BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design against Burkholderia pseudomallei
Abstract
1. Introduction
2. Materials and Methods
2.1. Reverse Vaccinology-Based B. pseudomallei Antigen Prediction
2.2. Cloning BPSL1626
2.3. BPSL1626 Expression and Production
2.4. Crystallization of BPSL1626
2.5. Data Collection, Processing, Molecular Replacement, Model Building and Refinement
2.6. Computational Epitope Predictions
2.7. Blood Sample Collection
2.8. Detection of IFN-γ Production in Whole Blood Culture
2.9. Measurement of Human Plasma IgG Antibody Levels
2.10. Statistical Analyses
2.11. Protein Microarray Assays
2.12. Rabbit Immunizations and Generation of Polyclonal Abs against BPSL1626
3. Results
3.1. In Silico Identification of BSL1626 as a Potential Antigen
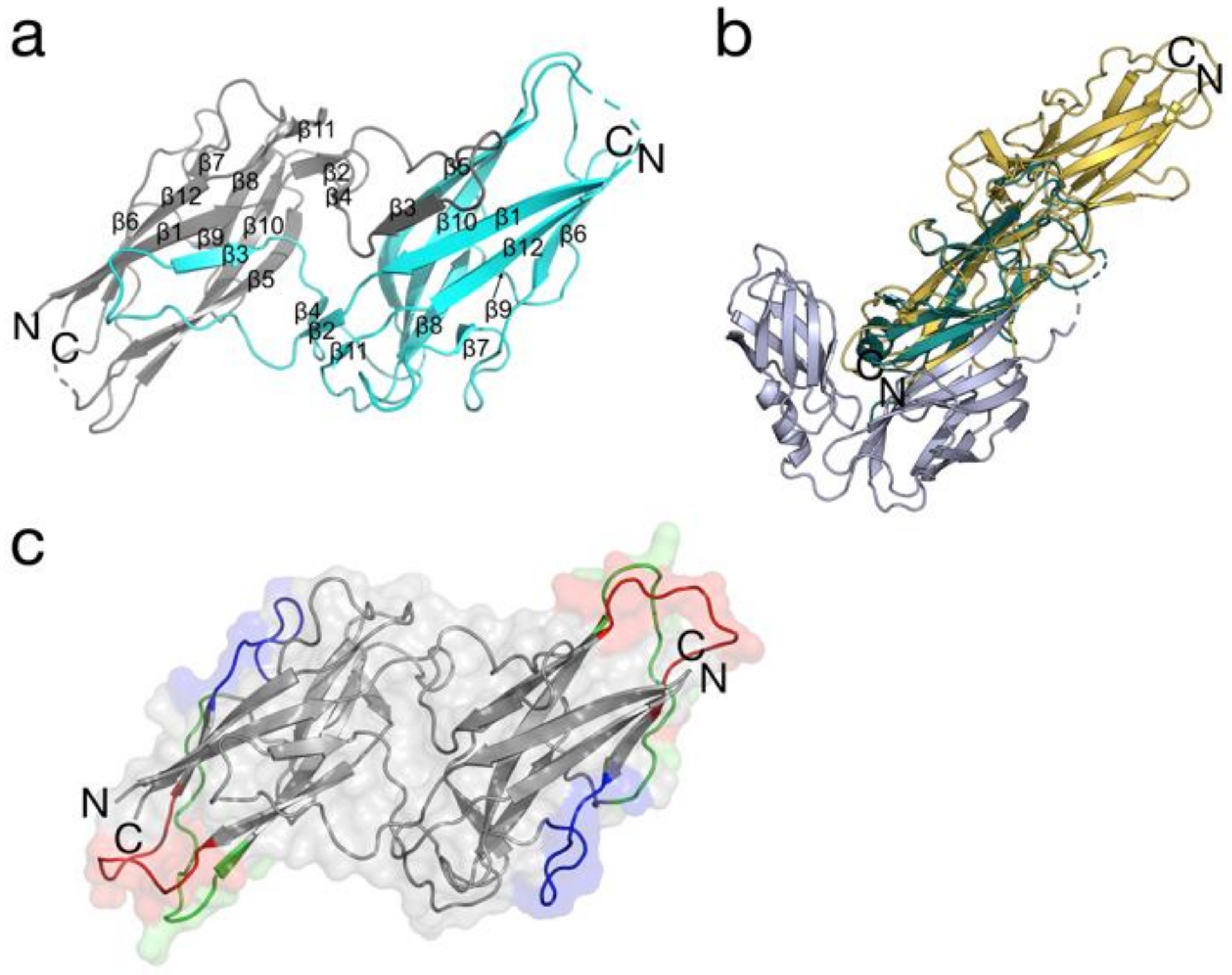
3.2. 3D Structure Analyses of BPSL1626
3.3. In Silico Epitope Predictions
3.4. Rabbit Immunizations with Recombinant BPSL1626
3.5. BPSL1626 Induces IFN-γ Production and Is Recognized by Human IgG Antibodies from Healthy Blood Samples Taken from Endemic Areas
3.6. Probing the Human Antibody Response to BPSL1626 in Burkholderia Affected Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wiersinga, W.J.; Currie, B.J.; Peacock, S.J. Melioidosis. N. Engl. J. Med. 2012, 367, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Limmathurotsakul, D.; Golding, N.; Dance, D.A.; Messina, J.P.; Pigott, D.M.; Moyes, C.L.; Rolim, D.B.; Bertherat, E.; Day, N.P.; Peacock, S.J.; et al. Predicted Global Distribution of Burkholderia Pseudomallei and Burden of Melioidosis. Nat. Microbiol. 2016, 1, 15008. [Google Scholar] [CrossRef] [PubMed]
- Titball, R.W.; Burtnick, M.N.; Bancroft, G.J.; Brett, P. Burkholderia Pseudomallei and Burkholderia Mallei Vaccines: Are We Close to Clinical Trials? Vaccine 2017, 35, 5981–5989. [Google Scholar] [CrossRef] [PubMed]
- Brett, P.J.; Mah, D.C.; Woods, D.E. Isolation and Characterization of Pseudomonas Pseudomallei Flagellin Proteins. Infect. Immun. 1994, 62, 1914–1919. [Google Scholar] [PubMed]
- Champion, O.L.; Gourlay, L.J.; Scott, A.E.; Lassaux, P.; Conejero, L.; Perletti, L.; Hemsley, C.; Prior, J.; Bancroft, G.; Bolognesi, M.; et al. Immunisation with Proteins Expressed During Chronic Murine Melioidosis Provides Enhanced Protection against Disease. Vaccine 2016, 34, 1665–1671. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y.; Mohamed, R.; Nathan, S. Immunogenic Burkholderia Pseudomallei Outer Membrane Proteins as Potential Candidate Vaccine Targets. PLoS ONE 2009, 4, e6496. [Google Scholar] [CrossRef] [PubMed]
- Nithichanon, A.; Gourlay, L.J.; Bancroft, G.J.; Ato, M.; Takahashi, Y.; Lertmemongkolchai, G. Boosting of Post-Exposure Human T-Cell and B-Cell Recall Responses in Vivo by Burkholderia Pseudomallei-Related Proteins. Immunology 2017, 151, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Rappuoli, R. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Callister, S.J.; McCue, L.A.; Turse, J.E.; Monroe, M.E.; Auberry, K.J.; Smith, R.D.; Adkins, J.N.; Lipton, M.S. Comparative Bacterial Proteomics: Analysis of the Core Genome Concept. PLoS ONE 2008, 3, e1542. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. Blat—The Blast-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Pruess, M.; Kersey, P.; Apweiler, R. The Integr8 Project—A Resource for Genomic and Proteomic Data. In Silico Biol. 2005, 5, 179–185. [Google Scholar] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. Psortb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. Netmhcpan, a Method for Mhc Class I Binding Prediction Beyond Humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Blicher, T.; Peters, B.; Sette, A.; Justesen, S.; Buus, S.; Lund, O. Quantitative Predictions of Peptide Binding to Any Hla-Dr Molecule of Known Sequence: Netmhciipan. PLoS Comput. Biol. 2008, 4, e1000107. [Google Scholar] [CrossRef] [PubMed]
- NCBI Resource Coordinators. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar]
- Gonzalez-Galarza, F.F.; Takeshita, L.Y.; Santos, E.J.; Kempson, F.; Maia, M.H.; da Silva, A.L.; Teles e Silva, A.L.; Ghattaoraya, G.S.; Alfirevic, A.; Jones, A.R.; et al. Allele Frequency Net 2015 Update: New Features for Hla Epitopes, Kir and Disease and Hla Adverse Drug Reaction Associations. Nucleic Acids Res. 2015, 43, D784–D788. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov Model Speed Heuristic and Iterative Hmm Search Procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: Hmmer3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. Integration, Scaling, Space-Group Assignment and Post-Refinement. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and Assessment of Data Quality. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Vagin, A.A.; Young, P.; Murshudov, G.N. Balbes: A Molecular-Replacement Pipeline. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. Bepipred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide Binding Predictions for Hla Dr, Dp and Dq Molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-Throughput Prediction of Protein Antigenicity Using Protein Microarray Data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. Ellipro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Sweredoski, M.J.; Baldi, P. Cobepro: A Novel System for Predicting Continuous B-Cell Epitopes. Protein Eng. Des. Sel. 2009, 22, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Nithichanon, A.; Rinchai, D.; Gori, A.; Lassaux, P.; Peri, C.; Conchillio-Sole, O.; Ferrer-Navarro, M.; Gourlay, L.J.; Nardini, M.; Vila, J.; et al. Sequence- and Structure-Based Immunoreactive Epitope Discovery for Burkholderia Pseudomallei Flagellin. PLoS Negl. Trop. Dis. 2015, 9, e0003917. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinform. 2014, 47. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.L.; Warner, J.; Melrose, W.; Durrheim, D.; Speare, R.; Reeder, J.C.; Ketheesan, N. Adaptive Immunity in Melioidosis: A Possible Role for T Cells in Determining Outcome of Infection with Burkholderia Pseudomallei. Clin. Immunol. 2004, 113, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.C.; O’Brien, M.; Freeman, K.; Lum, G.; Currie, B.J. Indirect Hemagglutination Assay in Patients with Melioidosis in Northern Australia. Am. J. Trop. Med. Hyg. 2006, 74, 330–334. [Google Scholar] [PubMed]
