Exploration and Modulation of Antibody Fragment Biophysical Properties by Replacing the Framework Region Sequences
Abstract
:1. Introduction
2. Results
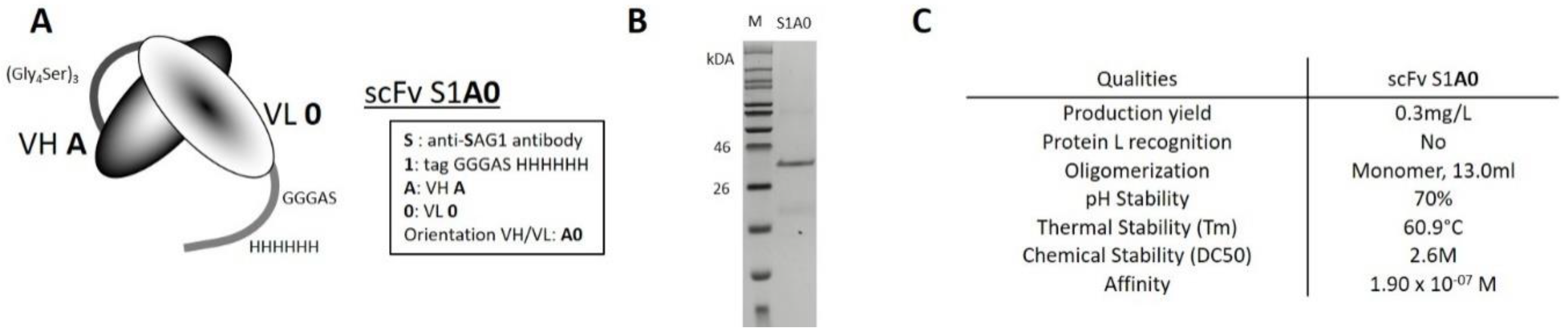
2.1. Biophysical Properties of Wild-Type scFv
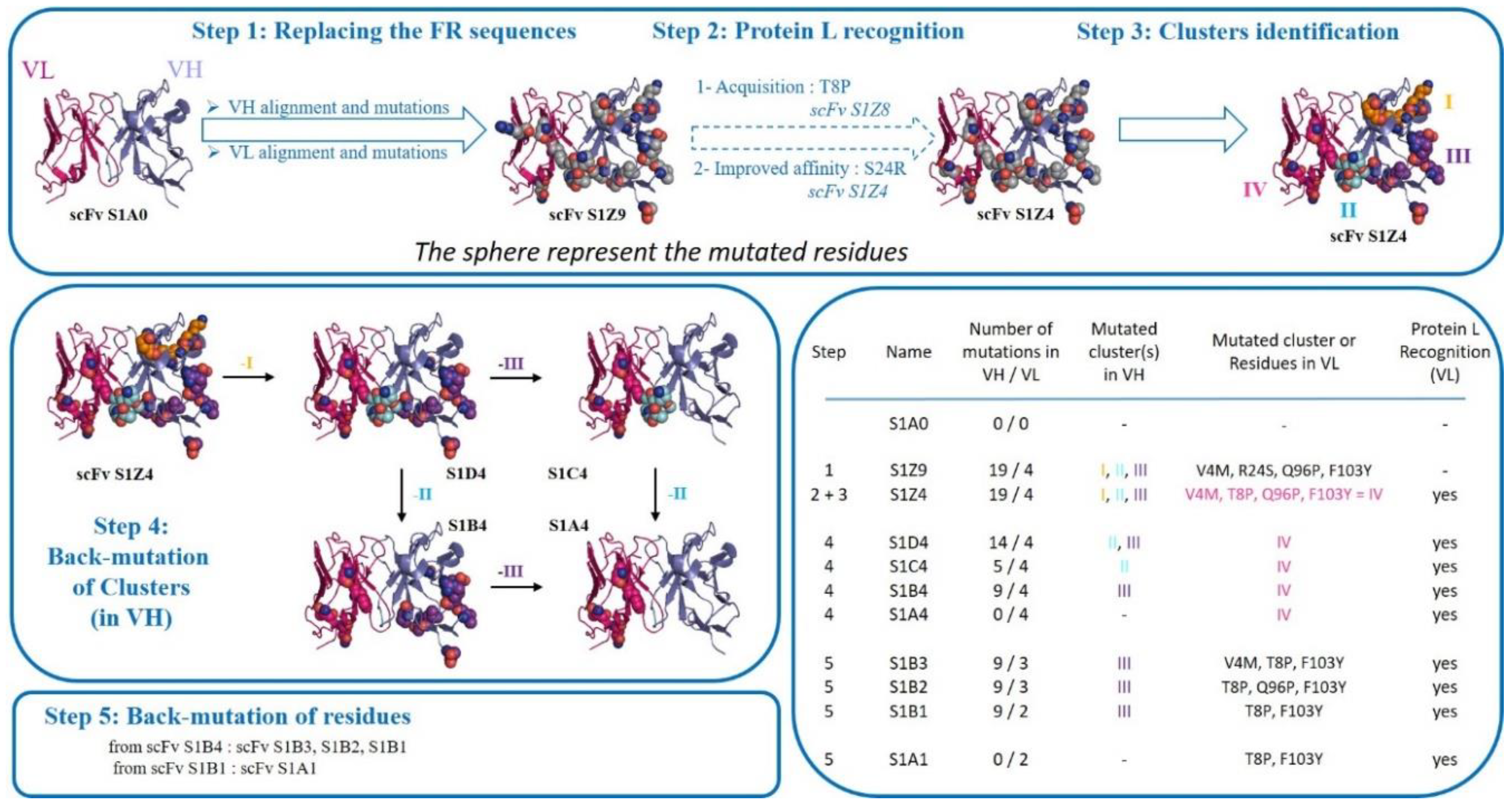
2.2. Methodology for Replacing the Framework (FR) Sequences
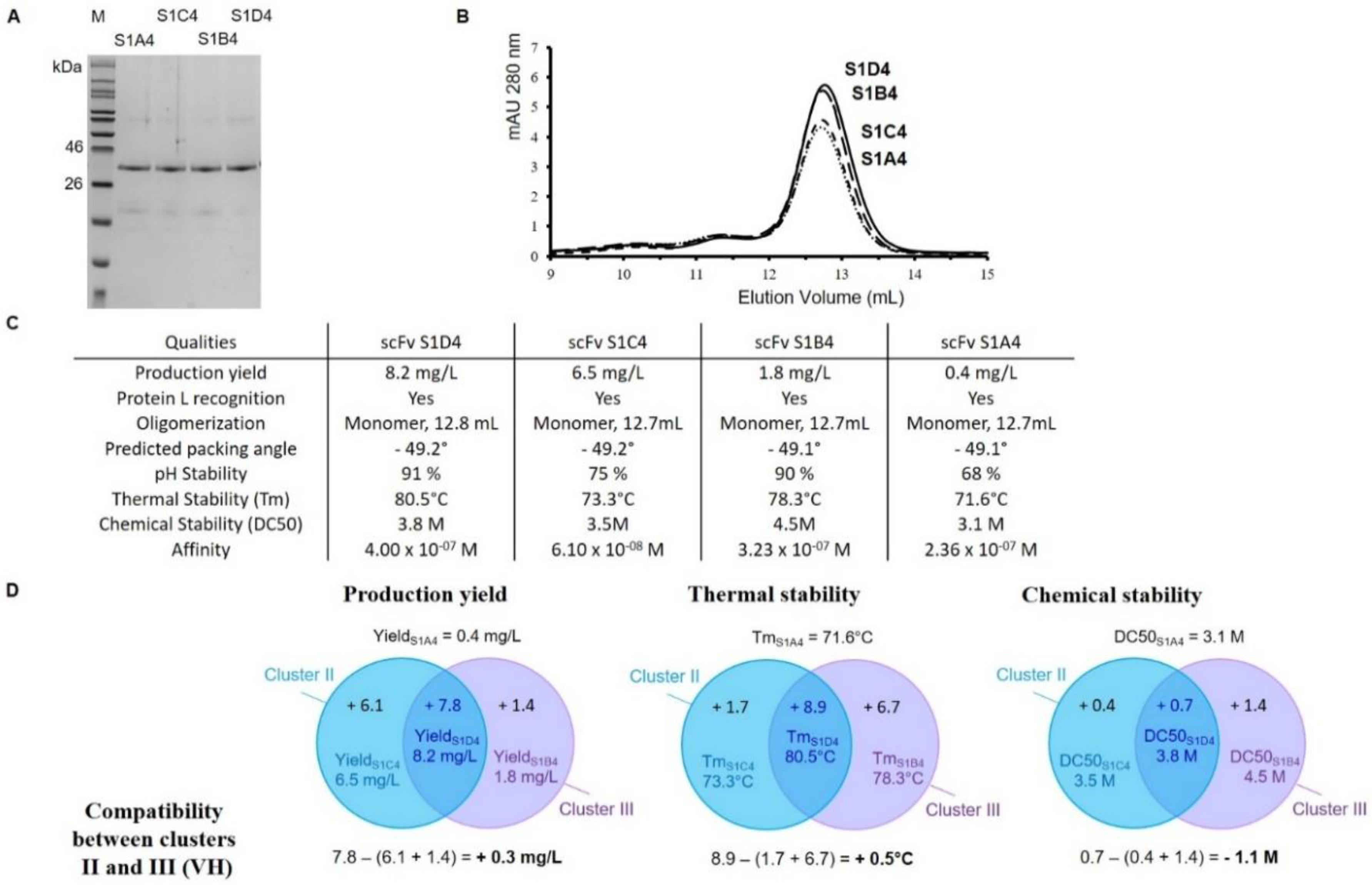
2.3. Functional Analysis
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Biochemical Characterization and scFv Integrity Analysis
4.3. Determination of Thermal and Chemical Stabilities
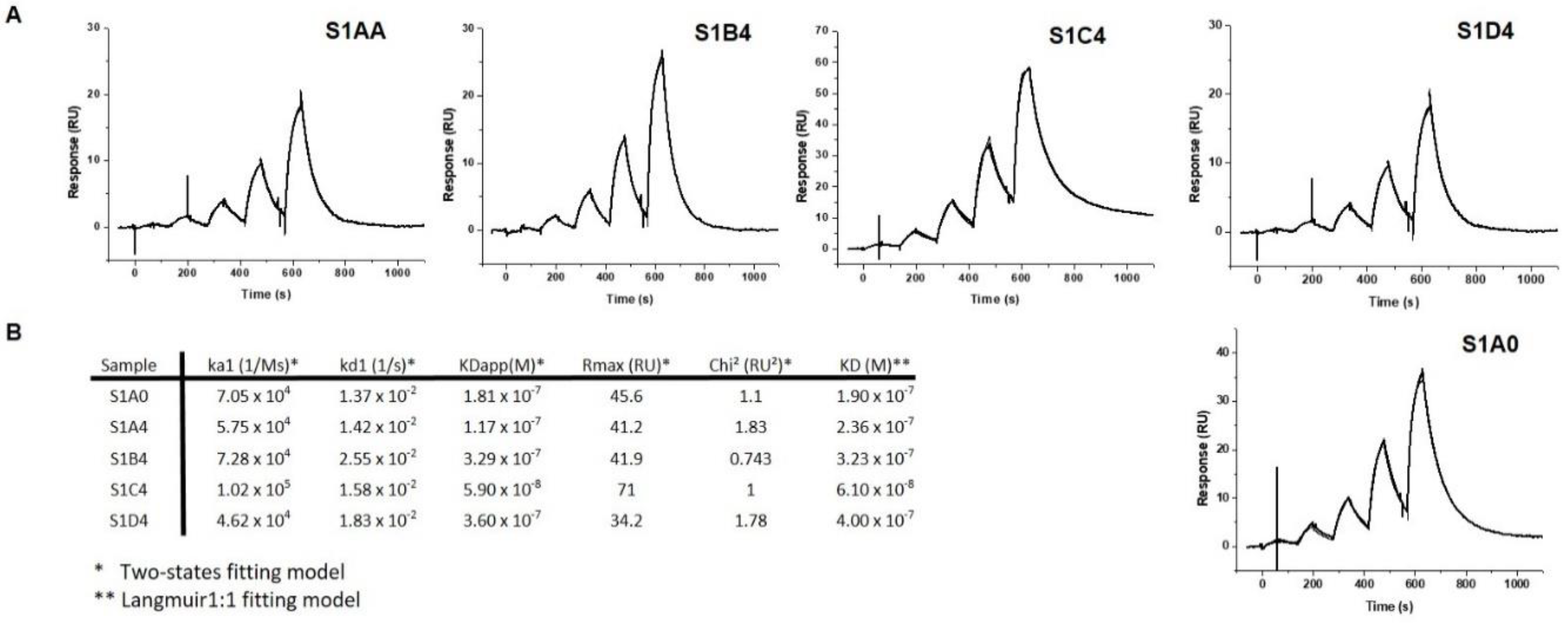
4.4. Affinity Analysis by Surface Plasmon Resonance
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations/Acronyms
| AA | Amino acid |
| CDR | complementary-determining regions |
| FR | Framework |
| Gly | glycine |
| GdnHCl | guanidinium chloride |
| PBS | phosphate-buffered saline |
| scFv | single-chain antibody variable fragment |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SEC | size exclusion chromatography |
| SPR | surface plasmon resonance |
| Tm | midpoint temperature |
| VH | heavy-chain variable |
| VL | light-chain variable |
| WT | Wild-type |
References
- Spiess, C.; Zhai, Q.; Carter, P.J. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol. Immunol. 2015, 67, 95–106. [Google Scholar] [CrossRef]
- Brinkmann, U.; Kontermann, R.E. The making of bispecific antibodies. MAbs 2017, 9, 182–212. [Google Scholar] [CrossRef]
- Chiu, M.L.; Goulet, D.R.; Teplyakov, A.; Gilliland, G.L. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126–1136. [Google Scholar] [CrossRef]
- El-Sayed, A.; Bernhard, W.; Barreto, K.; Gonzalez, C.; Hill, W.; Pastushok, L.; Fonge, H.; Geyer, C.R. Evaluation of antibody fragment properties for near-infrared fluorescence imaging of HER3-positive cancer xenografts. Theranostics 2018, 8, 4856–4869. [Google Scholar] [CrossRef]
