Abstract
There is an opinion in professional literature that edge-strands in β-sheet are critical to the processes of amyloid transformation. Propagation of fibrillar forms mainly takes place on the basis of β-sheet type interactions. In many proteins, the edge strands represent only a partially matched form to the β-sheet. Therefore, the edge-strand takes slightly distorted forms. The assessment of the level of arrangement can be carried out based on studying the secondary structure as well as the structure of the hydrophobic core. For this purpose, a fuzzy oil drop model was used to determine the contribution of each fragment with a specific secondary structure to the construction of the system being the effect of a certain synergy, which results in the construction of a hydrophobic core. Studying the participation of β-sheets edge fragments in the hydrophobic core construction is the subject of the current analysis. Statuses of these edge fragments in β-sheets in ferredoxin-like folds are treated as factors that disturb the symmetry of the system.
1. Introduction
The process of amyloid transformation is associated with the reproduction of structural forms of β-structure type [1]. The helical structure does not create conditions for propagation. On the one hand, the native protein structure guarantees stability but also prevents the formation of complexes in an uncontrolled manner [2]. In the proteins’ activity, there is a need for complexation, necessary for their activity, whose control protects against uncontrolled aggregation [3]. Amyloid transformation can be caused by the presence of mutations, but it also occurs with respect to the native form of proteins [4].
The activity of proteins is also accompanied by the possibility of dynamic structure change. The question concerns the limits of those changes that do not allow pathological changes [5,6]. Another condition that guarantees the activity of proteins is their solubility [7]. The critical external factor is the proper temperature [8,9]. In the light of all these conditions that may be relevant for aggregation, including uncontrolled aggregation, the status of the so-called edge β-strands in the β-sheets turns out to be critical. The investigation of the role of these fragments of polypeptide chain are studied in many works [10,11]. The role of these fragments was determined using targeted mutations introduced in edge-strands in the β-sheets [10,12,13].
In the present work, the assessment of the predisposition for aggregation (including the amyloid transformation) of the examined proteins is carried out based on the analysis of the structure of the hydrophobic core. For this purpose, a fuzzy oil drop model was used, with which it is possible to assess the degree of arrangement within a given protein and to identify local deviations from idealized distributions expressing the centric hydrophobic core as it is expected in spherical micele [14,15]. The 3D Gauss function is adopted to express this type of hydrophobicity distribution.
The subject of the analysis is a set of two proteins in which the role of two edge Beta-stands in Beta-sheet is discussed in the context of their participation in the amyloid transformation [8,16]. The analyzed elements were Acylphosphatase Drosphila Melanogaster and Sulfolobus solfataricus as well as 4 forms of mutation of the second one. The introduction of appropriate mutations is aimed at checking how the sequence of edge fragments influences dimer generation, which is a way of preventing complexation of more chains leading to the formation of an amyloid form.
2. Materials and Methods
2.1. Data
The subject of the analysis are acylphosphatase derived from Drosophila melanogaster (called AclDM in this paper) and Sulfolobus solfataricus (called AclSS in this paper) [8,16]. These proteins represent topology βαββαβ [8,11,17,18,19,20,21]. The unstructured N-terminal fragment with a length of 11 aa is absent in the AclSS structure (2BJD) in the form available in PDB—here absent in the pdb structure [8]. This unstructured segment by interacting with another molecule of this protein promotes the formation of amyloid-like structures. However, this enzyme is not identified as misfolded in any pathological process. The analysis focuses on the role of edge fragments—β-sheet components due to the possibility of interaction resulting in the formation of amyloid-like complexes.
Biotechnological modification of these edge-fragments aimed at protection against amyloid-like aggregation enabled the identification of the role of these fragments [22].
In comparison with SS, Dro2 shows much lower protection against the generation of amyloid-like forms [23]. The AclDM protein undergoes amyloid transformation in the presence of small amounts of 2,2,2-trifluoroethanol [22]. Using the edge-fragment modification of acylphosphataseSS, conditions for native-like aggregation in the form of dimers were checked as a way to prevent the formation of amyloid-like aggregates. Therefore, forms of this protein with the introduced mutations [10] are also the subject of the analysis. The analysis was carried out on the basis of structures available in PDB—as given in Table 1 [24].

Table 1.
Proteins constituting the subject of the analysis. Brief characteristics of these proteins are given. Secondary structure according to CATH (Class Architecture Topology/fold Homologous superfamily [25,26].
