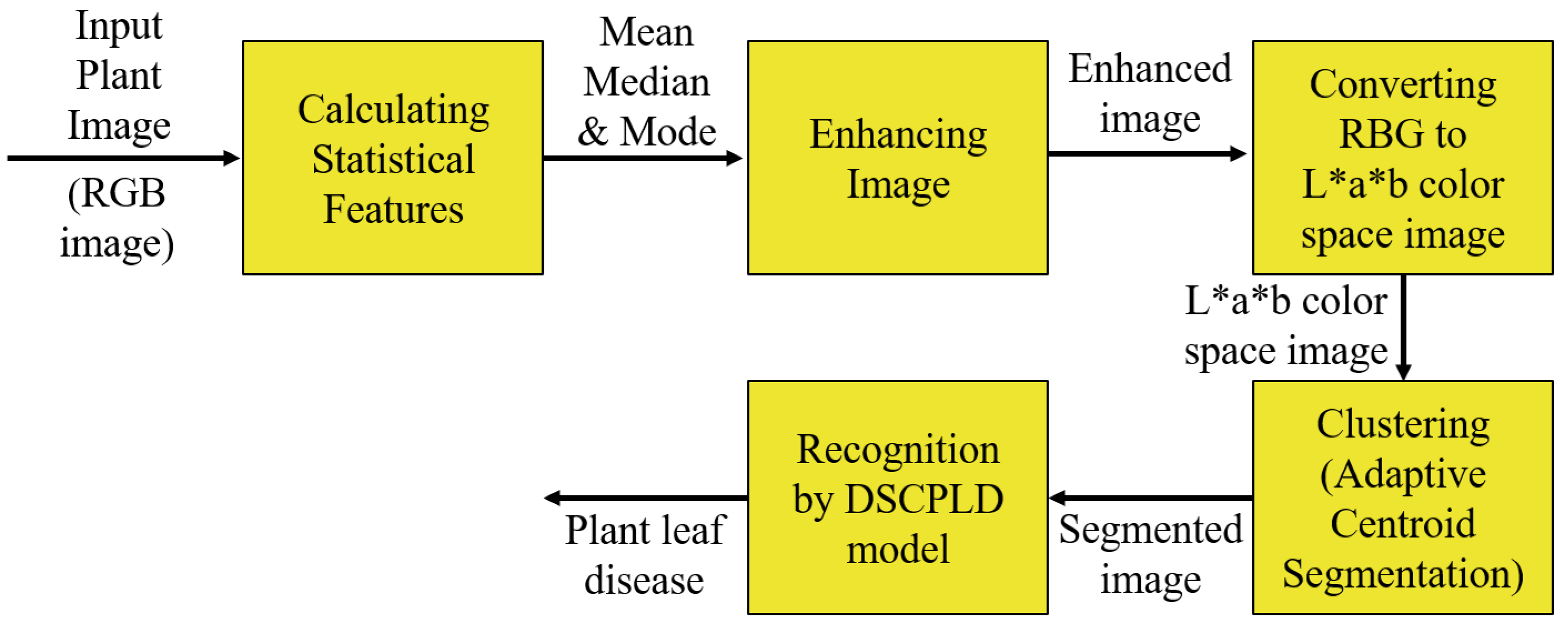
Figure 1.
The proposed framework for recognizing plant leaf disease.
Figure 1.
The proposed framework for recognizing plant leaf disease.
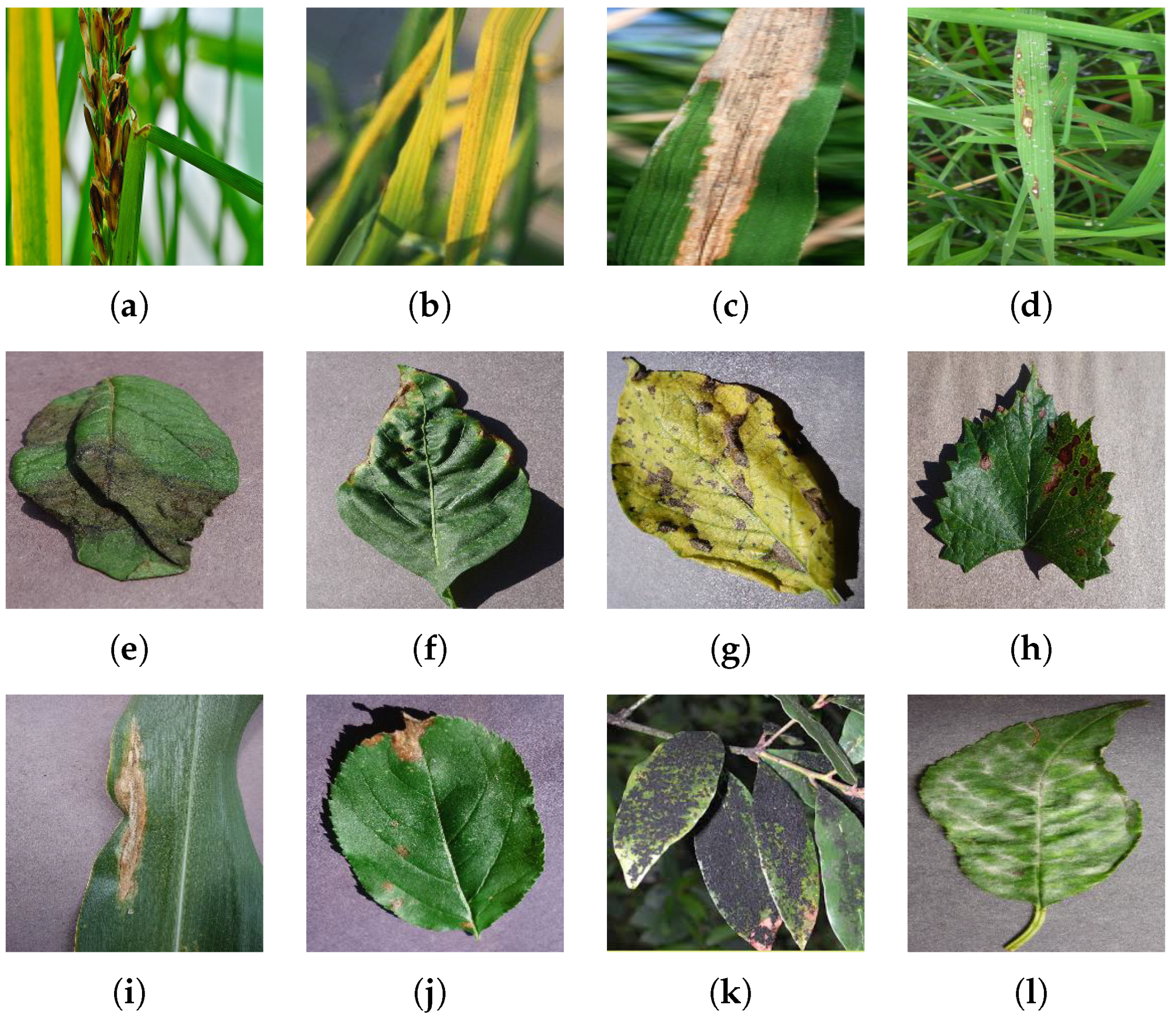
Figure 2.
Samples of plant leaf disease images under numerous health conditions in various backgrounds and having different symptoms: (a) Rice Sheath-rot, (b) Rice Tungro, (c) Rice Bacterial leaf-blight, (d) Rice Blast, (e) Potato Late-blight, (f) Pepper Bacterial-spot, (g) Potato Early-blight Pepper Bacterial-spot, (h) Grape Black-measles, (i) Corn Northern Leaf-blight, (j) Apple Black-rot, (k) Mango Sooty-mold, and (l) Cherry Powdery-mildew.
Figure 2.