- Wu, S.J.; Luo, J.; O’Neil, K.T.; Kang, J.; Lacy, E.R.; Canziani, G.; Baker, A.; Huang, M.; Tang, Q.M.; Raju, T.S.; et al. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng. Des. Sel. 2010, 23, 643–651. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.C.; Perchiacca, J.M.; Tessier, P.M. Toward aggregation-resistant antibodies by design. Trends Biotechnol. 2013, 31, 612–620. [Google Scholar] [CrossRef]
- Lakhrif, Z.; Pugnière, M.; Henriquet, C.; Di Tommaso, A.; Dimier-Poisson, I.; Billiald, P.; Juste, M.O.; Aubrey, N. A method to confer Protein L binding ability to any antibody fragment. MAbs 2016, 8, 379–388. [Google Scholar] [CrossRef] [Green Version]
- Lebozec, K.; Jandrot-Perrus, M.; Avenard, G.; Favre-Bulle, O.; Billiald, P. Quality and cost assessment of a recombinant antibody fragment produced from mammalian, yeast and prokaryotic host cells: A case study prior to pharmaceutical development. New Biotechnol. 2018, 44, 31–40. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y.; Park, J.; Liu, X.; Hu, Y.; Wang, T.; McFarland, K.; Betenbaugh, M.J. Design and Production of Bispecific Antibodies. Antibodies 2019, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Carter, P.; Presta, L.; Gorman, C.M.; Ridgway, J.B.; Henner, D.; Wong, W.L.; Rowland, A.M.; Kotts, C.; Carver, M.E.; Shepard, H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA 1992, 89, 4285–4289. [Google Scholar] [CrossRef] [Green Version]
- Ducancel, F.; Muller, B.H. Molecular engineering of antibodies for therapeutic and diagnostic purposes. MAbs 2012, 4, 445–457. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Hua, C.; Sentman, C.L.; Ackerman, M.E.; Bailey-Kellogg, C. Antibody humanization by structure-based computational protein design. MAbs 2015, 7, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Duan, C.F.; Qi, Y.H.; Dong, J.; Wang, G.N.; Zhao, G.X.; Wang, J.P.; Liu, J. Virtual mutation and directional evolution of anti-amoxicillin ScFv antibody for immunoassay of penicillins in milk. Anal. Biochem. 2017, 523, 44–45. [Google Scholar] [CrossRef]
