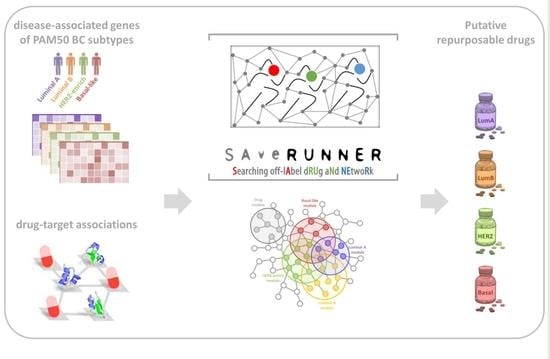
A Transcriptome- and Interactome-Based Analysis Identifies Repurposable Drugs for Human Breast Cancer Subtypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Network-Based Tools
2.1.1. SWIM
2.1.2. SAveRUNNER
2.2. Data Retrieval
2.3. In Silico Validatio: GSEA Analysis
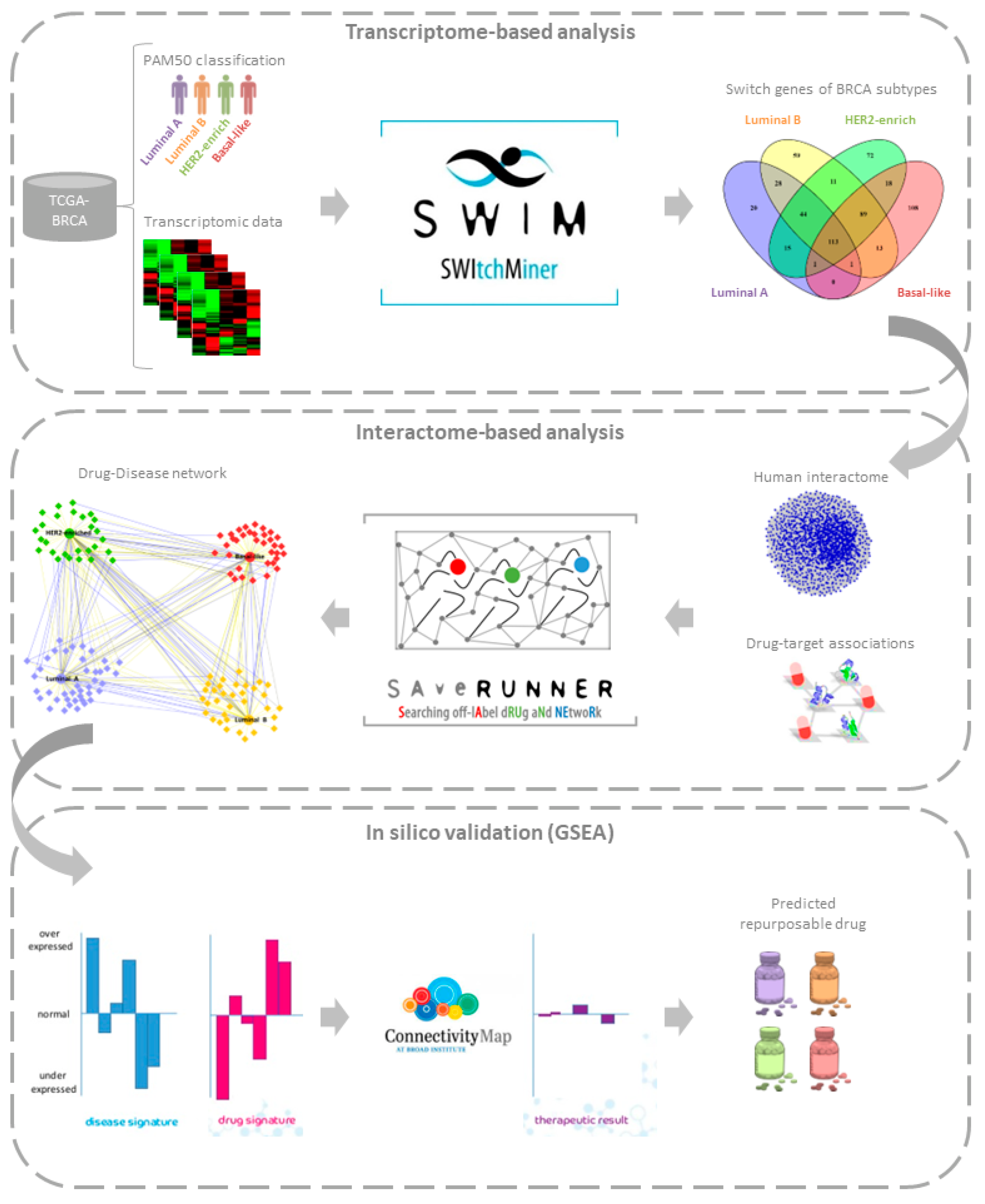
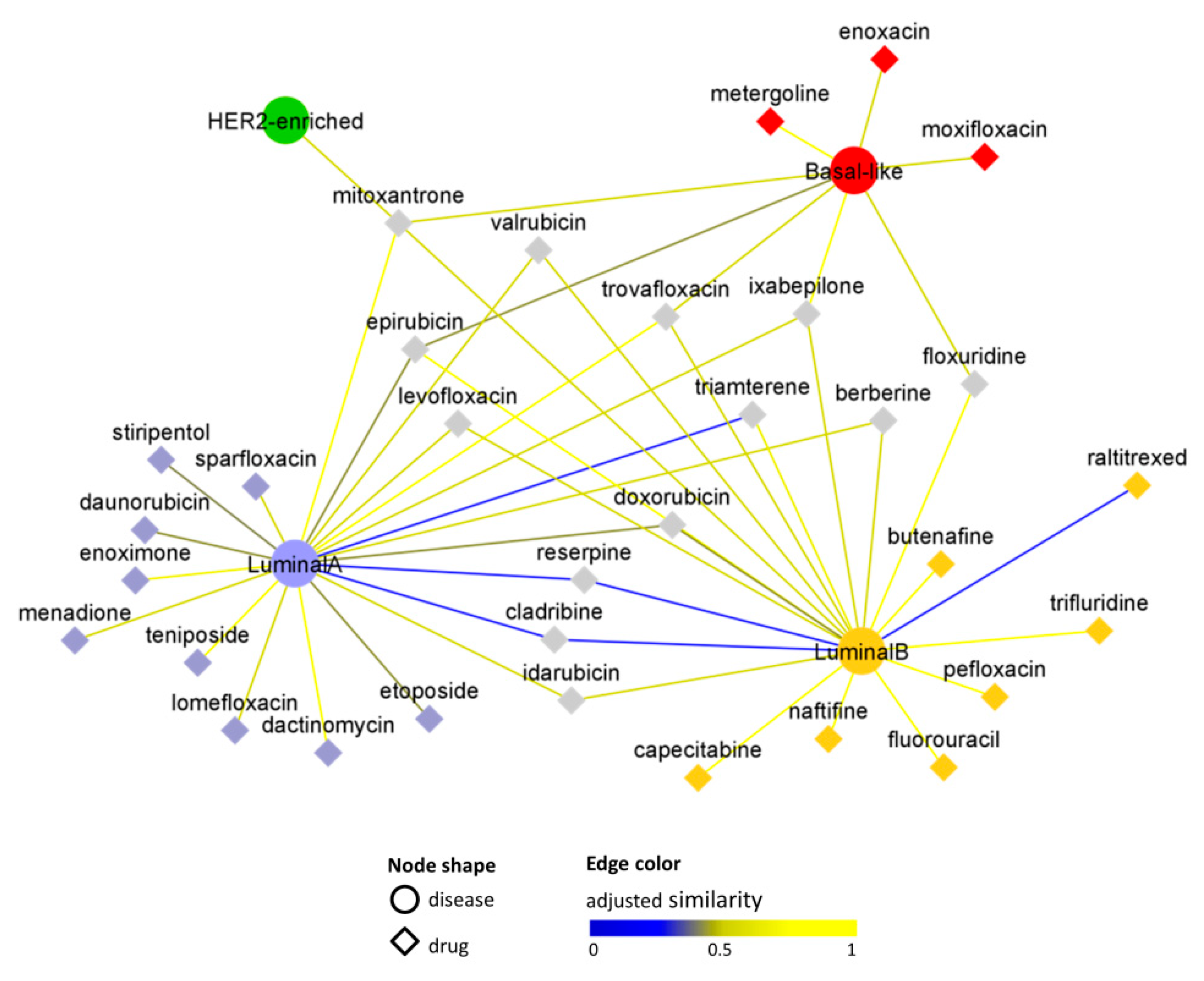
3. Results
3.1. Study Design
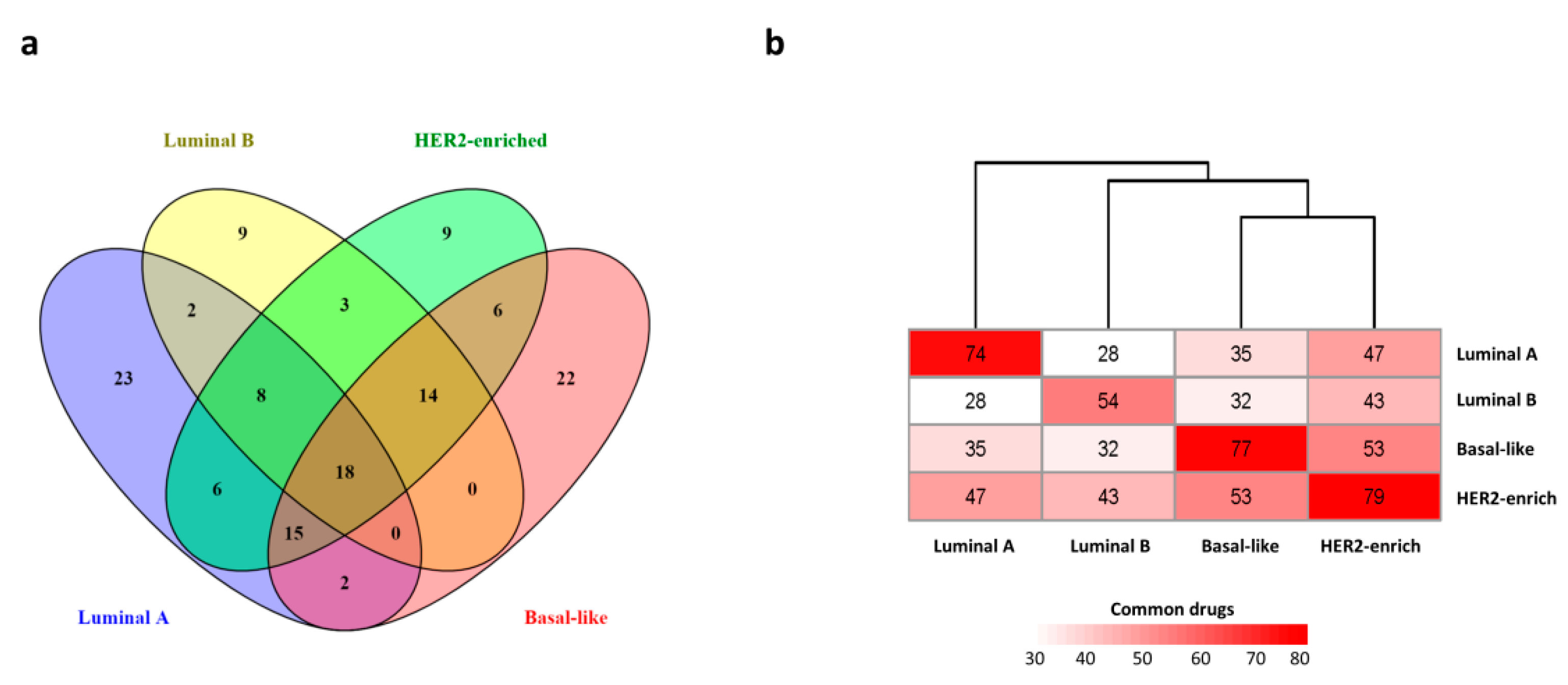
3.2. Transcriptome-Based Analysis
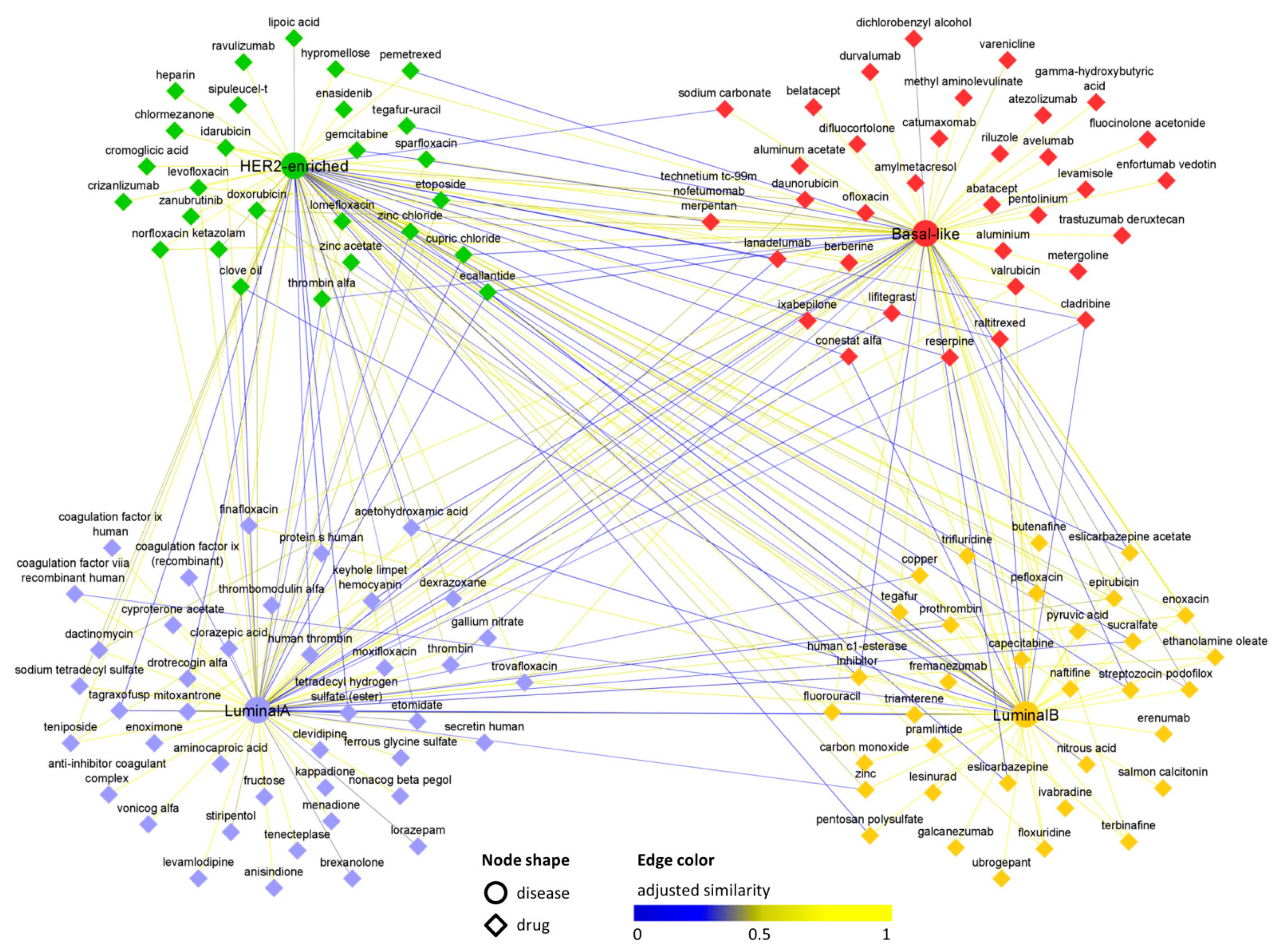
3.3. Interactome-Based Analysis
3.4. In Silico Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical Management of Breast Cancer Heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical Implications of the Intrinsic Molecular Subtypes of Breast Cancer. Breast Edinb. Scotl. 2015, 24 (Suppl. S2), S26–S35. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Caldera, M.; Buphamalai, P.; Müller, F.; Menche, J. Interactome-Based Approaches to Human Disease. Curr. Opin. Syst. Biol. 2017, 3, 88–94. [Google Scholar] [CrossRef]
- Firoozbakht, F.; Rezaeian, I.; Rueda, L.; Ngom, A. Computationally Repurposing Drugs for Breast Cancer Subtypes Using a Network-Based Approach. BMC Bioinform. 2022, 23, 143. [Google Scholar] [CrossRef]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.-L.; Loscalzo, J. Network-Based Approach to Prediction and Population-Based Validation of in Silico Drug Repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, C.; Jiang, J.; Lu, W.; Li, W.; Liu, G.; Zhou, W.; Huang, J.; Tang, Y. Prediction of Drug-Target Interactions and Drug Repositioning via Network-Based Inference. PLoS Comput. Biol. 2012, 8, e1002503. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Farina, L.; Paci, P. SAveRUNNER: A Network-Based Algorithm for Drug Repurposing and Its Application to COVID-19. PLoS Comput. Biol. 2021, 17, e1008686. [Google Scholar] [CrossRef]