Samples of plant leaf disease images under numerous health conditions in various backgrounds and having different symptoms: (a) Rice Sheath-rot, (b) Rice Tungro, (c) Rice Bacterial leaf-blight, (d) Rice Blast, (e) Potato Late-blight, (f) Pepper Bacterial-spot, (g) Potato Early-blight Pepper Bacterial-spot, (h) Grape Black-measles, (i) Corn Northern Leaf-blight, (j) Apple Black-rot, (k) Mango Sooty-mold, and (l) Cherry Powdery-mildew.
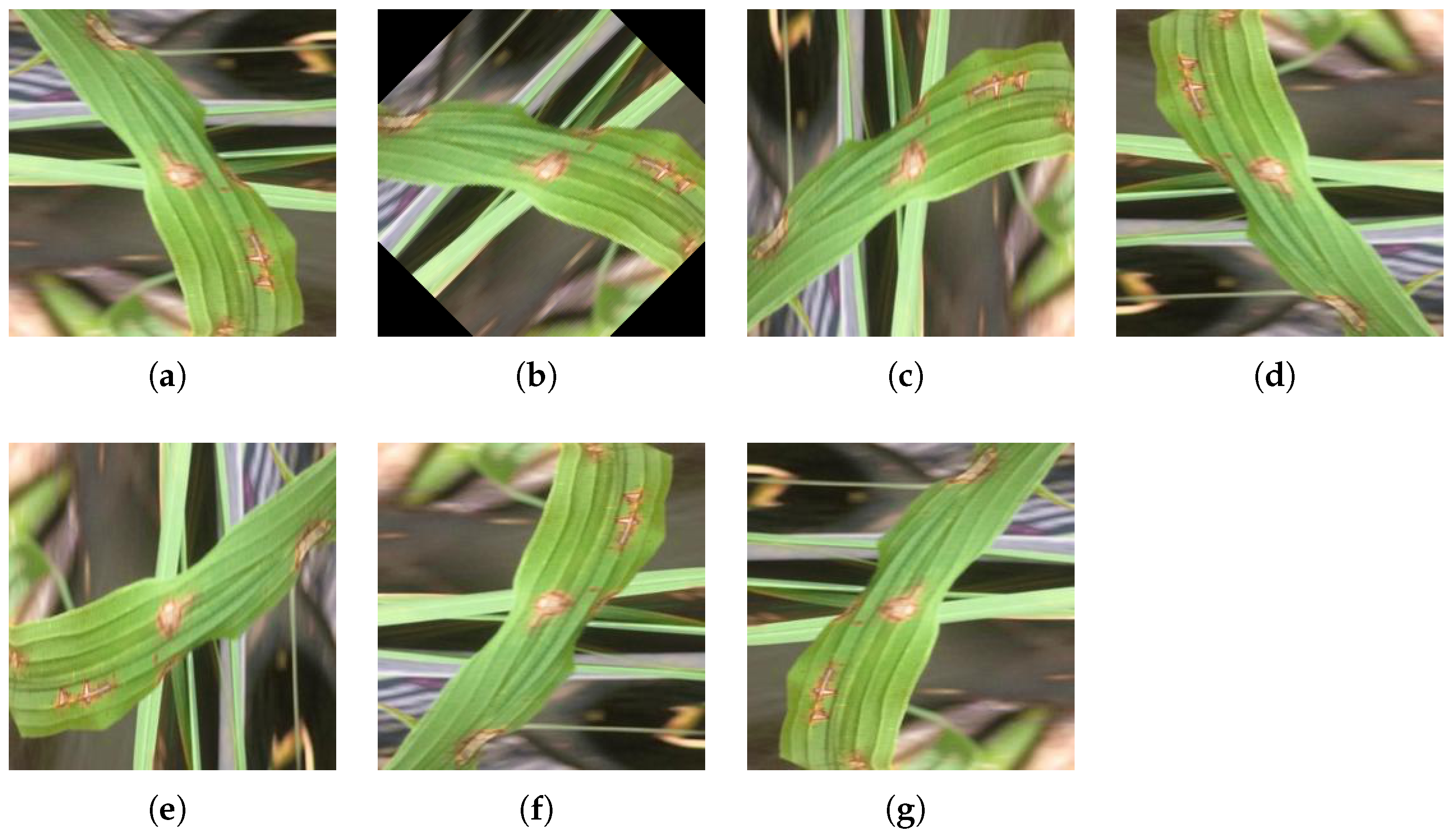
Figure 3.
Directional Disturbance: (a) Original Rice Blast image. (b) Rotated by 45°. (c) Rotated by 90°. (d) Rotated by 180°. (e) Rotated by 270°. (f) Horizontal mirror symmetry. (g) Vertical mirror symmetry.
Figure 3.
Directional Disturbance: (a) Original Rice Blast image. (b) Rotated by 45°. (c) Rotated by 90°. (d) Rotated by 180°. (e) Rotated by 270°. (f) Horizontal mirror symmetry. (g) Vertical mirror symmetry.
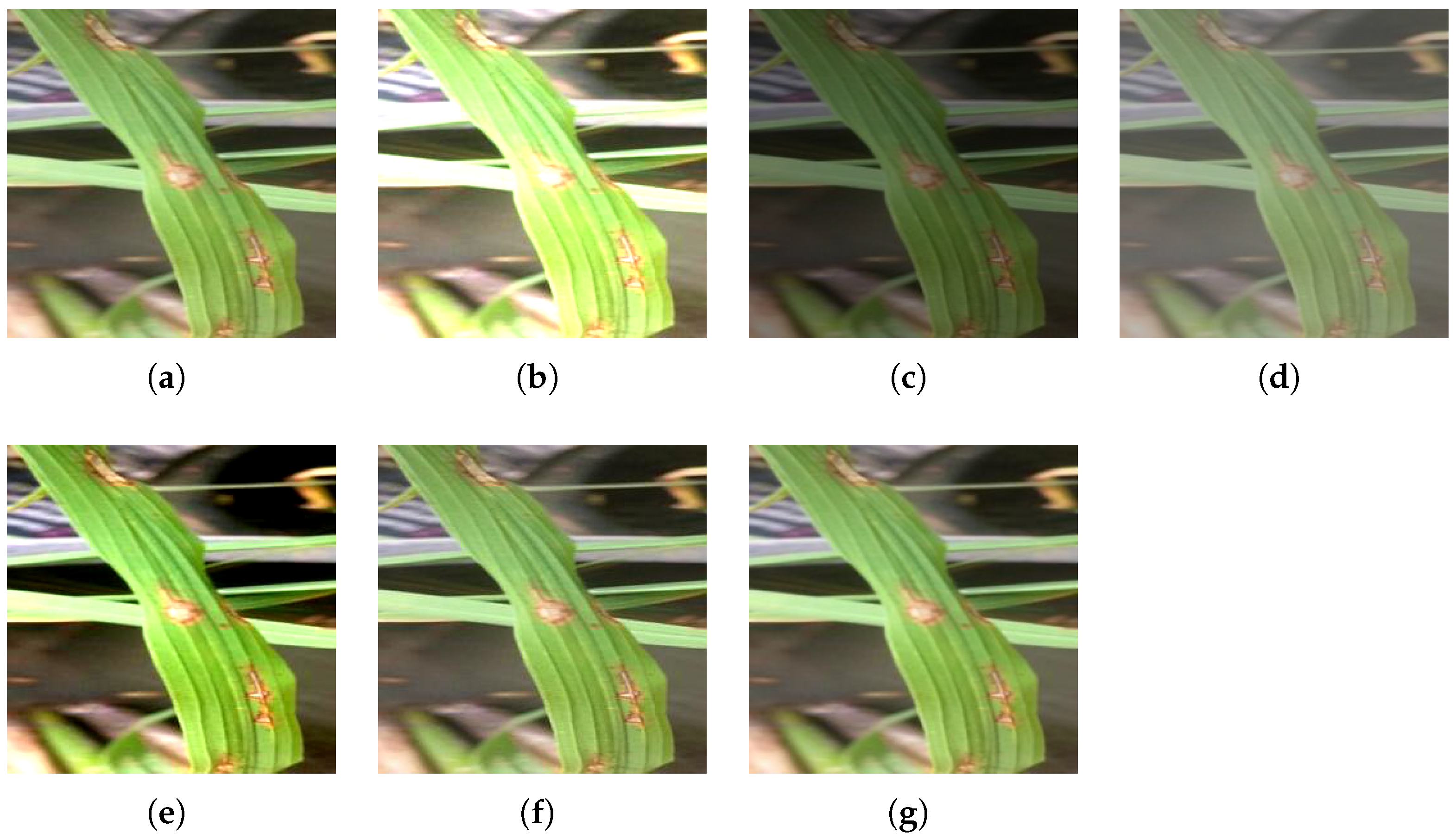
Figure 4.