- Lebozec, K.; Jandrot-Perrus, M.; Avenard, G.; Favre-Bulle, O.; Billiald, P. Design, development and characterization of ACT017, a humanized Fab that blocks platelet’s glycoprotein VI function without causing bleeding risks. MAbs 2017, 9, 945–958. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.F.; Ho, M. Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: Rationale and examples. MAbs 2017, 9, 419–429. [Google Scholar] [CrossRef] [Green Version]
- Yui, A.; Akiba, H.; Kudo, S.; Nakakido, M.; Nagatoishi, S.; Tsumoto, K. Thermodynamic analyses of amino acid residues at the interface of an antibody B2212A and its antigen roundabout homolog 1. J. Biochem. 2017, 162, 255–258. [Google Scholar] [CrossRef]
- Sun, W.; Yang, Z.; Lin, H.; Liu, M.; Zhao, C.; Hou, X.; Hu, Z.; Cui, B. Improvement in affinity and thermostability of a fully human antibody against interleukin-17A by yeast-display technology and CDR grafting. Acta Pharm. Sin. B 2019, 9, 960–972. [Google Scholar] [CrossRef]
- Tu, C.; Terraube, V.; Tam, A.S.P.; Stochaj, W.; Fennell, B.J.; Lin, L.; Stahl, M.; LaVallie, E.R.; Somers, W.; Finlay, W.J.J.; et al. A combination of structural and empirical analyses delineates the key contacts mediating stability and affinity increases in an optimized biotherapeutmrtic single-chain Fv (scFv). J. Biol. Chem. 2016, 291, 1267–1276. [Google Scholar] [CrossRef] [Green Version]
- Miller, B.R.; Demarest, S.J.; Lugovskoy, A.; Huang, F.; Wu, X.; Snyder, W.B.; Croner, L.J.; Wang, N.; Amatucci, A.; Michaelson, J.S.; et al. Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng. Des. Sel. 2010, 23, 549–557. [Google Scholar] [CrossRef]
- Unkauf, T.; Hust, M.; Frenzel, A. Antibody Affinity and Stability Maturation by Error-Prone PCR. In Methods in Molecular Biology; Hust, M., Lim, T.S., Eds.; Springer New York: New York, NY, USA, 2018; Volume 1701, pp. 393–407. [Google Scholar]