2.2. Fuzzy Oil Drop Model
This model has already been described in detail in the literature [14,15]. The assessment of the status of a given protein from the point of view of the hydrophobicity distribution is carried out based on the measurement of the degree of similarity of the distribution observed (O) in the protein molecule to the idealized distribution (T) expressed by the 3D Gauss function. The comparison of these two distributions, of which the T distribution is a reference distribution, is carried out using Kullback–Leibler entropy [27]. Interpretation of the divergence entropy value is possible after entering the second reference distribution (R). The R distribution is a uniform distribution where the level of hydrophobicity assigned to the residues is equal for all residues present in the protein and is equal to 1/N, where N is the number of amino acids in the chain. An RD (relative distance) parameter with a value lower than 0.5 means the proximity of the O and T distributions. Otherwise, the O distribution is similar to the R distribution.
The RD parameter can be used to evaluate both the hydrophobic core status of an entire protein molecule and the status of a selected fragment of the polypeptide chain in a given protein. In both cases, the calculation of the divergence entropy value is preceded by the operation of normalizing the distribution of T, O, and R.
A detailed description of the fuzzy oil drop model is available in [28].
In the present work, the discussed proteins will be analyzed making the status of the hydrophobic core a criterion for expressing the presence of a synergistic system covering all residues in the protein. It becomes possible to analyze the impact of the introduced mutations on the structure of a common hydrophobic core both in the monomer and dimers that these proteins form.
The value of the RD parameter is quite consistent with the value of the correlation coefficient for the relationship Ti to Oi. The high correlation value between the last two parameters can also be a measure of the compatibility of the O distribution with the T distribution. The higher RD the lower correlation coefficient can be seen in tables given in this paper. The very short fragments (like 3 aa) can be the exceptions. Values of correlation coefficient are given in the tables describing the status of discussed proteins and fragments with a specific secondary structure.
3. Results
A summary of the obtained RD values of monomers of the analyzed proteins as well as selected fragments of polypeptide chains is summarized in Table 2. including fragments representing sections with a specific secondary structure and immediate surroundings of catalytic residues.

Table 2.
RD parameter values for the whole molecule and for the selected fragments. Values close to are 0.5 underlined. CC—correlation coefficient for Ti versus Oi relation. Bold—lack of accordance between Ti and Oi.
The arrangement of β-structural segments is not identical in both compared proteins, although the high degree of similarity of the tertiary structure (often referred to as ferredoxin-like) expressed the same classification in CATH [25,26] justifies the possibility of comparative analysis. Interpretation of the results presented in Table 2 means that the compared proteins show a relatively high degree of similarity in the distribution of T and O. This, in turn, means that both proteins have a centrally located hydrophobic core and the surface of the proteins is covered with polar residues. Visualization of the relationship between T and O distributions reveals both the great similarity of these two profiles but also the local inconsistencies (Figure 1).
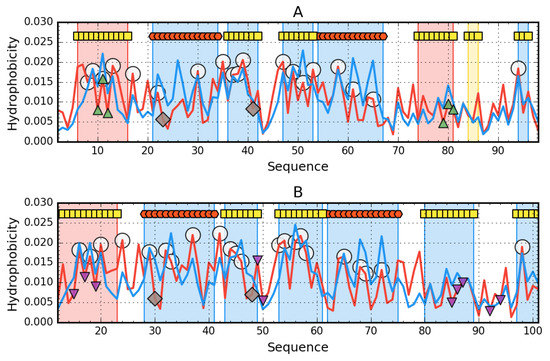
Figure 1.
Hydrophobicity density distribution profiles for 1URR (A) and chain A of 2BJD (B): theoretical (T—blue line) and observed (O—red line). Markers at the top denote secondary structure fragments (red hexagons—helices, yellow squares—sheets). Fragments marked by red shade exhibit RD (relative distance) > 0.5. Fragments marked by blue shade exhibit RD ≤ 0.5. Yellow region on A—RD close to 0.5. Green triangle markers (facing up) denote residues engaged in ligand binding. Purple triangles (facing down) denote protein-protein interaction. Catalytic residues are shown as brown rhombuses, while members of the hydrophobic core are marked by white circles.
The AclSS status (2BJD) shows the compatibility of the expected and observed distribution. Only 12-23 β-strand exhibits maladjustment. Residues involved in interacting with the second chain show the status compatible with the expected. This means that the presence of the second chain did not affect the arrangement within the monomer. The status of catalytic residues and their immediate environment shows an arrangement deviating from the expected one, although 48N slightly exceeds the critical value of 0.5. The 30R catalytic residue in AclSS has a status comparable to that of 23R in AclDM 1URR.