- Jourdan, J.-P.; Bureau, R.; Rochais, C.; Dallemagne, P. Drug Repositioning: A Brief Overview. J. Pharm. Pharmacol. 2020, 72, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Fiscon, G.; Paci, P. SAveRUNNER: An R-Based Tool for Drug Repurposing. BMC Bioinform. 2021, 22, 150. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, P.; Webster, R.M. The Breast Cancer Drug Market. Nat. Rev. Drug Discov. 2021, 20, 339–340. [Google Scholar] [CrossRef]
- Grimaldi, A.M.; Conte, F.; Pane, K.; Fiscon, G.; Mirabelli, P.; Baselice, S.; Giannatiempo, R.; Messina, F.; Franzese, M.; Salvatore, M.; et al. The New Paradigm of Network Medicine to Analyze Breast Cancer Phenotypes. Int. J. Mol. Sci. 2020, 21, 6690. [Google Scholar] [CrossRef]
- Paci, P.; Colombo, T.; Fiscon, G.; Gurtner, A.; Pavesi, G.; Farina, L. SWIM: A Computational Tool to Unveiling Crucial Nodes in Complex Biological Networks. Sci. Rep. 2017, 7, srep44797. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G. SWIMmeR: An R-Based Software to Unveiling Crucial Nodes in Complex Biological Networks. Bioinformatics 2022, 38, 586–588. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Paci, P. SWIM Tool Application to Expression Data of Glioblastoma Stem-like Cell Lines, Corresponding Primary Tumors and Conventional Glioma Cell Lines. BMC Bioinform. 2018, 19, 436. [Google Scholar] [CrossRef]