- Hsu, H.J.; Lee, K.H.; Jian, J.W.; Chang, H.J.; Yu, C.M.; Lee, Y.C.; Chen, I.C.; Peng, H.P.; Wu, C.Y.; Huang, Y.F.; et al. Antibody variable domain interface and framework sequence requirements for stability and function by high-throughput experiments. Structure 2014, 22, 22–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Rodríguez, E.R.; Ledezma-Candanoza, L.M.; Contreras-Ferrat, L.G.; Olamendi-Portugal, T.; Possani, L.D.; Becerril, B.; Riaño-Umbarila, L. A single mutation in framework 2 of the heavy variable domain improves the properties of a diabody and a related single-chain antibody. J. Mol. Biol. 2012, 423, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Hernández, V.; Del Pozo-Yauner, L.; Pedraza-Escalona, M.; Juárez-González, V.R.; Alcántara-Recillas, I.; Possani, L.D.; Becerril, B. Evaluation of three different formats of a neutralizing single chain human antibody against toxin Cn2: Neutralization capacity versus thermodynamic stability. Immunol. Lett. 2012, 143, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Proba, K.; Wörn, A.; Honegger, A.; Plückthun, A. Antibody scFv fragments without disulfide bonds made by molecular evolution. J. Mol. Biol. 1998, 275, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Montoliu-Gaya, L.; Murciano-Calles, J.; Martinez, J.C.; Villegas, S. Towards the improvement in stability of an anti-Aβ single-chain variable fragment, scFv-h3D6, as a way to enhance its therapeutic potential. Amyloid 2017, 24, 167–175. [Google Scholar] [CrossRef]
- Miklos, A.E.; Kluwe, C.; Der, B.S.; Pai, S.; Sircar, A.; Hughes, R.A.; Berrondo, M.; Xu, J.; Codrea, V.; Buckley, P.E.; et al. Brief Communication Structure-Based Design of Supercharged, Highly Thermoresistant Antibodies. Chem. Biol. 2012, 19, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Perchiacca, J.M.; Lee, C.C.; Tessier, P.M. Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold. Protein Eng. Des. Sel. 2014, 27, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Austerberry, J.I.; Dajani, R.; Panova, S.; Roberts, D.; Golovanov, A.P.; Pluen, A.; Van der Walle, C.F.; Uddin, S.; Warwicker, J.; Derrick, J.P.; et al. The effect of charge mutations on the stability and aggregation of a human single chain Fv fragment. Eur. J. Pharm. Biopharm. 