A low RD value for the whole molecule does not preclude local mismatch, as revealed by high RD values for chain fragments. Status of β-strand 6-16 shows locally high non-compatibility with the expected distribution. Similarly, β-strand 74-81 reveals a different hydrophobicity distribution than expected. The fragment 84-86 was also distinguished, whose status minimally exceeds the limit value of 0.5, which entitles the qualification of this fragment also to the status of non-compatible. The location of the highlighted fragments in the 3D presentation reveals the role of the discussed fragments as edge segments.
The N-terminal fragment in AclDM (1URR) has a local excess of hydrophobicity, which may potentially predispose it to interact with another protein or chain. Similarly, the edge segment (74-81 together with 84-86) shows different distribution, exposing high hydrophobicity in places where rather low level is expected. The status of catalytic residues and their immediate surroundings is varied. The surroundings of R23 is clearly not adapted to the expected distribution while that of N41 is well adapted to the expected distribution (Figure 2). The β-structured fragment in AclSS (2BJD), showing a different status than expected, is not the edge one. Its status is comparable to that in AclDM (1URR).
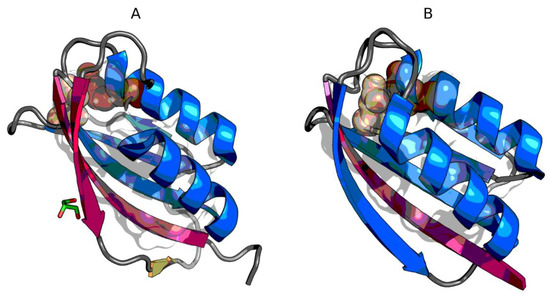
Figure 2.
3D presentation of 1URR (A) and chain A of 2BJD (B). Fragments marked by red color exhibit RD > 0.5. Fragments marked by blue color exhibit RD ≤ 0.5. Yellow fragment on A—RD close to 0.5. Catalytic residues are shown as spheres (darker color—23R in 1URR and 30R in 2BJD, lighter color—41N in 1URR and 48N in 2BJD). Member of the hydrophobic core are represented by semi-transparent surface inside each protein.
From the point of view of the structure of the hydrophobic core, the stability of AclSS (2BJD) turns out to be higher (assuming the presence of a hydrophobic core as a stabilizing factor) than AclDM 1URR. An additional difference is the fact that the edge fragment 74-86 in AclDM 1URR is unstable. If the assumption about the decisive role of edge fragments of β-plates in maintaining the stability of the whole protein is correct, then the AclDM protein should have a greater predisposition to structural changes than the AclSS protein.
3.1. The Presence of Mutations Limiting the Predisposition to Aggregation of AclSS
It should be noted that all the newly introduced amino acids in the area of the edge segment reduce the hydrophobicity of this segment (Table 3).

Table 3.
Mutant status and their secondary fragments. Positions with status greater than 0.5 for RD values in bold. Fragments with mutation present—underlined. Mutant—the position expressing status for the mutation position and the immediately adjacent segment from i-5 aa to i+5 aa. For the AclSS form (2BJD) it is the segment 79−91 covering both mutation positions. CC—correlation coefficient for Ti versus Oi relation.
The introduction of the mutation in all cases resulted in an increase in RD value, however slightly. In all versions (native and in mutants) the status of the N-terminal segment shows maladjustment to the expected distribution. Additional β-structural fragments appear in the mutants showing maladjustment status. However, this does not apply to the edge segment, which in all the discussed versions has the status of a high compatibility of the T and O distribution.
Characteristic for the Y86E and V84P forms is the additional maladjustment of the hydrophobicity distribution within the 28-41 helix. The β-structure fragment 43-49 also changes its status to showing maladjustment. The status of the complete β-sheet shows adaptation to the expected distribution, although in the case of V84D and Y86E some increase in the value of RD is noted. Profiles visualizing T and O distributions for the discussed mutants are given in Figure 3, where local differences and deviations from the T distribution are visible.