- Falcone, R.; Conte, F.; Fiscon, G.; Pecce, V.; Sponziello, M.; Durante, C.; Farina, L.; Filetti, S.; Paci, P.; Verrienti, A. BRAFV600E-Mutant Cancers Display a Variety of Networks by SWIM Analysis: Prediction of Vemurafenib Clinical Response. Endocrine 2019, 64, 406–413. [Google Scholar] [CrossRef]
- Palumbo, M.C.; Zenoni, S.; Fasoli, M.; Massonnet, M.; Farina, L.; Castiglione, F.; Pezzotti, M.; Paci, P. Integrated Network Analysis Identifies Fight-Club Nodes as a Class of Hubs Encompassing Key Putative Switch Genes That Induce Major Transcriptome Reprogramming during Grapevine Development. Plant Cell 2014, 26, 4617–4635. [Google Scholar] [CrossRef]
- Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Sirota, M.; Dudley, J.T.; Kim, J.; Chiang, A.P.; Morgan, A.A.; Sweet-Cordero, A.; Sage, J.; Butte, A.J. Discovery and Preclinical Validation of Drug Indications Using Compendia of Public Gene Expression Data. Sci. Transl. Med. 2011, 3, 96ra77. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-Based Drug Repurposing for Novel Coronavirus 2019-NCoV/SARS-CoV-2. Cell Discov. 2020, 6, 1–18. [Google Scholar] [CrossRef]
- Paci, P.; Fiscon, G.; Conte, F.; Wang, R.-S.; Farina, L.; Loscalzo, J. Gene Co-Expression in the Interactome: Moving from Correlation toward Causation via an Integrated Approach to Disease Module Discovery. Npj Syst. Biol. Appl. 2021, 7, 1–11. [Google Scholar] [CrossRef]
- Cheng, F.; Kovács, I.A.; Barabási, A.-L. Network-Based Prediction of Drug Combinations. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Fang, J.; Pieper, A.A.; Nussinov, R.; Lee, G.; Bekris, L.; Leverenz, J.B.; Cummings, J.; Cheng, F. Harnessing Endophenotypes and Using Network Medicine in Alzheimer’s Drug Repurposing. Med. Res. Rev. 2020, 40, 2386–2426. [Google Scholar] [CrossRef]
