block were effectively handled by partitioning the block further into k blocks. Since the size of each partition is prime number, when the hash function is modulo divided by the prime number the randomness increased and the collision decreased resulting in the lesser FPP. However, there is a slight increase in the FPP compared to the SBF and the difference is significantly less compared to the CBBF.
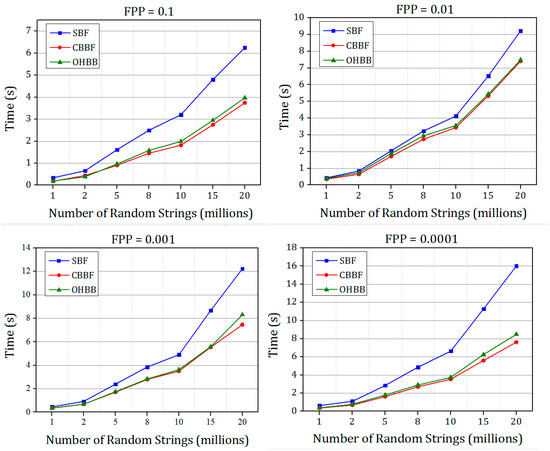
Figure 8.
The comparison of insertion speed of SBF, CBBF and OHBF with different false positive probabilities for Dataset 1.
The time taken for inserting the K-mer strings into the Bloom filter with varying FPP is shown in Figure 9. The insertion time of OHBB is better than the SBF. There is an increase in the insertion time compared to the CBBF because of the modulo operation in the partition sizes. When the hash value increases, the partition size also grows which in turn increases the number of modulo operations performed on the hash value, However, the difference is very much less because the modulo operation replaces the hash function carried out for all the k values in the SBF and CBBF.
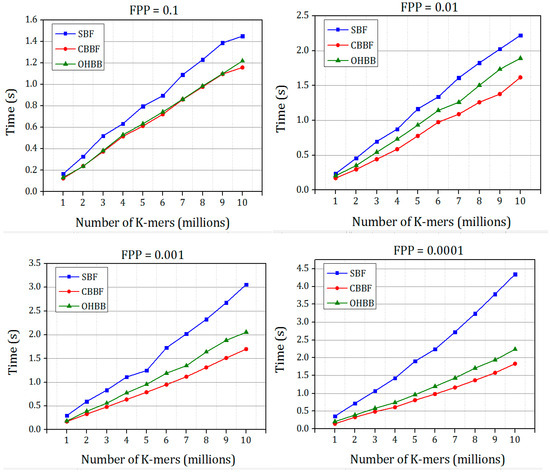
Figure 9.
The comparison of insertion speed of SBF, CBBF and OHBB with different false positive probabilities for Dataset 2.
4.4. Membership Check Speed
The querying speed of the SBF with different hash functions and CBBF with MSH are shown in Figure 10 for the different false positive probabilities. Random strings of different sizes were generated and inserted in the Bloom filter. Another set of unique random strings were generated and queried in the Bloom filter. The negative membership query is needed for evaluating the exact false positive probability of the system. In all the experiments, the theoretical false positive probability is fixed; by varying the number of inputs n, the size of the Bloom filter m has been calculated and then the memory is allocated. The query performance also shows promising results for all the range of false positive probability.
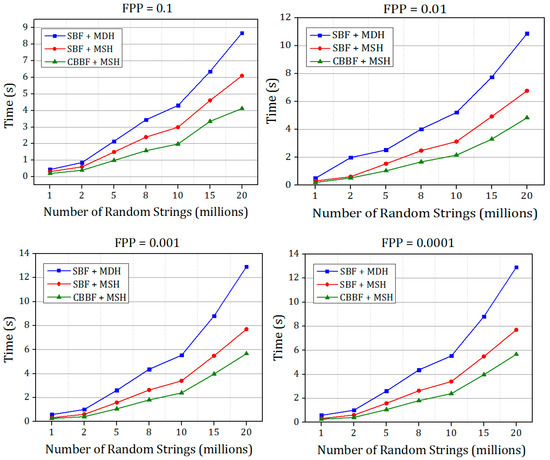
Figure 10.