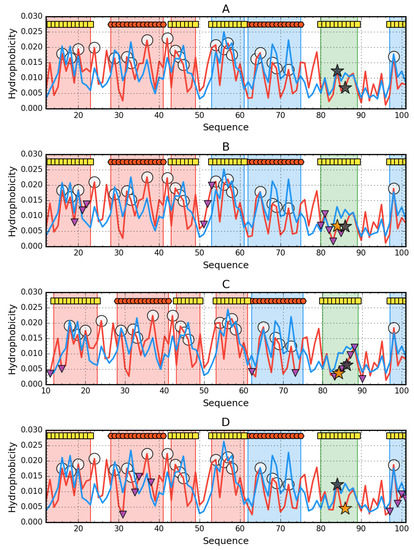
Figure 3.
Hydrophobicity density distribution profiles for monomers of 4OIX (A), 4OJ3 (B), 4OJG (C), 4OJH (D): theoretical (T—blue line) and observed (O—red line). Markers at the top denote secondary structure fragments (red hexagons—helices, yellow squares—sheets). Fragments marked by red shade exhibit RD > 0.5. Fragments marked by blue shade exhibit RD ≤ 0.5. Green fragments—β-strand with mutations. Purple triangles (facing down) denote protein-protein interaction. Orange stars mark the mutation in given protein, while black stars mark the mutations in other proteins.
Conclusions from the profiles in relation to the fragment 80-89 reveals a local hydrophobicity deficit that takes various forms. This is due to the introduction of residues with lower hydrophobicity compared to the native structure.
Visualization of the location of β-structured sections with the status RD > 0.5 is given in Figure 4.
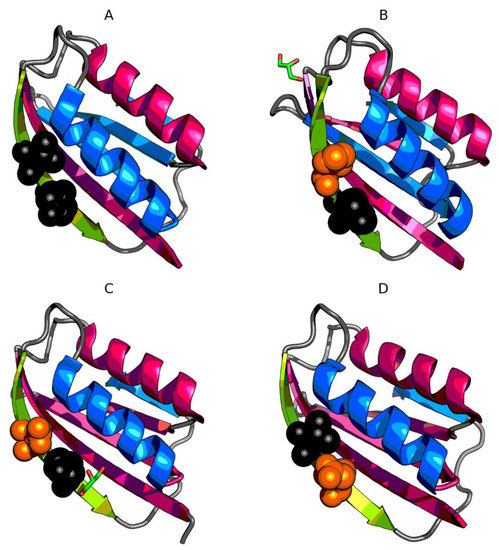
Figure 4.
3D presentation of monomers of 4OIX (A), 4OJ3 (B), 4OJG (C), 4OJH (D). Fragments marked by red color exhibit RD > 0.5. Fragments marked by blue color exhibit RD ≤ 0.5. Green fragments—β-strand with mutations. Orange spheres mark the mutation in given protein, while black spheres mark the mutations in other proteins.
3.2. The Status of Edge β-Strands in Complexes
The interface status in the complex showing RD < 0.5 means that interface is involved in the construction of the common core. If among the residues in the interface there are those belonging to the fragment of an edge, it means that the discussed edge fragment is involved in complexing the second chain. The only symmetrical form of the dimer was generated by V84D, where the interface has the form of propagating β-sheet. Dimer symmetry is visible on the profiles of Figure 5. Table 4 summarizes the parameters characterizing the edge fragment and its contribution to the interface structure. P–P means the protein–protein interaction zone, which was defined based on the distance criterion.
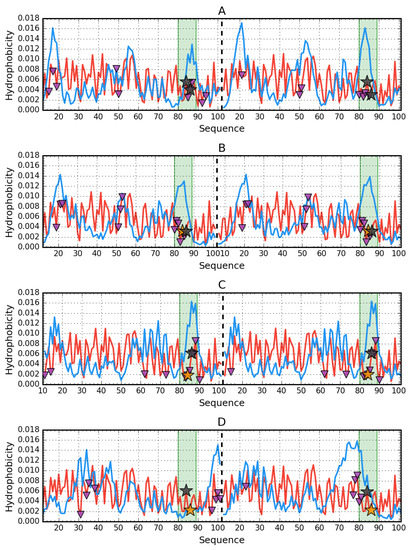
Figure 5.
Hydrophobicity density distribution profiles for dimers of 2BJD (A), 4OJ3 (B), 4OJG (C), 4OJH (D): theoretical (T—blue line) and observed (O—red line). Green fragments—β-strand with mutations. Purple triangles (facing down) denote protein-protein interaction. Orange stars mark the mutation in given protein, while black stars mark the mutations in other proteins. Dashed vertical line is the chain separator.

Table 4.