- Gourlay, L.J.; Peri, C.; Ferrer-Navarro, M.; Conchillo-Sole, O.; Gori, A.; Rinchai, D.; Thomas, R.J.; Champion, O.L.; Michell, S.L.; Kewcharoenwong, C.; et al. Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology. Chem. Biol. 2013, 20, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Pirri, G.; Damin, F.; Solinas, I.; Chiari, M. A New Polymeric Coating for Protein Microarrays. Anal. Biochem. 2004, 332, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Peri, C.; Gori, A.; Gagni, P.; Sola, L.; Girelli, D.; Sottotetti, S.; Cariani, L.; Chiari, M.; Cretich, M.; Colombo, G. Evolving Serodiagnostics by Rationally Designed Peptide Arrays: The Burkholderia Paradigm in Cystic Fibrosis. Sci. Rep. 2016, 6, 32873. [Google Scholar] [CrossRef] [PubMed]
- Muruato, L.A.; Tapia, D.; Hatcher, C.L.; Kalita, M.; Brett, P.J.; Gregory, A.E.; Samuel, J.E.; Titball, R.W.; Torres, A.G. The Use of Reverse Vaccinology in the Design and Construction of Nano-Glycoconjugate Vaccines against Burkholderia Pseudomallei. Clin. Vaccine Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A. Enhancing the Functional Annotation of Pdb Structures in Pdbsum Using Key Figures Extracted from the Literature. Bioinformatics 2007, 23, 1824–1827. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.; Titball, R.W.; Peacock, S.J.; Cerdeno-Tarraga, A.M.; Atkins, T.; Crossman, L.C.; Pitt, T.; Churcher, C.; Mungall, K.; Bentley, S.D.; et al. Genomic Plasticity of the Causative Agent of Melioidosis, Burkholderia Pseudomallei. Proc. Natl. Acad. Sci. USA 2004, 101, 14240–14245. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.D.; Puorger, C.; Scharer, M.A.; Eidam, O.; Grutter, M.G.; Capitani, G.; Glockshuber, R. Quality Control of Disulfide Bond Formation in Pilus Subunits by the Chaperone FimC. Nat. Chem. Biol. 2012, 8, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Watson, J.D.; Thornton, J.M. Profunc: A Server for Predicting Protein Function from 3d Structure. Nucleic Acids Res. 2005, 33, W89–W93. [Google Scholar] [CrossRef] [PubMed]
- Scarabelli, G.; Morra, G.; Colombo, G. Predicting Interaction Sites from the Energetics of Isolated Proteins: A New Approach to Epitope Mapping. Biophys. J. 2010, 98, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
- Santanirand, P.; Harley, V.S.; Dance, D.A.; Drasar, B.S.; Bancroft, G.J. Obligatory Role of Gamma Interferon for Host Survival in a Murine Model of Infection with Burkholderia Pseudomallei. Infect. Immun. 1999, 67, 3593–3600. [Google Scholar] [PubMed]
- Monterrubio-Lopez, G.P.; Merchand, Y.; Gonzalez, J.A.; Ribas-Aparicio, R.M. Identification of Novel Potential Vaccine Candidates against Tuberculosis Based on Reverse Vaccinology. Biomed. Res. Int. 2015, 2015, 483150. [Google Scholar] [CrossRef] [PubMed]