Illumination Disturbance: (a) Original Rice Blast image. (b) Brightened image. (c) Darkened image. (d) Less contrast image. (e) More contrast image. (f) Sharpened image. (g) Blur image.
Figure 4.
Illumination Disturbance: (a) Original Rice Blast image. (b) Brightened image. (c) Darkened image. (d) Less contrast image. (e) More contrast image. (f) Sharpened image. (g) Blur image.
Figure 5.
Effect of image enhancement on recognizing PLD: (a) rice blast disease image, and (b) apple black rot disease image. (c,d) are histogram of (a,b), respectively; (e,g) are the color segmentation results of (a,b), respectively, in traditional K-means clustering having extra noise without image enhancement, and (f,h) are the segmentation results of (a,b), respectively, in our modified color segmentation algorithm with image enhancement.
Figure 5.
Effect of image enhancement on recognizing PLD: (a) rice blast disease image, and (b) apple black rot disease image. (c,d) are histogram of (a,b), respectively; (e,g) are the color segmentation results of (a,b), respectively, in traditional K-means clustering having extra noise without image enhancement, and (f,h) are the segmentation results of (a,b), respectively, in our modified color segmentation algorithm with image enhancement.
Figure 6.
The effect of our modified segmentation technique under different critical environments: (a–e) are the RGB PLD samples. (f–j) are segmented regions of interest (ROIs) of (a–e) after implementing adaptive centroid-based segmentation.
Figure 6.
The effect of our modified segmentation technique under different critical environments: (a–e) are the RGB PLD samples. (f–j) are segmented regions of interest (ROIs) of (a–e) after implementing adaptive centroid-based segmentation.
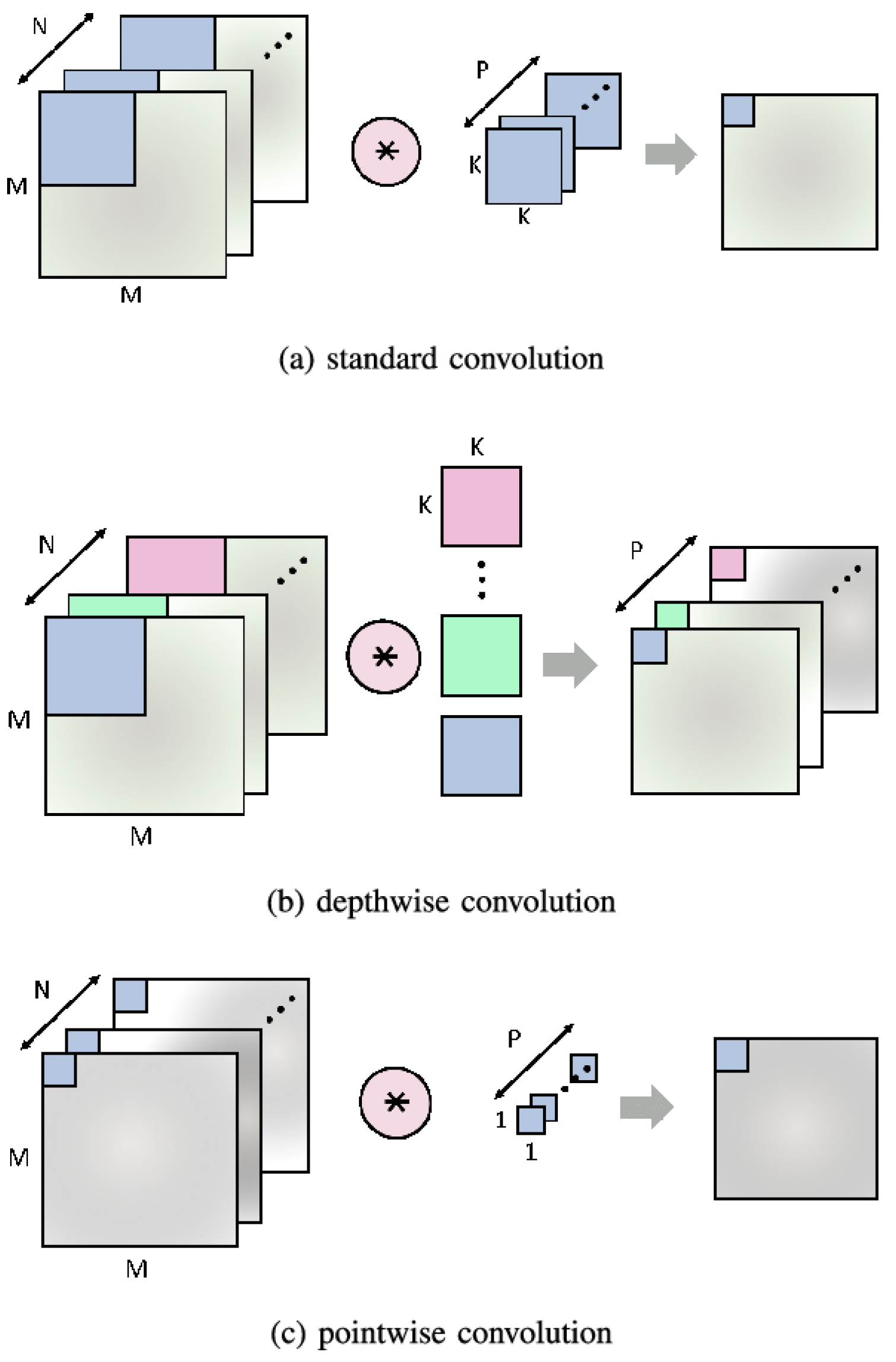
Figure 7.
Comparison among various convolutions.
Figure 7.
Comparison among various convolutions.
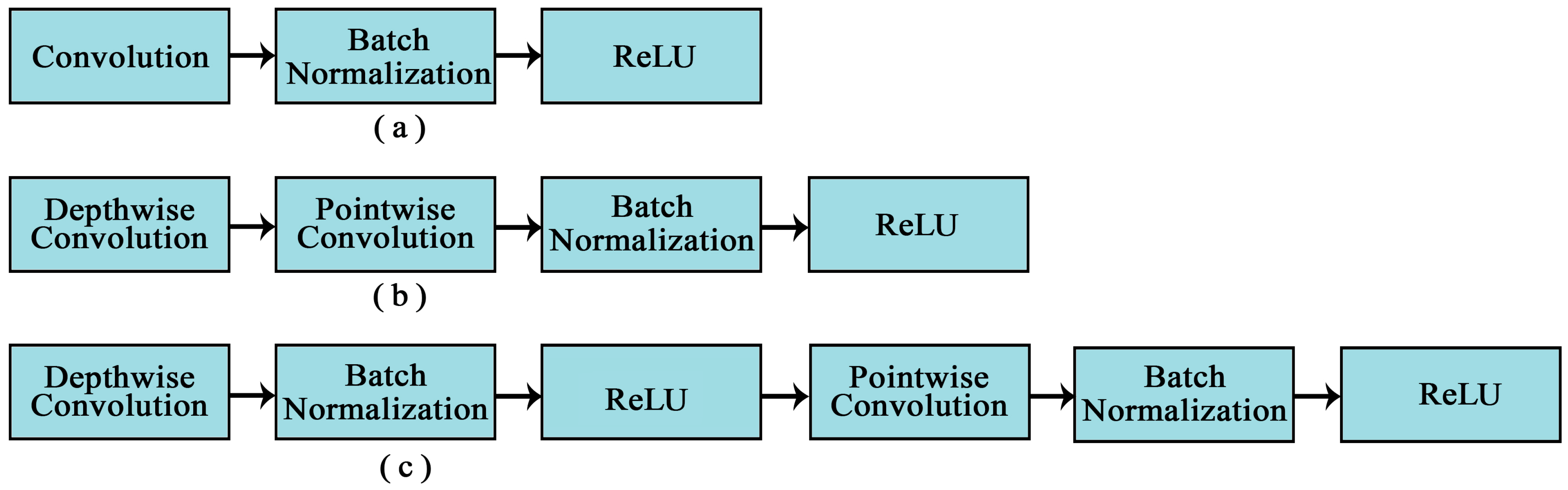
Figure 8.