Status of the complete complex, interface (protein–protein interaction, P–P), non-interface (No P–P) part of the complex, and edge-fragment status in complex. Status RD < 0.5 is underlined.
Analysis of the results presented in Table 4 reveals that the V84D dimer (4OJG) was created by generating a common hydrophobic core (very low RD value for P–P interaction). The RD values characterizing the edge sections in this complex also show values below 0.5, which means that their status is also compatible with the expected hydrophobicity distribution for the complex, where the common hydrophobic core should be located in the central part. The significant part of residues included in the edge fragments in the interface suggests that in the case of the V84D (4OJG) edge fragments are involved in the interface construction. The central hydrophobic core is mainly generated by the proximity of two Phe88 and Met16, as seen in the profiles of Figure 5.
The parameter set for V86E (4OJH) reveals the asymmetrical arrangement of monomers in the dimer due to the significant differences in the status of the edge fragment in both chains. The minimum number of residues of this fragment is involved in creating the interface, whose status is far from the ordering as expected for the complex (similarly in V84P (4OJ3)). None of these complexes show the presence of a common hydrophobic core for the dimer (high RD values for the complexes).
Summing up the results contained in Table 4. it can be stated that none of the dimers produced by the mutants recreated the structural form present in the native AclSS protein (2BJD). Have structures with less predisposition to aggregation than represented by AclSS (2BJD) been obtained? From the point of view of fuzzy oil drop analysis, only the V84D (4OJG) model shows a complexing mechanism based on hydrophobic interaction. The authors [10], introducing residues with lower hydrophobicity, expected the participation of charges in the interaction between monomers. In V86E (4OJH) edge, the fragment does not participate at all in creating the interface. In contrast, complexing in V84D (4OJG) is based on a hydrophobic interaction. Despite the introduction of a residue with a much lower intrinsic hydrophobicity, the interface status turns out to be compatible with the centric hydrophobic core. The proportion of this type of interaction appears to be higher than in the complex formed by two native monomers. In the construction of the interface in 4OJH (Y86E), a small number of residues originating from the edge fragment takes part.
V84 is rather involved in the formation of the core within the monomer (directed towards the center of the monomer). Y86 is rather exposed to the aquatic environment. Y86 is included in the interface only in the A chain (Figure 6).
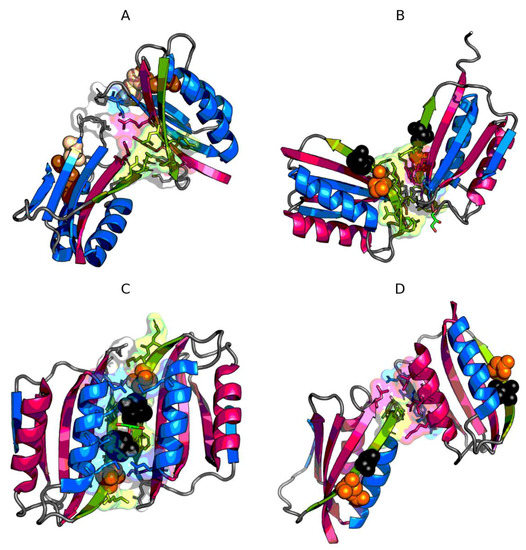
Figure 6.
3D presentation of dimers of 2BJD (A), 4OJ3 (B), 4OJG (C), 4OJH (D). Fragments marked by red color exhibit RD > 0.5. Fragments marked by blue color exhibit RD ≤ 0.5. Green fragments—β-strand with mutations. Orange spheres mark the mutation in given protein, while black spheres mark the mutations in other proteins. Residues engaged in protein-protein interaction have their side chains displayed as sticks and are surrounded by a semi-transparent surface.
3.3. Presence of Trifluoroethanol (TFE)
The low concentration (5% vol / vol) of trifluoroethanol (TFE) is enough for the AclDM (1URR) form to undergo rapid amyloid transformation [7]. This phenomenon based on the fuzzy oil drop model is interpreted as the effect of the influence of the presence of factors (as in this case TFE) on the structuring of water, which provides an external field for the process of folding polypeptide chains leading to the appearance of the native form of the protein. The amyloid form may be the result of a different structuring of water in the presence of such compounds as urea, which promotes the unfolding of the polypeptide chain. The adjustment of the hydrophobicity distribution to 3D Gauss function for globular proteins and in the amyloid proteins—to 2D Gauss function can be regarded as the effect of such a change in the field characteristics generated by water.