- Montigiani, S.; Falugi, F.; Scarselli, M.; Finco, O.; Petracca, R.; Galli, G.; Mariani, M.; Manetti, R.; Agnusdei, M.; Cevenini, R.; et al. Genomic Approach for Analysis of Surface Proteins in Chlamydia Pneumoniae. Infect. Immun. 2002, 70, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Moriel, D.G.; Bertoldi, I.; Spagnuolo, A.; Marchi, S.; Rosini, R.; Nesta, B.; Pastorello, I.; Corea, V.A.; Torricelli, G.; Cartocci, E.; et al. Identification of Protective and Broadly Conserved Vaccine Antigens from the Genome of Extraintestinal Pathogenic Escherichia Coli. Proc. Natl. Acad. Sci. USA 2010, 107, 9072–9077. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Rajan, G.S.; Kharb, R.; Biswas, S. Genome Wide Analysis of Chlamydia Pneumoniae for Candidate Vaccine Development. Curr. Comput. Aided Drug Des. 2016, 12, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, L.J.; Peano, C.; Deantonio, C.; Perletti, L.; Pietrelli, A.; Villa, R.; Matterazzo, E.; Lassaux, P.; Santoro, C.; Puccio, S.; et al. Selecting Soluble/Foldable Protein Domains through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Musson, J.A.; Reynolds, C.J.; Rinchai, D.; Nithichanon, A.; Khaenam, P.; Favry, E.; Spink, N.; Chu, K.K.; de Soyza, A.; Bancroft, G.J.; et al. Cd4+ T Cell Epitopes of Flic Conserved between Strains of Burkholderia: Implications for Vaccines against Melioidosis and Cepacia Complex in Cystic Fibrosis. J. Immunol. 2014, 193, 6041–6049. [Google Scholar] [CrossRef] [PubMed]
- Healey, G.D.; Elvin, S.J.; Morton, M.; Williamson, E.D. Humoral and Cell-Mediated Adaptive Immune Responses Are Required for Protection against Burkholderia Pseudomallei Challenge and Bacterial Clearance Postinfection. Infect. Immun. 2005, 73, 5945–5951. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Bolognesi, M.; Colombo, G.; Gourlay, L.J. Structural Vaccinology for Melioidosis Vaccine Design and Immunodiagnostics. Curr. Trop. Med. Rep. 2017, 4, 103–110. [Google Scholar] [CrossRef]
- Choudhury, D.; Thompson, A.; Stojanoff, V.; Langermann, S.; Pinkner, J.; Hultgren, S.J.; Knight, S.D. X-Ray Structure of the Fimc-Fimh Chaperone-Adhesin Complex from Uropathogenic Escherichia coli. Science 1999, 285, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capelli, R.; Peri, C.; Villa, R.; Nithichanon, A.; Conchillo-Solé, O.; Yero, D.; Gagni, P.; Chiari, M.; Lertmemongkolchai, G.; Cretich, M.; et al. BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design against Burkholderia pseudomallei. Antibodies 2018, 7, 26. https://doi.org/10.3390/antib7030026
Capelli R, Peri C, Villa R, Nithichanon A, Conchillo-Solé O, Yero D, Gagni P, Chiari M, Lertmemongkolchai G, Cretich M, et al. BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design against Burkholderia pseudomallei. Antibodies. 2018; 7(3):26. https://doi.org/10.3390/antib7030026
Chicago/Turabian StyleCapelli, Riccardo, Claudio Peri, Riccardo Villa, Arnone Nithichanon, Oscar Conchillo-Solé, Daniel Yero, Paola Gagni, Marcella Chiari, Ganjana Lertmemongkolchai, Marina Cretich, and et al. 2018. "BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design against Burkholderia pseudomallei" Antibodies 7, no. 3: 26. https://doi.org/10.3390/antib7030026
APA StyleCapelli, R., Peri, C., Villa, R., Nithichanon, A., Conchillo-Solé, O., Yero, D., Gagni, P., Chiari, M., Lertmemongkolchai, G., Cretich, M., Daura, X., Bolognesi, M., Colombo, G., & Gourlay, L. J. (2018). BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design against Burkholderia pseudomallei. Antibodies, 7(3), 26. https://doi.org/10.3390/antib7030026