The comparison of membership check speed of Bloom filter with MDH and Bloom filter with MSH and Cache Blocked Bloom filter with MSH for different false positive probabilities FPP = 0.1, 0.01, 0.001, 0.0001 for Dataset 1.
Figure 10 and Figure 11 show the membership query performance of the SBF and the CBBF for the different FPP. In all the four versions, CBBF shows an increase in the querying speed. When the same algorithm is applied for the real time datasets, for example human genome assembly, the performance improvement in the insertion and querying process is more significant because of the linear relationship between the input data and Bloom filter size.
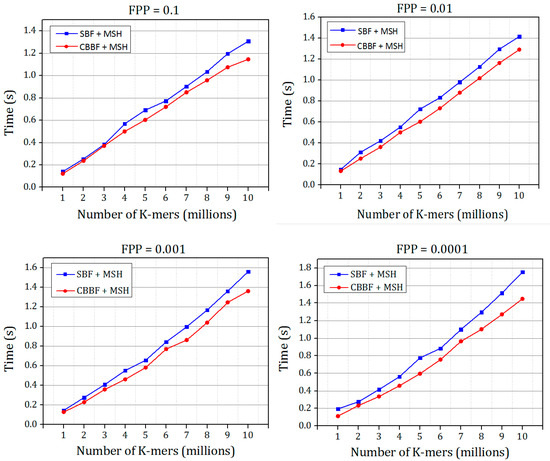
Figure 11.
The comparison of membership check speed of Bloom filter with MSH and Cache Blocked Bloom filter with MSH for different false positive probabilities for Dataset 2.
Figure 12 and Figure 13 show the membership check performance of the SBF, CBBF and OHBB for dataset 1 and dataset 2.
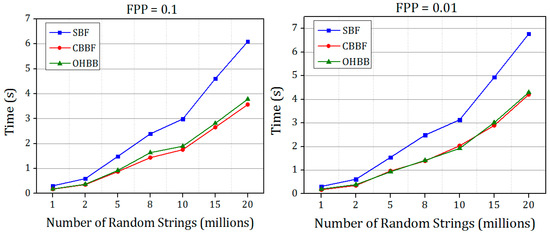
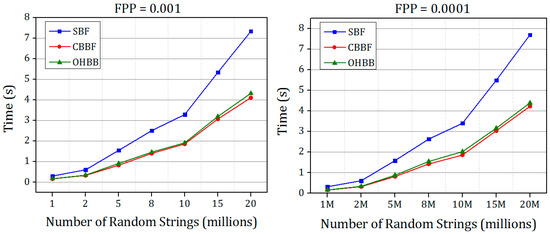
Figure 12.
The comparisons of membership check speed in SBF, CBBF and OHBB with different false positive probabilities for Dataset 1.
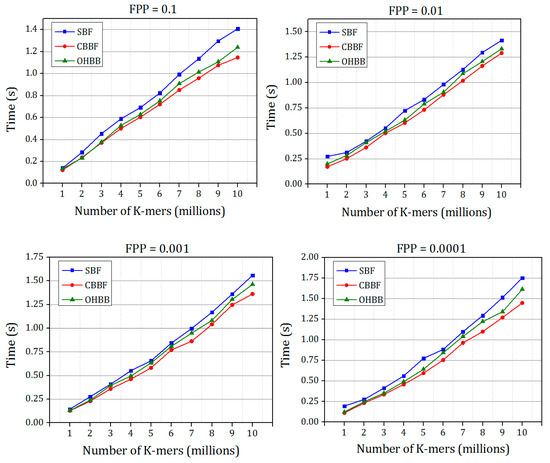
Figure 13.
The comparisons of membership check speed of SBF, CBBF and OHBB with different false positive probabilities for random strings Dataset 2.
It is shown in Figure 12 and Figure 13 that the query performance of the random strings is increased for all the false positive probabilities. There is a minimal difference in the speed compared to the CBBF. For Dataset 2, the performance lies in between the SBF and CBBF. The membership check speed for the random strings is almost same as the CBBF. The clock cycles taken for modulo operation in less compared to the clock cycles required for the hash function computation. The datasets used in experiments were simulated datasets. The performance comparison of the proposed Bloom filter and SBF on real time gene data is shown in Table A1. The results also supports that there is an improvement in the insertion and query time of the proposed Bloom filter compared to Standard Bloom filter (SBF).