For some reason a single chain selects the arrangement expressed by a 2D Gaussian hydrophobicity distribution, where one of sigma parameters for one of the directions in three-dimensional space is equal to zero (say σz → 0 for the orientation of the amyloid fibril, where the long axis of the fibril coincides with the Z-axis). Aggregation of many molecules into fibril forms a σz → ∞ system that strives for infinity. However, the fibril system is a consequence of 2D Gaussian structure for a single polypeptide chain [28,29,30].
4. Discussion
In the present study, the assessment of stability and potential aggregation abilities was carried out from the point of view of the characteristics of the hydrophobic core treated (see biochemistry textbooks) as a factor stabilizing tertiary structure. The experimental study aimed to demonstrate the role of β-edge in the potential for transformation, including amyloid AclDM (1URR). The focus was also on the analysis of the presence of the characteristic bulkier in the edge [10]. In the amyloid-like aggregation process of these proteins, segment 1-11 is important, which is not present in the crystal structures [10]. That treatment in [10] was aimed at highlighting the characteristics of the edge fragment, including the characteristic role of bulkier on aggregation abilities. Therefore, experimental research revealing the effect of introduced mutations within the edge fragment allowed to indicate the role of this fragment. Taking the structure of the hydrophobic core as a criterion for this stability, the monomeric forms of these proteins prove to be stable. In the AclDM (1URR) structure, the edge-β-sheet segment shows incompatibility to the core centric system in contrast to AclSS (2BJD), where its status is consistent with idealized distribution. The characteristic status of catalytic residues and their immediate surroundings shows incompatible distribution [31].
Despite the low RDs for the whole molecules, local disorder of distribution is observed. The number of β-strands varies from 1 to 3. In mutants, however, they are located in the central part of the β-sheet. Edge fragment with status incompatible with idealized distribution is present in AclDM (1URR). The aim of analysis is also the estimation of dimer stability. The ability to generate them promotes the method of preventing aggregation (amyloid forms). This means that no common core was generated in the dimers (RD values> 0.5 for dimers) (Table 4). On the other hand, in V84D, the status of the interface itself turns out to be as expected. This means that this version generates a common hydrophobic core. Edge sections take part in its construction, forming a continuing β-sheet system. In the construction of this core, Phe88 and Met16 have a large contribution. The status of the whole dimer (RD > 0.5) results from the disorder of the rest of the chains.
The contribution of TFE as a factor favoring amyloid transformation [10] seems to support the conclusions based on fuzzy oil drop model. The presence of TFE is assumed to rather influence the unknown structuralization of water playing the role of external force field for folding protein. According to the fuzzy oil drop model, it is also assumed that the aqueous environment (including 0.9% NaCl) generates an appropriate external field directing the process towards the concentration of hydrophobic residues in the center of the molecule with simultaneous exposure of hydrophilic residues on the surface. The disturbance of water structuring through for example shaking or the presence of factors like TFE—changes its interaction with the folding chain leading to a different type of synergy. Instead of the centric hydrophobic core expressed by means of 3D Gaussian functions, an arrangement is generated, which for a single chain being a component of fibril, adopts that compatible with 2D Gauss’s function. All amyloids available to date represent a flat system with a centrally located hydrophobic core [19]. Most likely, the presence of TFE has just such an influence. Other external factors, including shaking (increasing the presence of the water/air interphase system) affect water structuralization and the external field generated by it directs the folding process in a direction other than the native [32,33,34].
The phenomenon of solubility requires the detailed explanation even for simple molecules like monosacharides [35]. The phenomenon of protein folding shall take into consideration the mutual influence of water environment and soluble molecule.
5. Conclusions
This paper shows the role of edge β-strands in β-sheets relying in introduction a disorder of the ordering form present in β-sheets. The consequence of this arrangement is stopping the propagation present in an ordered β-sheet form, which theoretically can be continued unlimitedly.
The introduced mutations showed the highest efficiency of the V84D (4OJG) case, where the stability of the hydrophobic core turns out to be the highest. The interface consists of edge sections creating a joint continuation of the β-sheet, however limited to dimer. The freedom of amyloid transformation in the case of AclDM (1URR) in the presence of TFE is rather due to the effect of this factor on water structuring.