- Basu, A.; Singh, R.; Gupta, S. Bacterial Infections in Cancer: A Bilateral Relationship. WIREs Nanomed. Nanobiotechnology 2022, 14, e1771. [Google Scholar] [CrossRef] [PubMed]
- van Elsland, D.; Neefjes, J. Bacterial Infections and Cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Shi, W.; Huang, Z.; Xia, W.; Huang, J.; Su, Y.; Wang, S.; Shi, Y.; Bi, X.; et al. Efficacy of Moxifloxacin plus Treatment of Physician’s Choice in Patients with Metastatic Breast Cancer. Oncologist 2020, 25, e1439–e1445. [Google Scholar] [CrossRef] [PubMed]
- Patitungkho, S.; Adsule, S.; Dandawate, P.; Padhye, S.; Ahmad, A.; Sarkar, F.H. Synthesis, Characterization and Anti-Tumor Activity of Moxifloxacin–Copper Complexes against Breast Cancer Cell Lines. Bioorg. Med. Chem. Lett. 2011, 21, 1802–1806. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Talwar, P. Repositioning of Fluoroquinolones from Antibiotic to Anti-Cancer Agents: An Underestimated Truth. Biomed. Pharmacother. 2019, 111, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Jiang, S.; Ye, W.; Ren, X.; Wang, X.; Zhang, Y.; Yin, M.; Wang, K.; Tao, Y.; Yang, J.; et al. Combined Treatment of Mitoxantrone Sensitizes Breast Cancer Cells to Rapalogs through Blocking EEF-2K-Mediated Activation of Akt and Autophagy. Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Chu, B.; Shi, S.; Li, X.; Hu, L.; Shi, L.; Zhang, H.; Xu, Q.; Ye, L.; Lin, G.; Zhang, N.; et al. Preparation and Evaluation of Teniposide-Loaded Polymeric Micelles for Breast Cancer Therapy. Int. J. Pharm. 2016, 513, 118–129. [Google Scholar] [CrossRef]