We have not compared the memory utilization of the different Bloom filter approaches because: In improved CBBF, the entire Bloom filter memory is logically split into blocks where the block size is equal to the size of the cache line. By rounding off the Bloom filter memory bits to 512, the entire memory can be split into blocks of size 512. Therefore, the increase in the memory in CBBF is negligible compared to SBF. In OHBB, the blocks were further partitioned into prime number of blocks where the partition length is less than the block size. Thus, there is also a negligible increase in the memory of OHBB filter compared to SBF. Hence in all the above approaches, the memory utilization remains same.
4.5. False Positive Probability (FPP)
The simulated false positive probabilities of all the four versions of Bloom filter are shown in Figure 14 and Figure 15. The Bloom filter and its variants were designed based on the input elements by fixed FPP. The corresponding FPP values were recorded from the experiments. Each experiment is repeated five times and the average value is chosen to plot the figures.
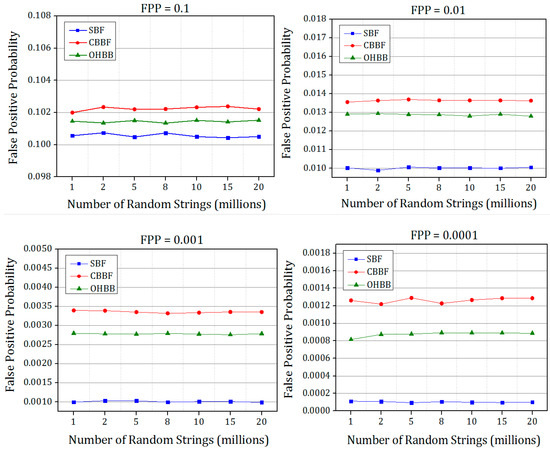
Figure 14.
The comparison of simulated false positive probability of SBF, CBBF and OHBB with different theoretical false positive probabilities for Dataset 1.
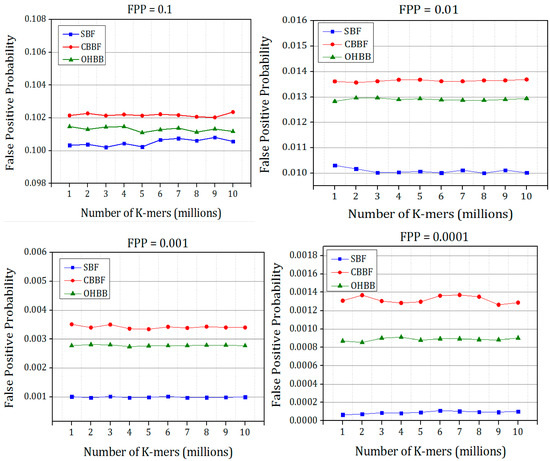
Figure 15.
The comparison of simulated false positive probability of SBF, CBBF and OHBB with different theoretical false positive probabilities for Dataset 2.
The comparison of the theoretical false positive probability and the simulated false positive probability is shown in Figure 16. From the figure it is clear that the simulation results match the theoretical results exactly. It is also clear from the figure that when the load of the Bloom filter increased, the false positive probability also increased.
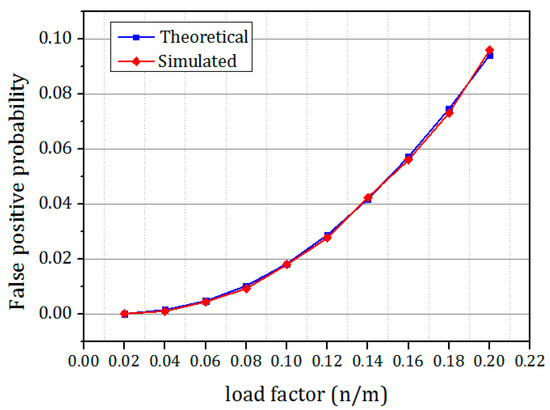
Figure 16.
The comparison of simulated false positive probability of OHBB with the theoretical false positive probability of OHBB filters for different load factor.