Primary modules for PLD recognition. (a) traditional convolutional layer, (b) quantization friendly depth-wise separable convolution, and (c) depth-wise separable convolution proposed in MobileNet.
Figure 8.
Primary modules for PLD recognition. (a) traditional convolutional layer, (b) quantization friendly depth-wise separable convolution, and (c) depth-wise separable convolution proposed in MobileNet.
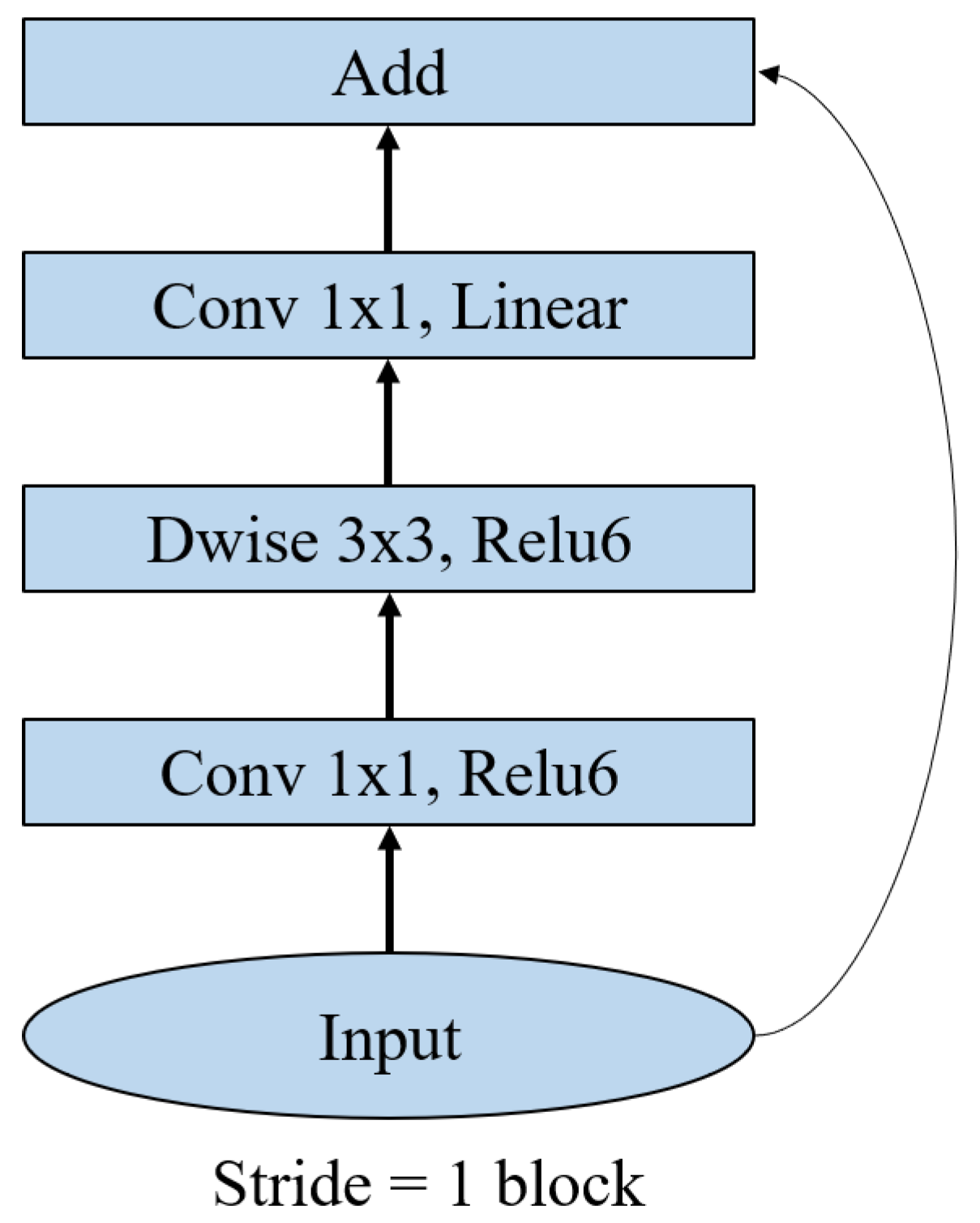
Figure 9.
Primary module of MobileNetV2 for PLD recognition.
Figure 9.