- Das, T.; Nair, R.R.; Green, R.; Padhee, S.; Howell, M.; Banerjee, J.; Mohapatra, S.S.; Mohapatra, S. Actinomycin D Down-Regulates SOX2 Expression and Induces Death in Breast Cancer Stem Cells. Anticancer Res. 2017, 37, 1655–1663. [Google Scholar] [CrossRef]
- Marchionatti, A.M.; Picotto, G.; Narvaez, C.J.; Welsh, J.; Tolosa de Talamoni, N.G. Antiproliferative Action of Menadione and 1,25(OH)2D3 on Breast Cancer Cells. J. Steroid Biochem. Mol. Biol. 2009, 113, 227–232. [Google Scholar] [CrossRef]
- Guizzardi, S.; Picotto, G.; Rodriguez, V.; Welsh, J.; Narvaez, C.; Bohl, L.; Tolosa de Talamoni, N. Combined Treatment of Menadione and Calcitriol Increases the Antiproliferative Effect by Promoting Oxidative/Nitrosative Stress, Mitochondrial Dysfunction, and Autophagy in Breast Cancer MCF-7 Cells. Can. J. Physiol. Pharmacol. 2020, 98, 548–556. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, H.-J.; Yu, Y.; Zhang, Y.; Li, R.-J.; Ju, R.-J.; Wang, X.-X.; Sun, M.-G.; Shi, J.-F.; Lu, W.-L. Mitochondrial Targeting Liposomes Incorporating Daunorubicin and Quinacrine for Treatment of Relapsed Breast Cancer Arising from Cancer Stem Cells. Biomaterials 2012, 33, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Giannone, G.; Milani, A.; Ghisoni, E.; Genta, S.; Mittica, G.; Montemurro, F.; Valabrega, G. Oral Etoposide in Heavily Pre-Treated Metastatic Breast Cancer: A Retrospective Series. Breast Edinb. Scotl. 2018, 38, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Alpsoy, A.; Yasa, S.; Gündüz, U. Etoposide Resistance in MCF-7 Breast Cancer Cell Line Is Marked by Multiple Mechanisms. Biomed. Pharmacother. 2014, 68, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Sledge, G.W., Jr. Etoposide in the Management of Metastatic Breast Cancer. Cancer 1991, 67, 266–270. [Google Scholar] [CrossRef]