It is evident from the results that there is a substantial improvement in the FPP compared to the CBBF. The improvement is achieved due to the randomness produced by the prime size partitions. The difference in FPP between the SBF and OHBB filter is very much less. The hash function k is chosen as 3 for the FPP 0.1 and for the lesser FPP the hash functions are fixed as 5. The number of partitions were fixed to five in each memory block for the lower FPP such as FPP = 0.01, 0.001, 0.0001 which exhibited the better result. The insertion and membership query results were taken for the four FPP values to check the performance of the different load values. For image processing applications [34], the speed and the memory of the Bloom filter is the design constraint and hence FPP is fixed as 0.1 whereas for gene applications, the FPP is fixed as 0.001, and for the networking applications the FPP is fixed as very less than 0.0001. To meet the requirements of all applications, the efficient Bloom filter variant has been designed. The proposed work paves a way to the researchers to choose appropriate hash function and the bloom filter variant for their application requirements.
5. Conclusions
In this paper, a new variant of the Bloom filter OHBB was proposed, implemented and evaluated. The number of memory access has been reduced by the CBBF where each block size was same as the cache size. The block-based Bloom filter used fewer hash functions to achieve improvement in the speed at the cost of increase in the false positive probability. The false positive probability is reduced by the proposed OHBB filter where the blocks were partitioned, and each partition size is chosen as the prime number. The requirement of number of hash function is reduced to one. The proposed OHBB filter was validated with the random strings dataset, simulated and real K-mers dataset. The OHBB filter achieved good performance in terms of the insertion speed and membership query speed. The proposed OHBB filter with FPP 0.1 would be suitable for image processing applications and the OHBB filter with FPP 0.0001 would be suitable for the network and the genome assembly applications. Though there was a trade-off between the false positive probability and speed, the performance of the OHBB filter had been relatively between the performances of CBBF and the SBF. This can be further improved by parallelizing the hash functions and the insertion and membership query process in the multicore and distributed environment.
Author Contributions
Conceptualization, A.M. and E.P.; methodology, E.P.; software, E.P.; validation, E.P. and A.M.; data curation, E.P.; writing—original draft preparation, E.P.; writing—review and editing, A.M. and E.P.; supervision, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
In DNA Sequencing, the K-mers were generated from the DNA reads. The generated K-mers were stored in the Bloom filter for querying and for the error correction. The gene sequences of Faragaria vesca (Wild strawberry) species were extracted from the database (ftp://ftp.ddbj.nig.ac.jp/ddbj_database accessed on 19 August 2022) and implemented in the proposed method. The input files are Fastq formatted files. There are 176,443 reads in the input file with an average read length of 341 DNA bases. The input DNA reads are split into K-mers of different length (50, 100, 150) and loaded into Bloom filter. The time taken for inserting the K-mers into the Bloom filter with different hash values was recorded. The query file is taken from the same species which contains 1,097,310 reads of average length 339. The reads from the query file is split into K-mers and queried in the Bloom filter. The time taken for insertion and querying the K-mers is shown in Table A1.

Table A1.
The insertion and querying time comparison of real time gene data in SBF and the proposed filter.
Table A1.
The insertion and querying time comparison of real time gene data in SBF and the proposed filter.