Primary module of MobileNetV2 for PLD recognition.
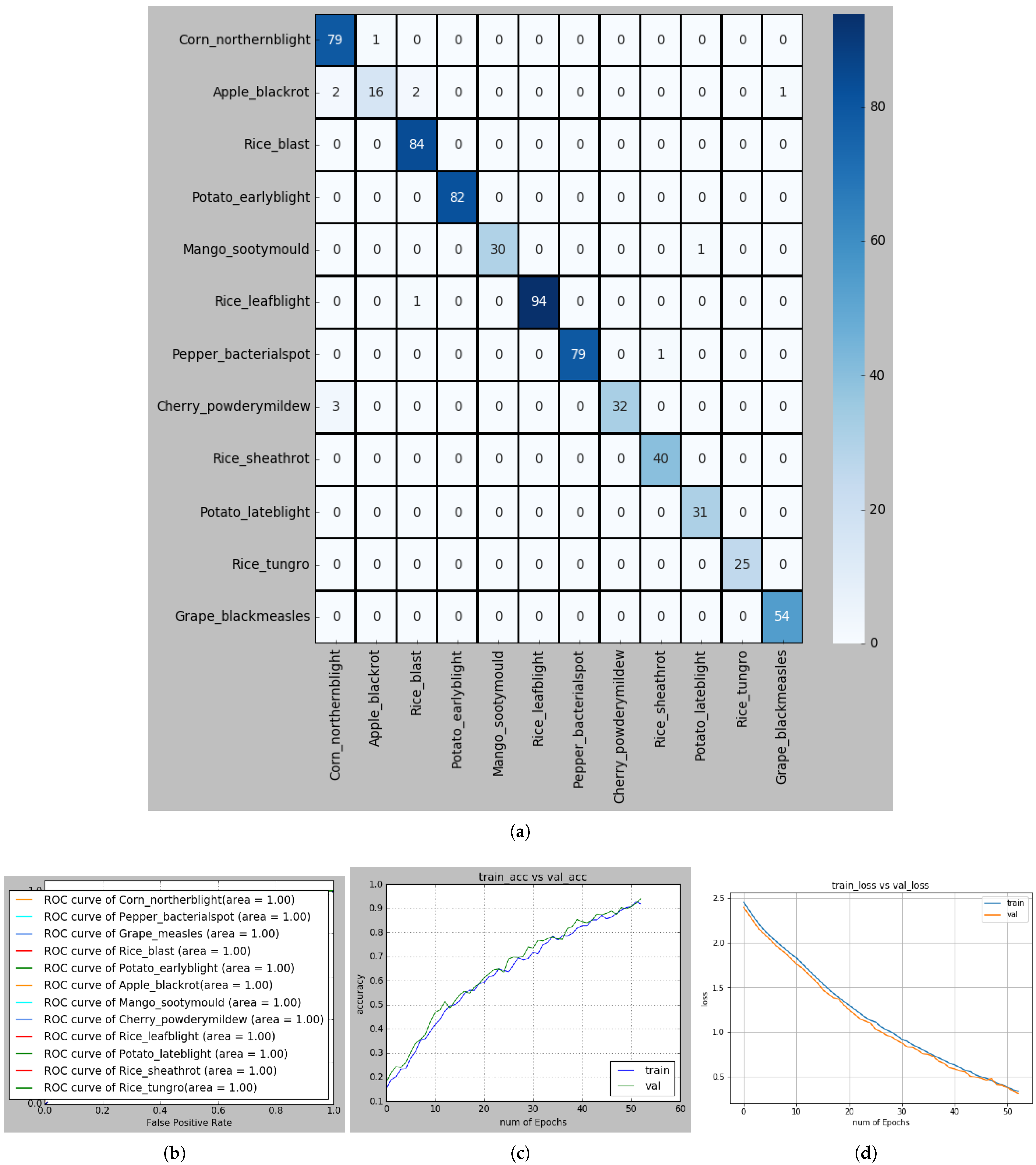
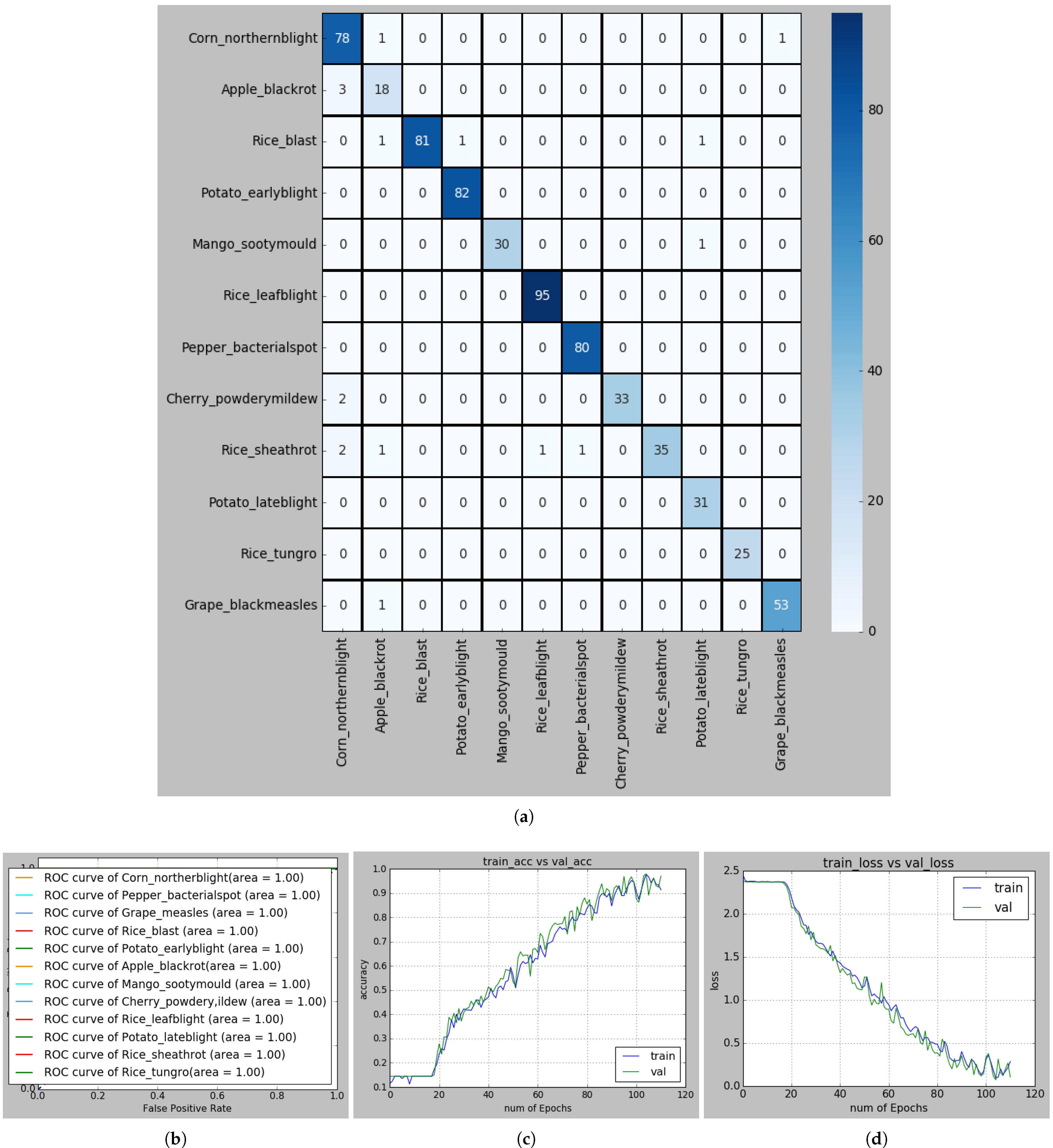
Figure 10.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-modified MobileNet-based recognition framework.
Figure 10.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-modified MobileNet-based recognition framework.
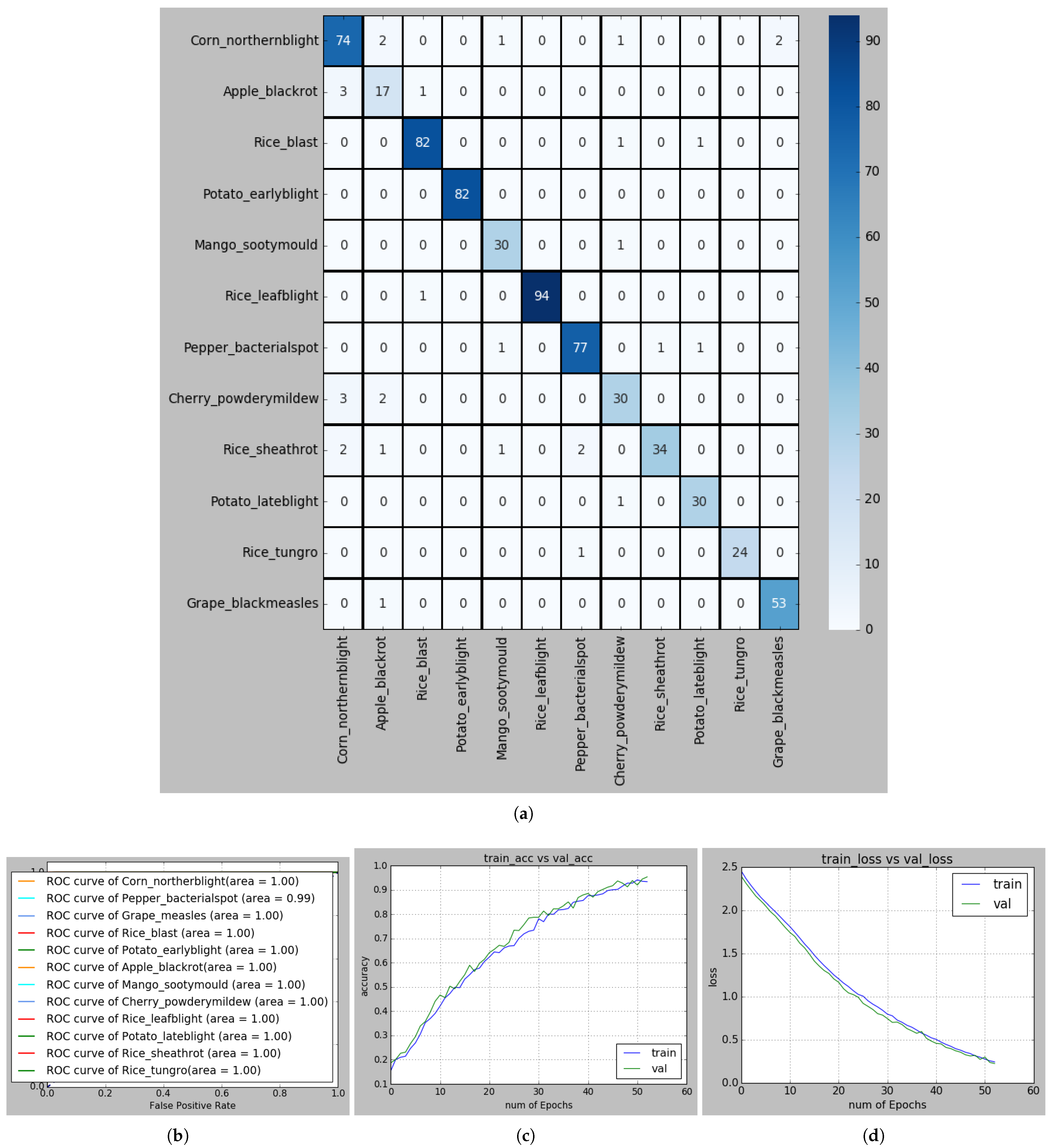
Figure 11.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-reduced MobileNet-based recognition framework.