- Hu, N.; Zhu, A.; Si, Y.; Yue, J.; Wang, X.; Wang, J.; Ma, F.; Xu, B.; Yuan, P. A Phase II, Single-Arm Study of Apatinib and Oral Etoposide in Heavily Pre-Treated Metastatic Breast Cancer. Front. Oncol. 2021, 10, 565384. [Google Scholar] [CrossRef]
- Bonuccelli, G.; De Francesco, E.M.; de Boer, R.; Tanowitz, H.B.; Lisanti, M.P. NADH Autofluorescence, a New Metabolic Biomarker for Cancer Stem Cells: Identification of Vitamin C and CAPE as Natural Products Targeting “Stemness”. Oncotarget 2017, 8, 20667–20678. [Google Scholar] [CrossRef]
- Mok, K.-C.; Tsoi, H.; Man, E.P.; Leung, M.-H.; Chau, K.M.; Wong, L.-S.; Chan, W.-L.; Chan, S.-Y.; Luk, M.-Y.; Chan, J.Y.W.; et al. Repurposing Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels as a Novel Therapy for Breast Cancer. Clin. Transl. Med. 2021, 11, e578. [Google Scholar] [CrossRef]
- Masuda, N.; Lee, S.-J.; Ohtani, S.; Im, Y.-H.; Lee, E.-S.; Yokota, I.; Kuroi, K.; Im, S.-A.; Park, B.-W.; Kim, S.-B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef]
- Bonadonna, G.; Valagussa, P.; Moliterni, A.; Zambetti, M.; Brambilla, C. Adjuvant Cyclophosphamide, Methotrexate, and Fluorouracil in Node-Positive Breast Cancer—The Results of 20 Years of Follow-Up. N. Engl. J. Med. 1995, 332, 901–906. [Google Scholar] [CrossRef]
- Badve, S.; Dabbs, D.J.; Schnitt, S.J.; Baehner, F.L.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Basal-like and Triple-Negative Breast Cancers: A Critical Review with an Emphasis on the Implications for Pathologists and Oncologists. Mod. Pathol. 2011, 24, 157–167. [Google Scholar] [CrossRef]