2017, 115, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Sakhnini, L.I.; Greisen, P.J.; Wiberg, C.; Bozoky, Z.; Lund, S.; Wolf Perez, A.M.; Karkov, H.S.; Huus, K.; Hansen, J.J.; Bülow, L.; et al. Improving the Developability of an Antigen Binding Fragment by Aspartate Substitutions. Biochemistry 2019, 58, 2750–2759. [Google Scholar] [CrossRef]
- Seeliger, D.; Schulz, P.; Litzenburger, T.; Spitz, J.; Hoerer, S.; Blech, M.; Enenkel, B.; Studts, J.M.; Garidel, P.; Karow, A.R. Boosting antibody developability through rational sequence optimization. MAbs 2015, 7, 505–515. [Google Scholar] [CrossRef]
- Graille, M.; Stura, E.A.; Bossus, M.; Muller, B.H.; Letourneur, O.; Battail-Poirot, N.; Sibaï, G.; Gauthier, M.; Rolland, D.; Le Du, M.-H.; et al. Crystal Structure of the Complex between the Monomeric Form of Toxoplasma gondii Surface Antigen 1 (SAG1) and a Monoclonal Antibody that Mimics the Human Immune Response. J. Mol. Biol. 2005, 354, 447–458. [Google Scholar] [CrossRef]
- Hannachi, E.; Bouratbine, A.; Mousli, M. Enhancing the detection of Toxoplasma gondii via an anti-SAG1 scFv-alkaline phosphatase immunoconjugate. Biotechnol. Rep. 2019, 23, e00360. [Google Scholar] [CrossRef]
- Ehrenmann, F.; Kaas, Q.; Lefranc, M.P. IMGT/3dstructure-DB and IMGT/domaingapalign: A database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MHcSF. Nucleic Acids Res. 2009, 38, 301–307. [Google Scholar] [CrossRef]
- Muzard, J.; Adi-Bessalem, S.; Juste, M.; Laraba-Djebari, F.; Aubrey, N.; Billiald, P. Grafting of protein L-binding activity onto recombinant antibody fragments. Anal. Biochem. 2009, 388, 331–338. [Google Scholar] [CrossRef]
- Zahid, M.; Loyau, S.; Bouabdelli, M.; Aubrey, N.; Jandrot-Perrus, M.; Billiald, P. Design and reshaping of an scFv directed against human platelet glycoprotein VI with diagnostic potential. Anal. Biochem. 2011, 417, 274–282. [Google Scholar] [CrossRef]