Figure 11.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-reduced MobileNet-based recognition framework.
Figure 12.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-extended MobileNet-based recognition framework.
Figure 12.
(a) Confusion matrix for recognizing PLDs; (b) ROC curve of each PLD; (c) Accuracy curve, and (d) Loss curve in S-extended MobileNet-based recognition framework.
Figure 13.
Processing steps of depth-wise separable convolutional PLD (DSCPLD) recognition framework using S-modified MobileNet: (a) Original Rice Blast image. (b) Segmented image after applying adaptive centroid-based segmentation (ACS). (c) Activations on the first CONV layer. (d) Activations on the first ReLU layer. (e) Activations on the first Max-pooling layer. (f) Activations on the first separable CONV layer. (g) Activations on the second separable CONV layer. (h) Activations on the second Max-pooling layer. (i) Activations on the second ReLU layer. (j) Activations on the third separable CONV layer. (k) Activations on the fourth separable CONV layer. (l) Activations on the third Max-pooling layer. (m) Activations on the third ReLU layer. (n) Activations on the fifth separable CONV layer. (o) Activations on the sixth separable CONV layer. (p) Activations on the fourth Max-pooling layer. (q) Activations on the fourth ReLU layer. and (r) Predicted result.
Figure 13.
Processing steps of depth-wise separable convolutional PLD (DSCPLD) recognition framework using S-modified MobileNet: (a) Original Rice Blast image. (b) Segmented image after applying adaptive centroid-based segmentation (ACS). (c) Activations on the first CONV layer. (d) Activations on the first ReLU layer. (e) Activations on the first Max-pooling layer. (f) Activations on the first separable CONV layer. (g) Activations on the second separable CONV layer. (h) Activations on the second Max-pooling layer. (i) Activations on the second ReLU layer. (j) Activations on the third separable CONV layer. (k) Activations on the fourth separable CONV layer. (l) Activations on the third Max-pooling layer. (m) Activations on the third ReLU layer. (n) Activations on the fifth separable CONV layer. (o) Activations on the sixth separable CONV layer. (p) Activations on the fourth Max-pooling layer. (q) Activations on the fourth ReLU layer. and (r) Predicted result.
Figure 14.
(a) Confusion matrix for recognizing PLDs and (b) ROC curve of each PLD in F-modified MobileNet-based recognition framework.
Figure 14.
(a) Confusion matrix for recognizing PLDs and (b) ROC curve of each PLD in F-modified MobileNet-based recognition framework.
Table 1.
Summary of some benchmark plant leaf disease (PLD) recognition frameworks.
Table 1.
Summary of some benchmark plant leaf disease (PLD) recognition frameworks.
| References | Data Collected from | Classes/Species | Number of Images | Data Augmentation | CNN Architecture | Accuracy |
|---|
| [6] | PlantVillage | 58/25 | 54,309 | Yes | VGG | 99.53% |
| [7] | PlantVillage | 38/14 | 54,306 | Yes | GoogleNet | 99.35% |
| [8] | Collected | 15/6 | 4483 | Yes | Modified CaffeNet | 96.30% |
| [10] | PlantVillage | 38/14 | 54,305 | Yes | DenseNet121 | 99.75% |
| [11] | Collected | 2/1 | 5808 | Yes | Custom | 95.83% |
| [13] | PlantVillage | 3/1 | 3700 | Yes | Modified LeNet | 92.88% |
| [14] | Collected | 9/1 | 1426 | Yes | Two stage CNN | 93.3% |
| [18] | Collected | 4/1 | 1053 | Yes | Modified AlexNet | 97.62% |
| [19] | PlantVillage, Collected | 42/12 | 79,265 | Yes | ResNet152 | 90.88% |
| [20] | PlantVillage, Collected | 10/1 | 17929 | N/A | F-CNN, S-CNN | 98.6% |
| [21] | Collected | 7/1 | 7905 | Yes | Custom | 90.16% |
| [26] | PlantVillage | 38/14 | 54,323 | Yes | InceptionV3 | 99.76% |
| [28] | Collected | 9/1 | 5000 | Yes | R-FCNN, ResNet50 | 85.98% |
| [29] | Collected | 56/14 | 1567 | Yes | GoogleNet | 94% |
| [30] | Collected | 10/1 | 500 | No | Custom | 95.48% |
| [31] | Collected | 6/1 | 6029 | Yes | DenseNet+RF | 97.59% |
Table 2.
Limitations of some benchmark PLD recognition frameworks.
Table 2.
Limitations of some benchmark PLD recognition frameworks.
| References | Fall in Accuracy | Complex Background | Multiple Diseases in a Sample | Train and Test Data from Same Dataset | Computational Complexity | Memory Restrictions |
|---|
| [6] | NR | NR | PR | NR | NR | NR |
| [7] | NR | NR | NR | NR | NR | NR |
| [8] | NR | R | R | NR | NR | NR |
| [10] | NR | NR | NR | NR | NR | NR |
| [11] | NR | NR | NR | NR | NR | NR |
| [13] | NR | NR | NR | NR | NR | NR |
| [14] | NR | PR | NR | NR | NR | R |
| [18] | R | NR | NR | NR | NR | NR |
| [19] | R | R | R | R | R | NR |
| [20] | R | R | R | R | NR | NR |
| [21] | NR | NR | NR | NR | NR | NR |
| [26] | NR | PR | NR | NR | NR | NR |
| [28] | PR | R | R | NR | NR | NR |
| [29] | R | PR | NR | NR | NR | NR |
| [30] | NR | PR | NR | NR | NR | NR |
| [31] | NR | R | PR | NR | NR | NR |
Table 3.
Dataset descriptions of plant leaf disease recognition.
Table 3.
Dataset descriptions of plant leaf disease recognition.
| Disease Class | #Org. Images | Distribution Techniques |
|---|
| Train | Validation | Test |
|---|
| Corn_northern_blight | 800 | 560 | 160 | 80 |
| Pepper_bacterial_spot | 800 | 560 | 160 | 80 |
| Grape_black_measles | 540 | 378 | 108 | 54 |
| Rice_blast | 840 | 588 | 168 | 84 |
| Rice_bacterial_leaf_blight | 950 | 665 | 190 | 95 |
| Rice_sheath_rot | 400 | 280 | 80 | 40 |
| Rice_Tugro | 250 | 175 | 50 | 25 |
| Potato_early_blight | 820 | 574 | 164 | 82 |
| Potato_late_blight | 310 | 217 | 62 | 31 |
| Apple_black_rot | 210 | 147 | 42 | 21 |
| Mango_sooty_mold | 310 | 217 | 62 | 31 |
| Cherry_powdery_mildew | 350 | 245 | 70 | 35 |
| Total | 6580 | 4606 | 1316 | 658 |
Table 4.