| K-mer Length | Total K-mers | Hash | Insertion Time | Total K-mers | Hash | Querying Time | ||
|---|---|---|---|---|---|---|---|---|
| SBF | OHBB | SBF | OHBB | |||||
| 50 | 29,200,531 | 1 | 0.778 | 0.7053 | 180,681,518 | 1 | 5.0857 | 4.8683 |
| 3 | 0.9916 | 0.7996 | 3 | 5.7096 | 5.289 | |||
| 5 | 1.2775 | 0.879 | 5 | 6.2855 | 5.5624 | |||
| 100 | 24,200,581 | 1 | 0.7741 | 0.6879 | 149,508,218 | 1 | 4.7607 | 4.4094 |
| 3 | 0.9728 | 0.7949 | 3 | 5.2385 | 4.6978 | |||
| 5 | 1.2089 | 0.8547 | 5 | 5.6906 | 4.9555 | |||
| 150 | 19,200,631 | 1 | 0.7468 | 0.6538 | 118,334,918 | 1 | 4.4184 | 3.9571 |
| 3 | 0.9028 | 0.7347 | 3 | 4.7343 | 4.181 | |||
| 5 | 1.0865 | 0.7858 | 5 | 5.0549 | 4.3824 | |||
References
- Byun, H.; Li, Q.; Lim, H. Vectored-Bloom filter for IP address lookup: Algorithm and hardware architectures. Appl. Sci. 2019, 9, 21. [Google Scholar] [CrossRef]
- Nour, B.; Khelifi, H.; Hussain, R.; Mastorakis, S.; Moungla, H. Access Control Mechanisms in Named Data Networks. ACM Comput. Surv. 2021, 54, 3. [Google Scholar] [CrossRef]
- Jang, S.; Byun, H.; Lim, H. Dynamically Allocated Bloom Filter-Based PIT Architectures. IEEE Access 2022, 10, 28165–28179. [Google Scholar] [CrossRef]
- Nayak, S.; Patgiri, R.; Borah, A. A survey on the roles of Bloom Filter in implementation of the Named Data Networking. Comput. Netw. 2021, 196, 108232. [Google Scholar] [CrossRef]
- Kim, J.; Ko, M.C.; Kim, J.; Shin, M.S. Route prefix caching using bloom filters in named data networking. Appl. Sci. 2020, 10, 7. [Google Scholar] [CrossRef]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 5, 768–777. [Google Scholar] [CrossRef]
- Chen, Y.L.; Chang, B.Y.; Yang, C.H.; Chiueh, T.D. A High-Throughput FPGA Accelerator for Short-Read Mapping of the Whole Human Genome. IEEE Trans. Parallel Distrib. Syst. 2021, 32, 1465–1478. [Google Scholar] [CrossRef]
- Solomon, B.; Kingsford, C. Fast search of thousands of short-read sequencing experiments. Nat. Biotechnol. 2016, 34, 300–302. [Google Scholar] [CrossRef]
- Jiang, M.; Zhao, C.; Mo, Z.; Wen, J. An improved algorithm based on Bloom filter and its application in bar code recognition and processing. Eurasip J. Image Video Process. 2018, 1, 1–12. [Google Scholar] [CrossRef]
- Shomaji, S.; Ghosh, P.; Ganji, F.; Woodard, D.; Forte, D. An Analysis of Enrollment and Query Attacks on Hierarchical Bloom Filter-Based Biometric Systems. IEEE Trans. Inf. Forensics Secur. 2021, 16, 5294–5309. [Google Scholar] [CrossRef]
- Kong, Q.; Lu, R.; Yin, F.; Cui, S. Blockchain-Based Privacy-Preserving Driver Monitoring for MaaS in the Vehicular IoT. IEEE Trans. Veh. Technol. 2021, 70, 3788–3799. [Google Scholar] [CrossRef]
- Stephens, Z.D.; Lee, S.Y.; Faghri, F.; Campbell, R.H.; Zhai, C.; Efron, M.J.; Iyer, R.; Schatz, M.C.; Sinha, S.; Robinson, G.E. Big data: Astronomical or genomical? PLoS Biol. 2015, 13, e1002195. [Google Scholar] [CrossRef]
- Bankevich, A.; Bzikadze, A.V.; Kolmogorov, M.; Antipov, D.; Pevzner, P.A. Multiplex de Bruijn graphs enable genome assembly from long, high-fidelity reads. Nat. Biotechnol. 2022, 40, 1075–1081. [Google Scholar] [CrossRef]
- Compeau, P.E.C.; Pevzner, P.A.; Tesler, G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011, 29, 987–991. [Google Scholar] [CrossRef]
- Surendar, A.; Arun, M. FPGA based multi-level architecture for next generation DNA sequencing. Biomed. Res. India 2016, 27, S75–S79. [Google Scholar]