- Di Tommaso, A.; Juste, M.O.; Martin-Eauclaire, M.-F.; Dimier-Poisson, I.; Billiald, P.; Aubrey, N. Diabody mixture providing full protection against experimental scorpion envenoming with crude Androctonus australis venom. J. Biol. Chem. 2012, 287, 14149–14156. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo, G.; Gruvegård, M.; Van Alstine, J. Antibody Fragments and Their Purification by Protein L Affinity Chromatography. Antibodies 2015, 4, 259–277. [Google Scholar] [CrossRef] [Green Version]
- Puligedda, R.D.; Vigdorovich, V.; Kouiavskaia, D.; Sather, D.N.; Dessain, S.K. Human IgA Monoclonal Antibodies That Neutralize Poliovirus, Produced by Hybridomas and Recombinant Expression. Antibodies 2020, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Pommié, C.; Levadoux, S.; Sabatier, R.; Lefranc, G.; Lefranc, M.P. IMGT standardized criteria for statistical analysis of immunoglobulin V-Region amino acid properties. J. Mol. Recognit. 2004, 17, 17–32. [Google Scholar] [CrossRef]
- Lee, J.; Kim, M.; Seo, Y.; Lee, Y.; Park, H.; Byun, S.J.; Kwon, M.H. The catalytic activity of a recombinant single chain variable fragment nucleic acid-hydrolysing antibody varies with fusion tag and expression host. Arch. Biochem. Biophys. 2017, 633, 110–117. [Google Scholar] [CrossRef]
- Zhang, K.; Geddie, M.L.; Kohli, N.; Kornaga, T.; Kirpotin, D.B.; Jiao, Y.; Rennard, R.; Drummond, D.C.; Nielsen, U.B.; Xu, L.; et al. Comprehensive optimization of a single-chain variable domain antibody fragment as a targeting ligand for a cytotoxic nanoparticle. MAbs 2015, 0862, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Weatherill, E.E.; Cain, K.L.; Heywood, S.P.; Compson, J.E.; Heads, J.T.; Adams, R.; Humphreys, D.P. Towards a universal disulphide stabilised single chain Fv format: Importance of interchain disulphide bond location and vLvH orientation. Protein Eng. Des. Sel. 2012, 25, 321–329. [Google Scholar] [CrossRef]