Author Contributions
Conceptualization, I.R. and L.K.; methodology, M.B.; software, M.B.; validation, I.R., formal analysis, K.S., MP-K; investigation, I.R.; resources, M.B.; data curation, P.F. M.P.-K.; writing—original draft preparation, I.R.; writing—review and editing, I.R. and M.B.; visualization, M.B.; supervision, I.R.; project administration, M.B.; funding acquisition, I.R. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Jagiellonian University—Medical College, grant number N41/DBS/000568.
Acknowledgments
Many thanks to Anna Zaremba-Śmietańska and Zdzisław Wiśniowski for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Selkoe, D. Folding proteins in fatal ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef]
- Richardson, J.S.; Richardson, D.C. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA 2002, 99, 2754–2759. [Google Scholar] [CrossRef]
- Bemporad, F.; Calloni, G.; Campioni, S.; Plakoutsi, G.; Taddei, N.; Chiti, F. Sequence and Structural Determinants of Amyloid Fibril Formation. Accounts Chem. Res. 2006, 39, 620–627. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Amyloid formation by globular proteins under native conditions. Nat. Methods 2008, 5, 15–22. [Google Scholar] [CrossRef]
- Bahar, I.; Cheng, M.H.; Lee, J.Y.; Kaya, C.; Zhang, S. Structure-Encoded Global Motions and Their Role in Mediating Protein-Substrate Interactions. Biophys. J. 2015, 109, 1101–1109. [Google Scholar] [CrossRef]
- Ferrolino, M.C.; Zhuravleva, A.; Budyak, I.L.; Krishnan, B.; Gierasch, L.M. Delicate Balance between Functionally Required Flexibility and Aggregation Risk in a β-Rich Protein. Biochemistry 2013, 52, 8843–8854. [Google Scholar] [CrossRef] [PubMed]
- De Simone, A.; Dhulesia, A.; Soldi, G.; Vendruscolo, M.; Hsu, S.-T.D.; Chiti, F.; Dobson, C.M. Experimental free energy surfaces reveal the mechanisms of maintenance of protein solubility. Proc. Natl. Acad. Sci. USA 2011, 108, 21057–21062. [Google Scholar] [CrossRef]
- Corazza, A.; Rosano, C.; Pagano, K.; Alverdi, V.; Esposito, G.; Capanni, C.; Bemporad, F.; Plakoutsi, G.; Stefani, M.; Chiti, F.; et al. Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus. Proteins: Struct. Funct. Bioinform. 2005, 62, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Plakoutsi, G.; Bemporad, F.; Monti, M.; Pagnozzi, D.; Pucci, P.; Chiti, F.; Pucci, P. Exploring the Mechanism of Formation of Native-like and Precursor Amyloid Oligomers for the Native Acylphosphatase from Sulfolobus solfataricus. Structure 2006, 14, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, M.; Bemporad, F.; Pellegrino, S.; Chiti, F.; Bolognesi, M.; Ricagno, S. Edge strand engineering prevents native-like aggregation in Sulfolobus solfataricus acylphosphatase. FEBS J. 2014, 281, 4072–4084. [Google Scholar] [CrossRef]
- Cheung, Y.-Y.; Lam, S.Y.; Chu, W.-K.; Allen, M.D.; Bycroft, M.; Wong, K.-B. Crystal Structure of a Hyperthermophilic Archaeal Acylphosphatase fromPyrococcus horikoshiiStructural Insights into Enzymatic Catalysis, Thermostability, and Dimerization†, ‡. Biochemistry 2005, 44, 4601–4611. [Google Scholar] [CrossRef] [PubMed]
- Bemporad, F.; De Simone, A.; Chiti, F.; Dobson, C.M. Characterizing Intermolecular Interactions That Initiate Native-Like Protein Aggregation. Biophys. J. 2012, 102, 2595–2604. [Google Scholar] [CrossRef] [PubMed]
- Chiti, T.; Dobson, C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Konieczny, L.; Brylinski, M.; Roterman, I. Gauss-function-Based model of hydrophobicity density in proteins. Silico Boil. 2006, 6, 15–22. [Google Scholar]
- Kalinowska, B.; Banach, M.; Konieczny, L.; Roterman, A.I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17, 1477–1507. [Google Scholar] [CrossRef]
- Zuccotti, S.; Rosano, C.; Ramazzotti, M.; Degl’Innocenti, D.; Stefani, M.; Manao, G.; Bolognesi, M. Three-dimensional structural characterization of a novel Drosophila melanogaster acylphosphatase. Acta Crystallogr. Sect. D Boil. Crystallogr. 2004, 60, 1177–1179. [Google Scholar] [CrossRef]
