Comparative Evaluation of Three Primary Antibody Clones for p16 Immunohistochemistry in Gynecologic Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Study Design
2.2. IHC Procedure
3. Interpretation
Statistical Analysis
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahajan, A. Practical issues in the application of p16 immunohistochemistry in diagnostic pathology. Hum. Pathol. 2016, 51, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Darragh, T.M.; Colgan, T.J.; Thomas Cox, J.; Heller, D.S.; Henry, M.R.; Luff, R.D.; McCalmont, T.; Nayar, R.; Palefsky, J.M.; Stoler, M.H.; et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: Background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Int. J. Gynecol. Pathol. 2013, 32, 76–115. [Google Scholar] [CrossRef]
- Khleif, S.N.; DeGregori, J.; Yee, C.L.; Otterson, G.A.; Kaye, F.J.; Nevins, J.R.; Howley, P.M. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc. Natl. Acad. Sci. USA 1996, 93, 4350–4354. [Google Scholar] [CrossRef]
- Kim, W.Y.; Sharpless, N.E. The regulation of INK4/ARF in cancer and aging. Cell 2006, 127, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiu, K.; Ren, J.; Zhao, Y.; Cheng, P. Roles of human papillomavirus in cancers: Oncogenic mechanisms and clinical use. Signal Transduct. Target. Ther. 2025, 10, 44. [Google Scholar] [CrossRef]
- Sano, T.; Oyama, T.; Kashiwabara, K.; Fukuda, T.; Nakajima, T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am. J. Pathol. 1998, 153, 1741–1748. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Crum, C.P.; Münger, K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. USA 2011, 108, 2130–2135. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef]
- Genovés, J.; Alameda, F.; Mancebo, G.; Solé, J.M.; Bellosillo, B.; Lloveras, B.; Agramunt, S.; Baró, M.T.; Muset, M.; Casado, B.; et al. Human papillomavirus detection and p16INK4a expression in cervical lesions: A comparative study. Hum. Pathol. 2014, 45, 826–833. [Google Scholar] [CrossRef]
- Nogueira-Rodrigues, A.; Oonk, M.H.M.; Lorusso, D.; Slomovitz, B.; Leitão, M.M., Jr.; Baiocchi, G. Comprehensive management of vulvovaginal cancers. CA Cancer J. Clin. 2025; early review. [Google Scholar] [CrossRef]
- Milea, A.; George, S.H.; Matevski, D.; Jiang, H.; Madunic, M.; Berman, H.K.; Gauthier, M.L.; Gallie, B.; Shaw, P.A. Retinoblastoma pathway deregulatory mechanisms determine clinical outcome in high-grade serous ovarian carcinoma. Mod. Pathol. 2014, 27, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Yemelyanova, A.; Ji, H.; Shih Ie, M.; Wang, T.L.; Wu, L.S.; Ronnett, B.M. Utility of p16 expression for distinction of uterine serous carcinomas from endometrial endometrioid and endocervical adenocarcinomas: Immunohistochemical analysis of 201 cases. Am. J. Surg. Pathol. 2009, 33, 1504–1514. [Google Scholar] [CrossRef]
- Saad, R.S.; Mashhour, M.; Noftech-Mozes, S.; Ismiil, N.; Dubé, V.; Ghorab, Z.; Faragalla, H.; Khalifa, M.A. P16INK4a expression in undifferentiated carcinoma of the uterus does not exclude its endometrial origin. Int. J. Gynecol. Pathol. 2012, 31, 57–65. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; Knudsen, K.E.; Dicker, A.P.; Knudsen, E.S. The meaning of p16ink4a expression in tumors. Cell Cycle 2011, 10, 2497–2503. [Google Scholar] [CrossRef]
- Ennis, C.M.; Rohrbach, M.R.; Schwalbe, M.; Mahajan, A.; Hartig, G.K. A Comparison of E6H4 and G175-405 p16-specific Monoclonal Antibodies in Oropharyngeal Squamous Cell Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 290–295. [Google Scholar] [CrossRef] [PubMed]
- NordiQC Assessment Run 72 2024 p16ink4a (p16). Available online: https://www.nordiqc.org/downloads/assessments/189_59.pdf (accessed on 5 June 2025).