S-modified MobileNet architecture for PLD recognition.
Table 4.
S-modified MobileNet architecture for PLD recognition.
| Function | Filter/Pool | #Filters | Output | #Parameters |
|---|
| Input | - | - | | 0 |
| Convolution | | 32 | | 896 |
| Max pooling | | - | | 0 |
| Separable Convolution | | 64 | | 2400 |
| Separable Convolution | | 64 | | 4736 |
| Max pooling | | - | | 0 |
| Separable Convolution | | 128 | | 8896 |
| Separable Convolution | | 128 | | 17,664 |
| Max pooling | | - | | 0 |
| Separable Convolution | | 256 | | 34,176 |
| Separable Convolution | | 256 | | 68,096 |
| Max pooling | | - | | 0 |
| Global Average Pooling | - | - | | 0 |
| Dense | - | - | | 263,168 |
| Dense | - | - | | 12,300 |
| Softmax | - | - | | 0 |
Table 5.
S-reduced MobileNet architecture for PLD recognition.
Table 5.
S-reduced MobileNet architecture for PLD recognition.
| Function | Filter/Pool | #Filters | Output | #Parameters |
|---|
| Input | - | - | | 0 |
| Convolution | | 32 | | 896 |
| Depth-wise Convolution | | 32 | | 32,800 |
| Point-wise Convolution | | 64 | | 2112 |
| Depth-wise Convolution | | 64 | | 262,208 |
| Point-wise Convolution | | 128 | | 8320 |
| Global Average Pooling | - | - | | 0 |
| Dense | - | - | | 1548 |
| Softmax | - | - | | 0 |
Table 6.
S-extended MobileNet architecture for PLD recognition.
Table 6.
S-extended MobileNet architecture for PLD recognition.
| Function | Filter/Pool | #Filters | Output | #Parameters |
|---|
| Input | - | - | | 0 |
| Convolution | | 32 | | 896 |
| Depth-wise Convolution | | 32 | | 32,800 |
| Point-wise Convolution | | 64 | | 2112 |
| Depth-wise Convolution | | 64 | | 262,208 |
| Point-wise Convolution | | 128 | | 8320 |
| Max pooling | | - | | 0 |
| Dense | - | - | | 5,25,312 |
| Dense | - | - | | 12,300 |
| Softmax | - | - | | 0 |
Table 7.
Hyper-parameters used in various models for PLD recognition.
Table 7.
Hyper-parameters used in various models for PLD recognition.
| Hyper-Parameters | SGD | Adam | RMSprop |
|---|
| Epochs | 50–150 | 50–150 | 50–150 |
| Batch size | 32, 64 | 32, 64 | 32, 64 |
| Learning rate | 0.001 | 0.001, 0.0001 | 0.0001 |
| - | 0.9 | - |
| - | 0.999 | - |
| Momentum | 0.8, 0.9 | - | - |
Table 8.
Source-wise dataset distribution summary.
Table 8.
Source-wise dataset distribution summary.
| Sources | Species | Diseases | No. of Training Images | No. of Validation Images | No. of Test Images | No. of Training Images (Source-Wise) | No. of Validation Images (Source-Wise) | No. of Test Images (Source-Wise) |
|---|
| PlantVillage | pepper | Bacterial-spot | 560 | 160 | 80 | 2898 | 1459 | 414 |
| Potato | Early-blight | 574 | 164 | 82 |
| Late-blight | 217 | 62 | 31 |
| Corn | Northern-blight | 560 | 160 | 80 |
| Mango | Sooty-mold | 217 | 62 | 31 |
| Apple | Black-rot | 147 | 42 | 21 |
| Cherry | Powdery-mildew | 245 | 70 | 35 |
| Grape | Black-measles | 378 | 108 | 54 |
| Kaggle | Rice | Blast | 588 | 168 | 84 | 1253 | 358 | 179 |
| Bacterial leaf-blight | 665 | 190 | 95 |
IRRI/BRRI/
other sources | Rice | Sheath-rot | 280 | 80 | 40 | 455 | 130 | 65 |
| Tungro | 175 | 50 | 25 |
| Total images | | | 4606 | 1316 | 658 | | | |
Table 9.
A concrete representation of accuracies and mean F1-score of various PLD recognition models using segmented images.
Table 9.
A concrete representation of accuracies and mean F1-score of various PLD recognition models using segmented images.
| Models | Training Accuracy | Validation Accuracy | Mean Test Accuracy | Mean F1-Score |
|---|
| VGG16 | 99.91% | 99.53% | 99.21% | 96.74% |
| VGG19 | 99.93% | 99.53% | 99.39% | 96.91% |
| AlexNet | 99.07% | 98.82% | 98.78% | 96.31% |
| MobileNetV1 | 99.93% | 99.41% | 99.24% | 95.67% |
| MobileNetV2 | 99.96% | 99.82% | 99.41% | 96.07% |
| MobileNetV3 | 100% | 99.89% | 99.55% | 96.97% |
| S-extended MobileNet | 99.78% | 99.31% | 98.37% | 95.92% |
| S-reduced MobileNet | 99.93% | 99.70% | 99.41% | 96.93% |
| S-modified MobileNet | 100% | 99.70% | 99.55% | 97.07% |
Table 10.
A concrete representation of computational latency and model size of various PLD recognition models using segmented images.
Table 10.
A concrete representation of computational latency and model size of various PLD recognition models using segmented images.
| Models | Image Size | FLOPs | MACC | # Parameters |
|---|
| VGG16 | | 213.5 M | 106.75 M | 15.2 M |
| VGG19 | | 287.84 M | 143.92 M | 20.6 M |
| AlexNet | | 127.68 M | 63.84 M | 6.4 M |
| MobileNetV1 | | 83.87 M | 41.93 M | 3.2 M |
| MobileNetV2 | | 81.91 M | 40.96 M | 1.61 M |
| MobileNetV3 | | 59.8 M | 29.90 M | 3.2 M |
| S-extended MobileNet | | 16.86 M | 8.43 M | 0.84 M |
| S-reduced MobileNet | | 3.70 M | 2.15 M | 0.31 M |
| S-modified MobileNet | | 5.78 M | 2.89 M | 0.41 M |
Table 11.
Various accuracies and mean F1-score of PLD models using full leaf images.
Table 11.
Various accuracies and mean F1-score of PLD models using full leaf images.
| Models | Training Accuracy | Validation Accuracy | Mean Test Accuracy | Mean F1-Score |
|---|
| VGG16 | 99.78% | 99.39% | 98.78% | 96.32% |
| VGG19 | 99.78% | 99.41% | 99.01% | 96.54% |
| AlexNet | 98.71% | 98.64% | 98.34% | 95.89% |
| MobileNetV1 | 99.81% | 99.43% | 98.79% | 96.54% |
| MobileNetV2 | 99.89% | 99.53% | 98.99% | 96.56% |
| MobileNetV3 | 99.91% | 99.53% | 99.05% | 96.58% |
| F-extended MobileNet | 99.58% | 99.21% | 98.14% | 95.22% |
| F-reduced MobileNet | 99.91% | 99.58% | 99.07% | 96.60% |
| F-modified MobileNet | 99.91% | 99.63% | 99.10% | 96.63% |
Table 12.
Performance comparison of each disease using S-modified MobileNet and F-modified MobileNet.
Table 12.