- Fusco, G.; De Simone, A.; Hsu, S.-T.D.; Bemporad, F.; Vendruscolo, M.; Chiti, F.; Dobson, C.M. 1H, 13C and 15N resonance assignments of human muscle acylphosphatase. Biomol. NMR Assign. 2011, 6, 27–29. [Google Scholar] [CrossRef]
- Pagano, K.; Ramazzotti, M.; Viglino, P.; Esposito, G.; Degl’Innocenti, D.; Taddei, N.; Corazza, A. NMR solution structure of the acylphosphatase from Escherichia coli. J. Biomol. NMR 2006, 36, 199–204. [Google Scholar] [CrossRef]
- Pastore, A.; Saudek, V.; Ramponi, G.; Williams, R.J. Three-dimensional structure of acylphosphatase. J. Mol. Boil. 1992, 224, 427–440. [Google Scholar] [CrossRef]
- Thunnissen, M.M.; Taddei, N.; Liguri, G.; Ramponi, G.; Nordlund, P. Crystal structure of common type acylphosphatase from bovine testis. Structure 1997, 5, 69–79. [Google Scholar] [CrossRef]
- Zuccotti, S.; Rosano, C.; Bemporad, F.; Stefani, M.; Bolognesi, M. Preliminary characterization of two different crystal forms of acylphosphatase from the hyperthermophile archaeon Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Boil. Cryst. Commun. 2004, 61, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Soldi, G.; Plakoutsi, G.; Taddei, N.; Chiti, F. Stabilization of a Native Protein Mediated by Ligand Binding Inhibits Amyloid Formation Independently of the Aggregation Pathway. J. Med. Chem. 2006, 49, 6057–6064. [Google Scholar] [CrossRef] [PubMed]
- Bemporad, F.; Ramazzotti, M. From the Evolution of Protein Sequences Able to Resist Self-Assembly to the Prediction of Aggregation Propensity. In International Review of Cell and Molecular Biology; Elsevier BV: Amsterdam, The Netherlands, 2017; pp. 1–47. [Google Scholar]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Homepage of CATH Database. Available online: http://www.cathdb.info/ (accessed on 4 May 2011).
- Dawson, N.L.; Lewis, T.E.; Das, S.; Lees, J.G.; Lee, D.; Ashford, P.; Orengo, C.A.; Sillitoe, I. CATH: An expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2016, 45, D289–D295. [Google Scholar] [CrossRef]
- Kullback, S.; Leibler, R.A. On Information and Sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Banach, M.; Fabian, P.; Stapor, K.; Konieczny, L.; Roterman, I. Structure of the Hydrophobic Core Determines the 3D Protein Structure—Verification by Single Mutation Proteins. Biomolecules 2020, 10, 767. [Google Scholar] [CrossRef]
- Banach, M.; Konieczny, L.; Roterman, A.I. The Amyloid as a Ribbon-Like Micelle in Contrast to Spherical Micelles Represented by Globular Proteins. Molecules 2019, 24, 4395. [Google Scholar] [CrossRef]
- Fabian, P.; Banach, M.; Stapor, K.; Konieczny, L.; Roterman, I. Structure of amyloid versus structure of globular proteins. Int. J. Mol. Sci. 2020, in press.
- Prymula, K.; Jadczyk, T.; Roterman, A.I. Catalytic residues in hydrolases: Analysis of methods designed for ligand-binding site prediction. J. Comput. Mol. Des. 2010, 25, 117–133. [Google Scholar] [CrossRef]
- Mohanta, D.; Jana, M. Can 2,2,2-trifluoroethanol be an efficient protein denaturant than methanol and ethanol under thermal stress? Phys. Chem. Chem. Phys. 2018, 20, 9886–9896. [Google Scholar] [CrossRef]
- Khan, M.V.; Rabbani, G.; Ishtikhar, M.; Khan, S.; Saini, G.; Khan, R. Non-fluorinated cosolvents: A potent amorphous aggregate inducer of metalloproteinase-conalbumin (ovotransferrin). Int. J. Boil. Macromol. 2015, 78, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Molla, A.R.; Mandal, D.K. Trifluoroethanol-induced conformational change of tetrameric and monomeric soybean agglutinin: Role of structural organization and implication for protein folding and stability. Biochim. 2013, 95, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Jäntschi, L. Structure–property relationships for solubility of monosaccharides. Appl. Water Sci. 2019, 9, 38. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).