- Egan, T.J.; Diem, D.; Weldon, R.; Neumann, T.; Meyer, S.; Urech, D.M. Novel multispecific heterodimeric antibody format allowing modular assembly of variable domain fragments. MAbs 2017, 9, 68–84. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.D.; Weaver-Feldhaus, J.; Gray, S.A.; Siegel, R.W.; Feldhaus, M.J. Production, purification, and characterization of human scFv antibodies expressed in Saccharomyces cerevisiae, Pichia pastoris, and Escherichia coli. Protein Expr. Purif. 2005, 42, 255–267. [Google Scholar] [CrossRef]
- Aubrey, N.; Devaux, C.; Sizaret, P.Y.; Rochat, H.; Goyffon, M.; Billiald, P. Design and evaluation of a diabody to improve protection against a potent scorpion neurotoxin. Cell. Mol. Life Sci. 2003, 60, 617–628. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cnudde, T.; Lakhrif, Z.; Bourgoin, J.; Boursin, F.; Horiot, C.; Henriquet, C.; di Tommaso, A.; Juste, M.O.; Jiacomini, I.G.; Dimier-Poisson, I.; et al. Exploration and Modulation of Antibody Fragment Biophysical Properties by Replacing the Framework Region Sequences. Antibodies 2020, 9, 9. https://doi.org/10.3390/antib9020009
Cnudde T, Lakhrif Z, Bourgoin J, Boursin F, Horiot C, Henriquet C, di Tommaso A, Juste MO, Jiacomini IG, Dimier-Poisson I, et al. Exploration and Modulation of Antibody Fragment Biophysical Properties by Replacing the Framework Region Sequences. Antibodies. 2020; 9(2):9. https://doi.org/10.3390/antib9020009
Chicago/Turabian StyleCnudde, Thomas, Zineb Lakhrif, Justine Bourgoin, Fanny Boursin, Catherine Horiot, Corinne Henriquet, Anne di Tommaso, Matthieu Olivier Juste, Isabella Gizzi Jiacomini, Isabelle Dimier-Poisson, and et al. 2020. "Exploration and Modulation of Antibody Fragment Biophysical Properties by Replacing the Framework Region Sequences" Antibodies 9, no. 2: 9. https://doi.org/10.3390/antib9020009
APA StyleCnudde, T., Lakhrif, Z., Bourgoin, J., Boursin, F., Horiot, C., Henriquet, C., di Tommaso, A., Juste, M. O., Jiacomini, I. G., Dimier-Poisson, I., Pugnière, M., Mévélec, M.-N., & Aubrey, N. (2020). Exploration and Modulation of Antibody Fragment Biophysical Properties by Replacing the Framework Region Sequences. Antibodies, 9(2), 9. https://doi.org/10.3390/antib9020009