- Cınar, V.; Hamurcu, Z.; Guler, A.; Nurdinov, N.; Ozpolat, B. Serotonin 5-HT7 Receptor Is a Biomarker Poor Prognostic Factor and Induces Proliferation of Triple-Negative Breast Cancer Cells through FOXM1. Breast Cancer Tokyo Jpn. 2022, 29, 1106–1120. [Google Scholar] [CrossRef] [PubMed]
- Blyufer, A.; Lhamo, S.; Tam, C.; Tariq, I.; Thavornwatanayong, T.; Mahajan, S.S. Riluzole: A Neuroprotective Drug with Potential as a Novel Anti-cancer Agent (Review). Int. J. Oncol. 2021, 59, 95. [Google Scholar] [CrossRef] [PubMed]
- Speyer, C.L.; Nassar, M.A.; Hachem, A.H.; Bukhsh, M.A.; Jafry, W.S.; Khansa, R.M.; Gorski, D.H. Riluzole Mediates Anti-Tumor Properties in Breast Cancer Cells Independent of Metabotropic Glutamate Receptor-1. Breast Cancer Res. Treat. 2016, 157, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Dolfi, S.C.; Medina, D.J.; Kareddula, A.; Paratala, B.; Rose, A.; Dhami, J.; Chen, S.; Ganesan, S.; Mackay, G.; Vazquez, A.; et al. Riluzole Exerts Distinct Antitumor Effects from a Metabotropic Glutamate Receptor 1-Specific Inhibitor on Breast Cancer Cells. Oncotarget 2017, 8, 44639–44653. [Google Scholar] [CrossRef] [PubMed]
- Speyer, C.L.; Bukhsh, M.A.; Jafry, W.S.; Sexton, R.E.; Bandyopadhyay, S.; Gorski, D.H. Riluzole Synergizes with Paclitaxel to Inhibit Cell Growth and Induce Apoptosis in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2017, 166, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic Target Database 2020: Enriched Resource for Facilitating Research and Early Development of Targeted Therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef] [PubMed]
| Luminal A | Luminal B | HER2 Enriched | Basal-Like | |