- Cristadoro, G.; Esposti, M.D.; Altmann, E.G. The common origin of symmetry and structure in genetic sequences. Sci. Rep. 2018, 8, 15817. [Google Scholar] [CrossRef]
- Shporer, S.; Chor, B.; Rosset, S.; Horn, D. Inversion symmetry of DNA K-mer counts: Validity and deviations. BMC Genomics 2016, 17, 696. [Google Scholar] [CrossRef]
- Shirazi, N.; Benyamin, D.; Luk, W.; Cheung, P.Y.K.; Guo, S. Quantitative analysis of FPGA-based database searching. J. VLSI Signal Process. Syst. Signal Image. Video Technol. 2001, 28, 85–96. [Google Scholar] [CrossRef]
- Dharmapurikar, S.; Krishnamurthy, P.; Taylor, D.E. Longest Prefix Matching Using Bloom Filters. IEEE/ACM Trans. Netw. 2006, 14, 397–409. [Google Scholar] [CrossRef]
- Jain, N.; Dahlin, M.; Tewari, R. Using Bloom filters to refine web search results. In Proceedings of the WebDB04: 7th International Workshop on the Web and Databases, Paris, France, 17–18 June 2004; pp. 25–30. [Google Scholar]
- Park, J.H.; Park, J.H. Blockchain security in cloud computing: Use cases, challenges, and solutions. Symmetry 2017, 9, 1–13. [Google Scholar] [CrossRef]
- Bloom, B.H. Space/Time Trade-offs in Hash Coding with Allowable Errors. Commun. ACM 1970, 13, 7. [Google Scholar] [CrossRef]
- Broder, A.; Mitzenmacher, M. Network Applications of Bloom Filters: A Survey. Internet Math. 2004, 1, 485–509. [Google Scholar] [CrossRef]
- Singh, A.; Garg, S.; Kaur, R.; Batra, S.; Kumar, N.; Zomaya, A.Y. Probabilistic data structures for big data analytics: A comprehensive review. Knowl. Based Syst. 2020, 188, 104987. [Google Scholar] [CrossRef]
- Putze, F.; Sanders, P.; Singler, J. Cache-, hash-, and space-efficient bloom filters. ACM J. Exp. Algorithmics 2009, 14, 4. [Google Scholar] [CrossRef]
- Qiao, Y.; Li, T.; Chen, S. Fast bloom filters and their generalization. IEEE Trans. Parallel Distrib. Syst. 2014, 25, 93–103. [Google Scholar] [CrossRef]
- Lu, J.; Wan, Y.; Li, Y.; Zhang, C.; Dai, H.; Wang, Y.; Zhang, G.; Liu, B. Ultra-Fast Bloom Filters using SIMD Techniques. IEEE Trans. Parallel Distrib. Syst. 2019, 30, 953–964. [Google Scholar] [CrossRef]
- Estébanez, C.; Saez, Y.; Recio, G.; Isasi, P. Performance of the most common non-cryptographic hash functions. Softw. Pract. Exp. 2014, 44, 681–698. [Google Scholar] [CrossRef]
- Aappleby, “Murmur Hash3.” [Online]. Available online: https://github.com/aappleby/smhasher/blob/master/src/MurmurHash3.cpp (accessed on 19 July 2022).
- Kirsch, A.; Mitzenmacher, M. Less hashing, same performance: Building a better bloom filter. Random Struct. Algorithms 2008, 33, 187–218. [Google Scholar] [CrossRef]
- Gou, X.; Zhao, C.; Yang, T.; Zou, L.; Zhou, Y.; Yan, Y.; Li, X.; Cui, B. Single Hash: Use One Hash Function to Build Faster Hash Based Data Structures. In Proceedings of the International Conference on Big Data and Smart Computing (BigComp), Shanghai, China, 15–17 January 2018; pp. 278–285. [Google Scholar]
- Reviriego, P.; Christensen, K.; Maestro, J.A. A Comment on ‘Fast Bloom Filters and Their Generalization. IEEE Trans. Parallel Distrib. Syst. 2016, 27, 303–304. [Google Scholar] [CrossRef]
- Lu, J.; Yang, T.; Wang, Y.; Dai, H.; Jin, L.; Song, H.; Liu, B. One-Hashing Bloom Filter. In Proceedings of the 23rd International Symposium on Quality of Service (IWQoS), Portland, OR, USA, 15–16 June 2015; pp. 289–298. [Google Scholar]
- Elgohary, H.M.; Darwish, S.M. Improving Uncertainty in Chain of Custody for Image Forensics Investigation Applications. IEEE Access 2022, 10, 14669–14679. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).