Performance comparison of each disease using S-modified MobileNet and F-modified MobileNet.
| Class | S-modified MobileNet | F-modified MobileNet |
|---|
| Accuracy (%) | F1-Score (%) | Accuracy (%) | F1-Score (%) |
|---|
| Corn_northern_blight | 99.08 | 96.34 | 98.18 | 92.77 |
| Pepper_bacterial_spot | 99.85 | 99.37 | 99.39 | 97.50 |
| Grape_black_measles | 99.85 | 99.08 | 99.39 | 96.30 |
| Rice_blast | 99.54 | 98.24 | 99.24 | 96.93 |
| Potato_early_blight | 100 | 100 | 99.70 | 98.87 |
| Apple_black_rot | 99.08 | 84.24 | 98.63 | 80 |
| Mango_sooty_mold | 99.85 | 98.36 | 99.39 | 93.75 |
| Cherry_powdery_mildew | 99.54 | 95.52 | 98.78 | 87.88 |
| Rice_bacterial_leaf_blight | 99.85 | 99.45 | 99.85 | 99.45 |
| Potato_late_blight | 99.85 | 98.41 | 99.24 | 91.80 |
| Rice_sheath_rot | 99.85 | 98.76 | 98.94 | 91.02 |
| Rice_Tugro | 100 | 100 | 99.85 | 97.96 |
| Total | 99.55 | 97.07 | 99.10 | 96.63 |
Table 13.
Performance comparison of each disease using S-reduced MobileNet and F-reduced MobileNet.
Table 13.
Performance comparison of each disease using S-reduced MobileNet and F-reduced MobileNet.
| Class | S-reduced MobileNet | F-reduced MobileNet |
|---|
| Accuracy (%) | F1-Score (%) | Accuracy (%) | F1-Score (%) |
|---|
| Corn_northern_blight | 98.63 | 94.54 | 98.18 | 92.77 |
| Pepper_bacterial_spot | 99.08 | 99.38 | 99.39 | 97.50 |
| Grape_black_measles | 99.54 | 98.15 | 99.39 | 96.30 |
| Rice_blast | 99.54 | 98.18 | 99.24 | 96.93 |
| Potato_early_blight | 99.85 | 99.40 | 99.70 | 98.87 |
| Apple_black_rot | 98.94 | 83.72 | 98.63 | 80 |
| Mango_sooty_mold | 99.85 | 98.36 | 99.39 | 93.75 |
| Cherry_powdery_mildew | 98.63 | 97.07 | 98.78 | 97.88 |
| Rice_bacterial_leaf_blight | 99.85 | 99.48 | 99.85 | 99.45 |
| Potato_late_blight | 99.54 | 96.88 | 99.24 | 91.80 |
| Rice_sheath_rot | 98.93 | 93.33 | 97.85 | 98.05 |
| Rice_Tugro | 100 | 100 | 99.85 | 97.96 |
| Total | 99.41 | 96.93 | 99.07 | 96.60 |
Table 14.
Performance comparison of each disease using S-extended MobileNet and F-extended MobileNet.
Table 14.
Performance comparison of each disease using S-extended MobileNet and F-extended MobileNet.
| Class | S-extended MobileNet | F-extended MobileNet |
|---|
| Accuracy (%) | F1-Score (%) | Accuracy (%) | F1-Score (%) |
|---|
| Corn_northern_blight | 97.87 | 91.36 | 97.18 | 90.67 |
| Pepper_bacterial_spot | 99.08 | 96.25 | 98.79 | 97.50 |
| Grape_black_measles | 99.54 | 97.25 | 99.39 | 96.30 |
| Rice_blast | 99.39 | 97.62 | 99.24 | 96.93 |
| Potato_early_blight | 100 | 100 | 99.70 | 98.87 |
| Apple_black_rot | 98.48 | 73.06 | 97.03 | 70.03 |
| Mango_sooty_mold | 99.39 | 95.24 | 99.39 | 93.75 |
| Cherry_powdery_mildew | 98.63 | 86.96 | 97.78 | 87.88 |
| Rice_bacterial_leaf_blight | 99.85 | 99.47 | 99.85 | 99.45 |
| Potato_late_blight | 99.54 | 95.24 | 99.24 | 90.67 |
| Rice_sheath_rot | 98.93 | 90.67 | 97.85 | 88.05 |
| Rice_Tugro | 99.84 | 97.96 | 99.85 | 97.96 |
| Total | 98.37 | 95.92 | 98.14 | 95.22 |
Table 15.
A concrete representation of experiments on MobileNetV3 with width multipliers.
Table 15.
A concrete representation of experiments on MobileNetV3 with width multipliers.
| Models | Mean Test Accuracy | Mean F1-Score | FLOPs | MACC | # Parameters |
|---|
| 0.25 MobileNetV3-224 | 95.48% | 93.39% | 4.30 M | 2.15 M | 0.38 M |
| 0.5 MobileNetV3-224 | 97.78% | 95.01% | 15.66 M | 7.83 M | 0.99 M |
| 0.75 MobileNetV3-224 | 98.81% | 95.64% | 34.16 M | 17.08 M | 1.98 M |
| 1.0 MobileNetV3-224 | 99.55% | 96.97% | 59.8 M | 29.90 M | 3.2 M |
Table 16.
A concrete representation of experiments on MobileNetV3 with resolutions.
Table 16.
A concrete representation of experiments on MobileNetV3 with resolutions.
| Models | Mean Test Accuracy | Mean F1-Score | FLOPs | MACC | # Parameters |
|---|
| 1.0 MobileNetV3-128 | 96.88% | 95.39% | 19.55 M | 9.77 M | 3.2 M |
| 1.0 MobileNetV3-160 | 99.08% | 95.78% | 30.48 M | 15.24 M | 3.2 M |
| 1.0 MobileNetV3-192 | 99.31% | 96.64% | 43.93 M | 21.97 M | 3.2 M |
| 1.0 MobileNetV3-224 | 99.55% | 96.97% | 59.8 M | 29.90 M | 3.2 M |
Table 17.
Performance evaluation trained on our dataset using S-modified MobileNet and test on different datasets using various optimizers.
Table 17.
Performance evaluation trained on our dataset using S-modified MobileNet and test on different datasets using various optimizers.
| Dataset | SGD | Adam | RMSprop |
|---|
| Rice dataset | 98.25% | 97.05% | 98.53% |
| Our PLD dataset | 99.31% | 99.39% | 99.55% |
Table 18.
Performance evaluation trained on our dataset using F-modified MobileNet and test on different datasets using various optimizers.
Table 18.
Performance evaluation trained on our dataset using F-modified MobileNet and test on different datasets using various optimizers.
| Dataset | SGD | Adam | RMSprop |
|---|
| Rice dataset | 90.65% | 92.25% | 95.53% |
| Our PLD dataset | 98.39% | 98.53% | 99.10% |
Table 19.
Comparison among some benchmark PLD recognition frameworks.
Table 19.
Comparison among some benchmark PLD recognition frameworks.
| References | Classes/Species | CNN Architecture | Fall in Accuracy | Computational Complexity | Memory Restriction | Accuracy |
|---|
| [6] | 58/25 | VGG | NR | NR | NR | 99.53% |
| [7] | 38/14 | GoogleNet | NR | NR | NR | 99.35% |
| [8] | 15/6 | Modified CaffeNet | NR | NR | NR | 96.30% |
| [11] | 2/1 | Custom | NR | NR | NR | 95.83% |
| [13] | 3/1 | Modified LeNet | NR | NR | NR | 92.88% |
| [14] | 9/1 | Two stage CNN | NR | NR | R | 93.3% |
| [18] | 4/1 | Modified AlexNet | R | NR | NR | 97.62% |
| [19] | 42/12 | ResNet152 | R | R | NR | 90.88% |
| [20] | 10/1 | F-CNN, S-CNN | R | NR | NR | 98.6% |
| [21] | 7/1 | Custom | NR | NR | NR | 90.16% |
| [28] | 9/1 | R-FCNN, ResNet50 | PR | NR | NR | 85.98% |
| [29] | 56/14 | GoogleNet | R | NR | NR | 94% |
| [30] | 10/1 | Custom | NR | NR | NR | 95.48% |
| [31] | 6/1 | DenseNet+RF | NR | NR | NR | 97.59% |
| Our work | 12/8 | S-modified MobileNet | R | R | R | 99.55% |