|---|---|---|---|---|
| Samples | ||||
| # normal | 111 | 111 | 111 | 111 |
| # tumour | 229 | 120 | 58 | 98 |
| switch genes | ||||
| # total | 222 | 358 | 363 | 343 |
| Input | Source | Publication Year/Version | Description |
|---|---|---|---|
| PAM50 switch genes | Grimaldi et al. [15] | 2020 | see Table 1 |
| Human interactome | Cheng et al. [8] | 2018 | 217,160 physical interactions connecting 15,970 biomolecules |
| Drug–target associations | DrugBank [22] | version 5.1.6 released on 22 April 2020 | 2165 genes interacting with 1873 drugs |
| Drug original indications | Therapeutic Target Database (TTD) [23] | version released on 1 June 2020 | 5059 drugs associated with 1136 diseases |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, F.; Sibilio, P.; Fiscon, G.; Paci, P. A Transcriptome- and Interactome-Based Analysis Identifies Repurposable Drugs for Human Breast Cancer Subtypes. Symmetry 2022, 14, 2230. https://doi.org/10.3390/sym14112230
Conte F, Sibilio P, Fiscon G, Paci P. A Transcriptome- and Interactome-Based Analysis Identifies Repurposable Drugs for Human Breast Cancer Subtypes. Symmetry. 2022; 14(11):2230. https://doi.org/10.3390/sym14112230
Chicago/Turabian StyleConte, Federica, Pasquale Sibilio, Giulia Fiscon, and Paola Paci. 2022. "A Transcriptome- and Interactome-Based Analysis Identifies Repurposable Drugs for Human Breast Cancer Subtypes" Symmetry 14, no. 11: 2230. https://doi.org/10.3390/sym14112230
APA StyleConte, F., Sibilio, P., Fiscon, G., & Paci, P. (2022). A Transcriptome- and Interactome-Based Analysis Identifies Repurposable Drugs for Human Breast Cancer Subtypes. Symmetry, 14(11), 2230. https://doi.org/10.3390/sym14112230