- Eba, J.; Nakamura, K. Overview of the ethical guidelines for medical and biological research involving human subjects in Japan. Jpn. J. Clin. Oncol. 2022, 52, 539–544. [Google Scholar] [CrossRef]
- Evaluation of Automatic Class III Designation for CINtec Histology Decision Summary. Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN160019.pdf (accessed on 1 June 2025).
- Goldsmith, J.D.; Troxell, M.L.; Roy-Chowdhuri, S.; Colasacco, C.F.; Edgerton, M.E.; Fitzgibbons, P.L.; Fulton, R.; Haas, T.; Kandalaft, P.L.; Kalicanin, T.; et al. Principles of Analytic Validation of Immunohistochemical Assays: Guideline Update. Arch. Pathol. Lab. Med. 2024, 148, e111–e153. [Google Scholar] [CrossRef]
- Shelton, J.; Purgina, B.M.; Cipriani, N.A.; Dupont, W.D.; Plummer, D.; Lewis, J.S., Jr. p16 immunohistochemistry in oropharyngeal squamous cell carcinoma: A comparison of antibody clones using patient outcomes and high-risk human papillomavirus RNA status. Mod. Pathol. 2017, 30, 1194–1203. [Google Scholar] [CrossRef]
- Thavaraj, S.; Robinson, M.; Dayal, S.; Bowen, C. Performance analysis of Leica Biosystems p16 monoclonal antibody in oropharyngeal squamous cell carcinoma. Diagn. Pathol. 2025, 20, 9. [Google Scholar] [CrossRef]
- Angelico, G.; Santoro, A.; Inzani, F.; Straccia, P.; Spadola, S.; Arciuolo, D.; Valente, M.; D’Alessandris, N.; Benvenuto, R.; Travaglino, A.; et al. An Emerging Anti-p16 Antibody-BC42 Clone as an Alternative to the Current E6H4 for Use in the Female Genital Tract Pathological Diagnosis: Our Experience and a Review on p16ink4a Functional Significance, Role in Daily-Practice Diagnosis, Prognostic Potential, and Technical Pitfalls. Diagnostics 2021, 11, 713. [Google Scholar] [CrossRef]
- Klaes, R.; Benner, A.; Friedrich, T.; Ridder, R.; Herrington, S.; Jenkins, D.; Kurman, R.J.; Schmidt, D.; Stoler, M.; von Knebel Doeberitz, M. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am. J. Surg. Pathol. 2002, 26, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Stoler, M.H.; Wright, T.C., Jr.; Ferenczy, A.; Ranger-Moore, J.; Fang, Q.; Kapadia, M.; Ridder, R. Routine Use of Adjunctive p16 Immunohistochemistry Improves Diagnostic Agreement of Cervical Biopsy Interpretation: Results from the CERTAIN Study. Am. J. Surg. Pathol. 2018, 42, 1001–1009. [Google Scholar] [CrossRef]
- Bean, S.M.; Meara, R.S.; Vollmer, R.T.; Conner, M.G.; Crowe, D.R.; Novak, L.; Eltoum, I.A.; Robboy, S.J.; Chhieng, D.C. p16 Improves interobserver agreement in diagnosis of anal intraepithelial neoplasia. J. Low. Genit. Tract. Dis. 2009, 13, 145–153. [Google Scholar] [CrossRef]
- Tao, A.S.; Zuna, R.; Darragh, T.M.; Grabe, N.; Lahrmann, B.; Clarke, M.A.; Wentzensen, N. Interobserver reproducibility of cervical histology interpretation with and without p16 immunohistochemistry. Am. J. Clin. Pathol. 2024, 162, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Baez-Navarro, X.; van Bockstal, M.R.; Nawawi, D.; Broeckx, G.; Colpaert, C.; Doebar, S.C.; Hogenes, M.C.H.; Koop, E.; Lambein, K.; Peeters, D.J.E.; et al. Interobserver Variation in the Assessment of Immunohistochemistry Expression Levels in HER2-Negative Breast Cancer: Can We Improve the Identification of Low Levels of HER2 Expression by Adjusting the Criteria? An International Interobserver Study. Mod. Pathol. 2023, 36, 100009. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.A.; Russell, P.A.; Cherian, M.; Duhig, E.E.; Godbolt, D.; Jessup, P.J.; Khoo, C.; Leslie, C.; Mahar, A.; Moffat, D.F.; et al. Intra- and Interobserver Reproducibility Assessment of PD-L1 Biomarker in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4569–4577. [Google Scholar] [CrossRef] [PubMed]
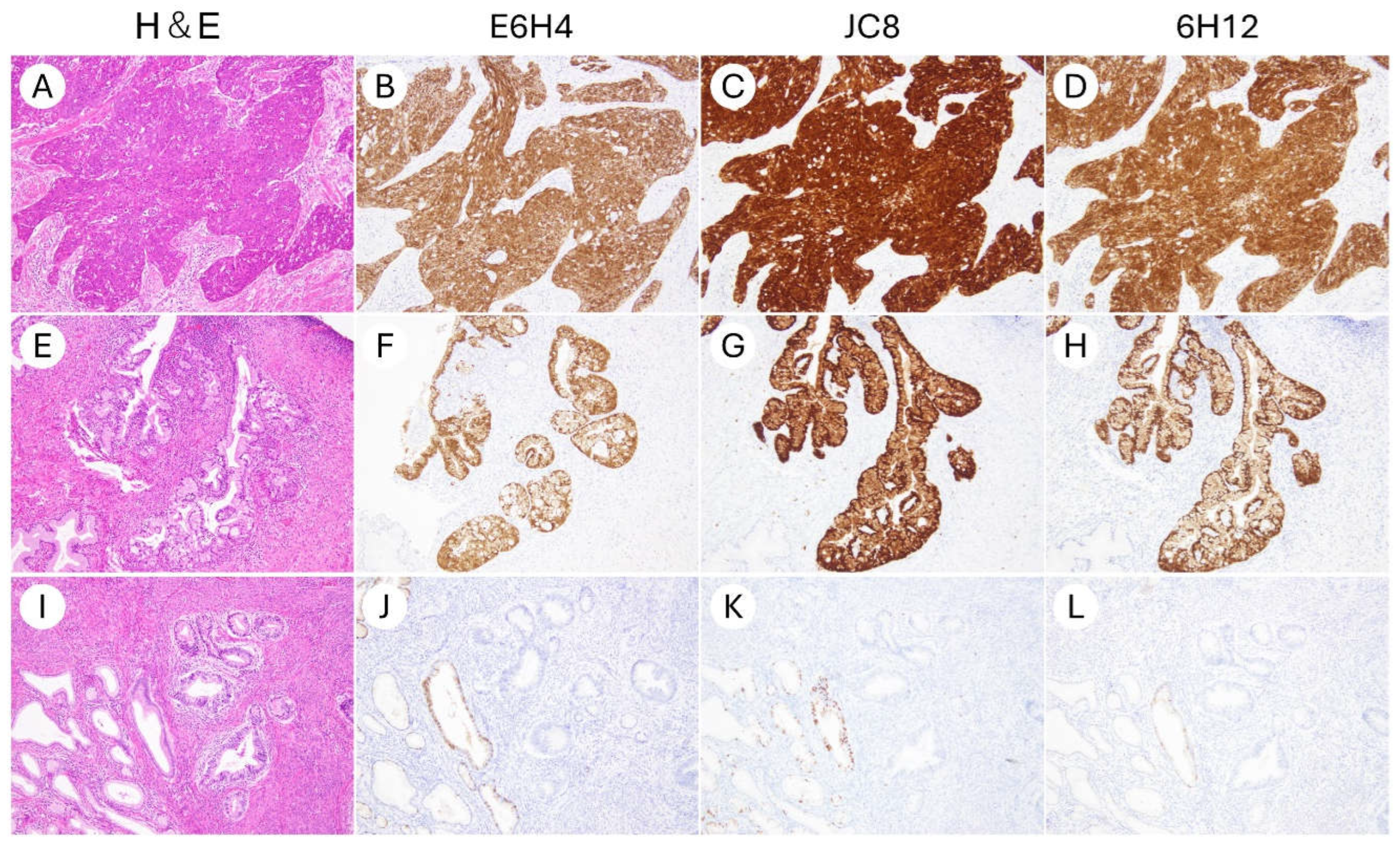
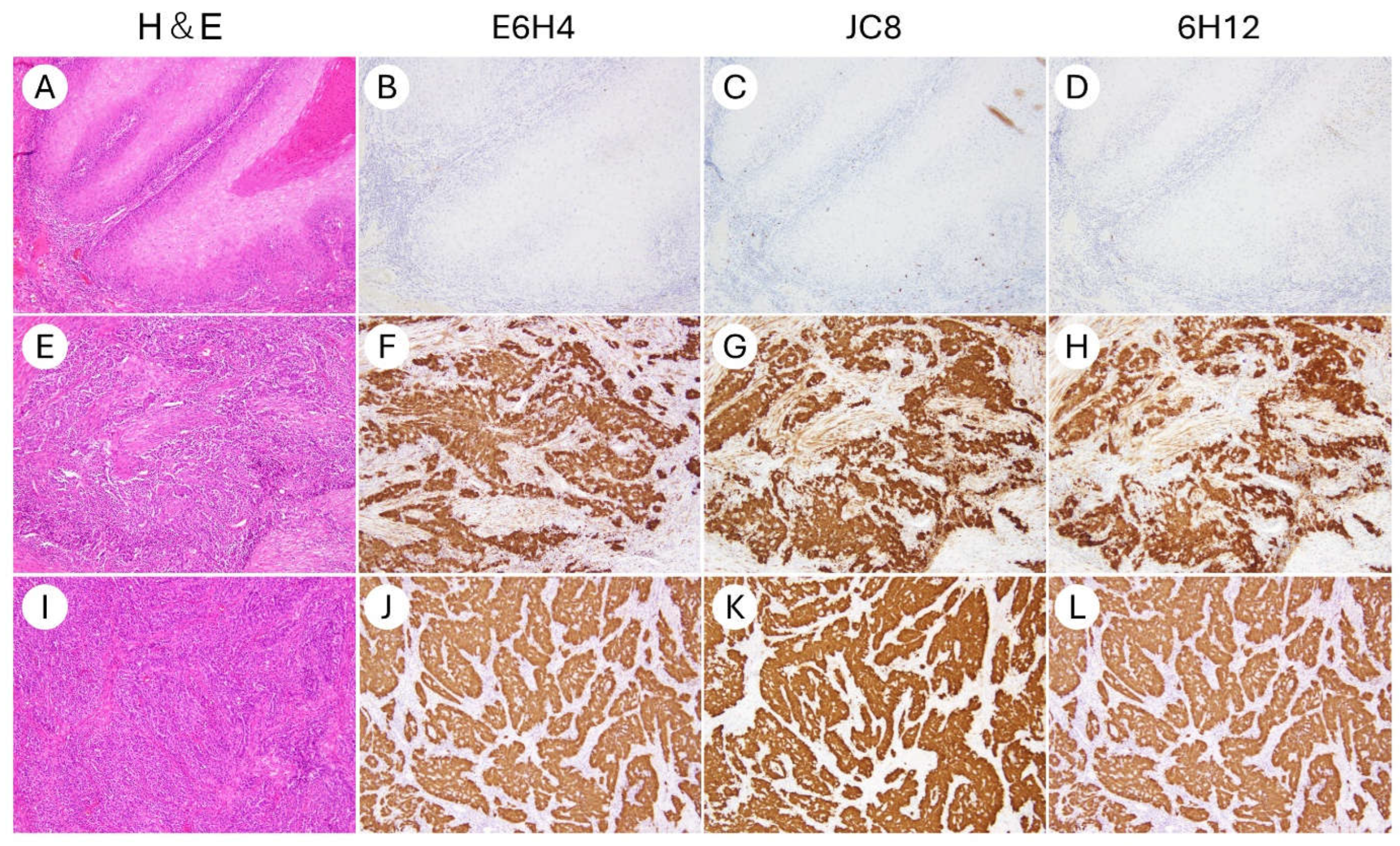
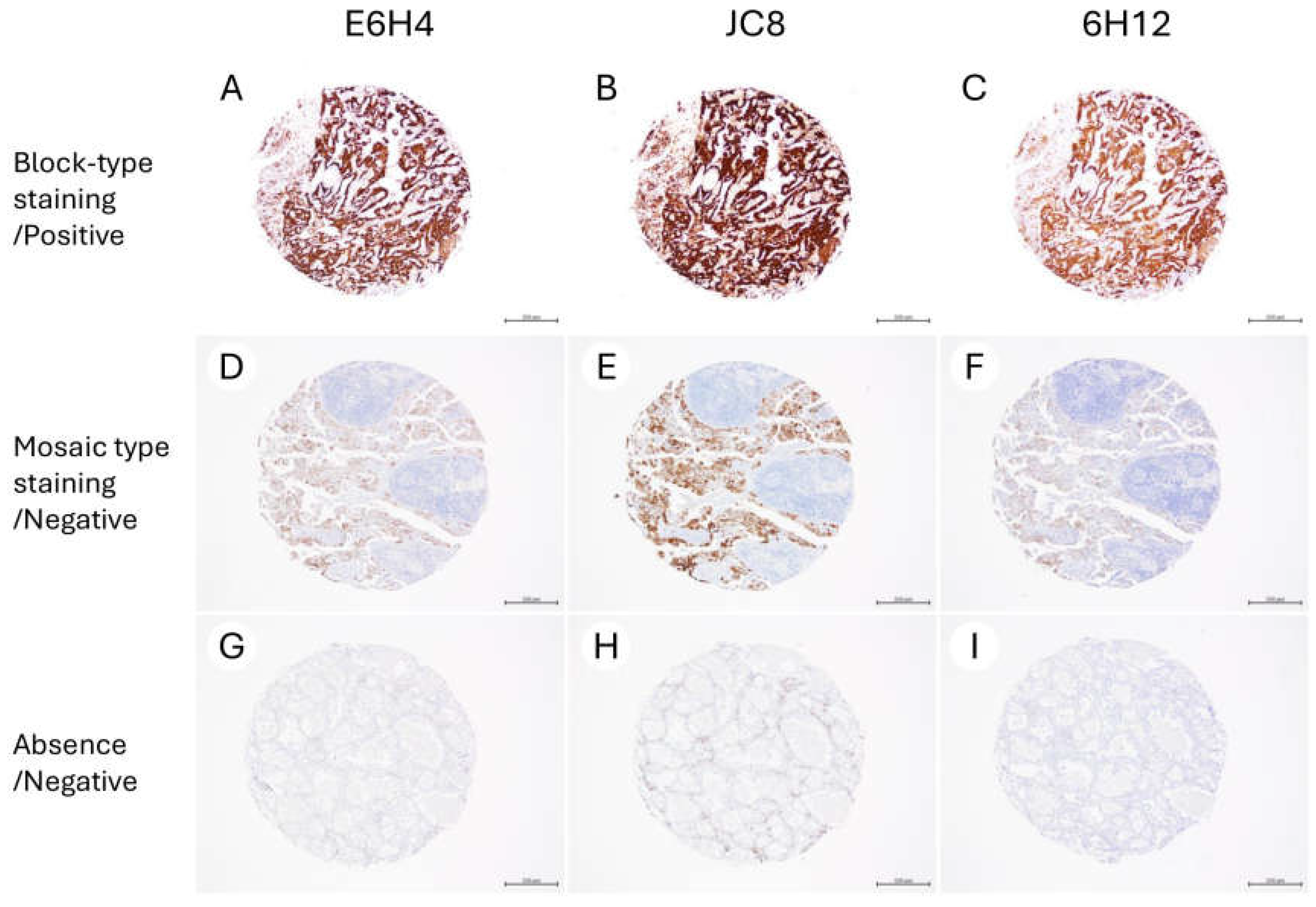
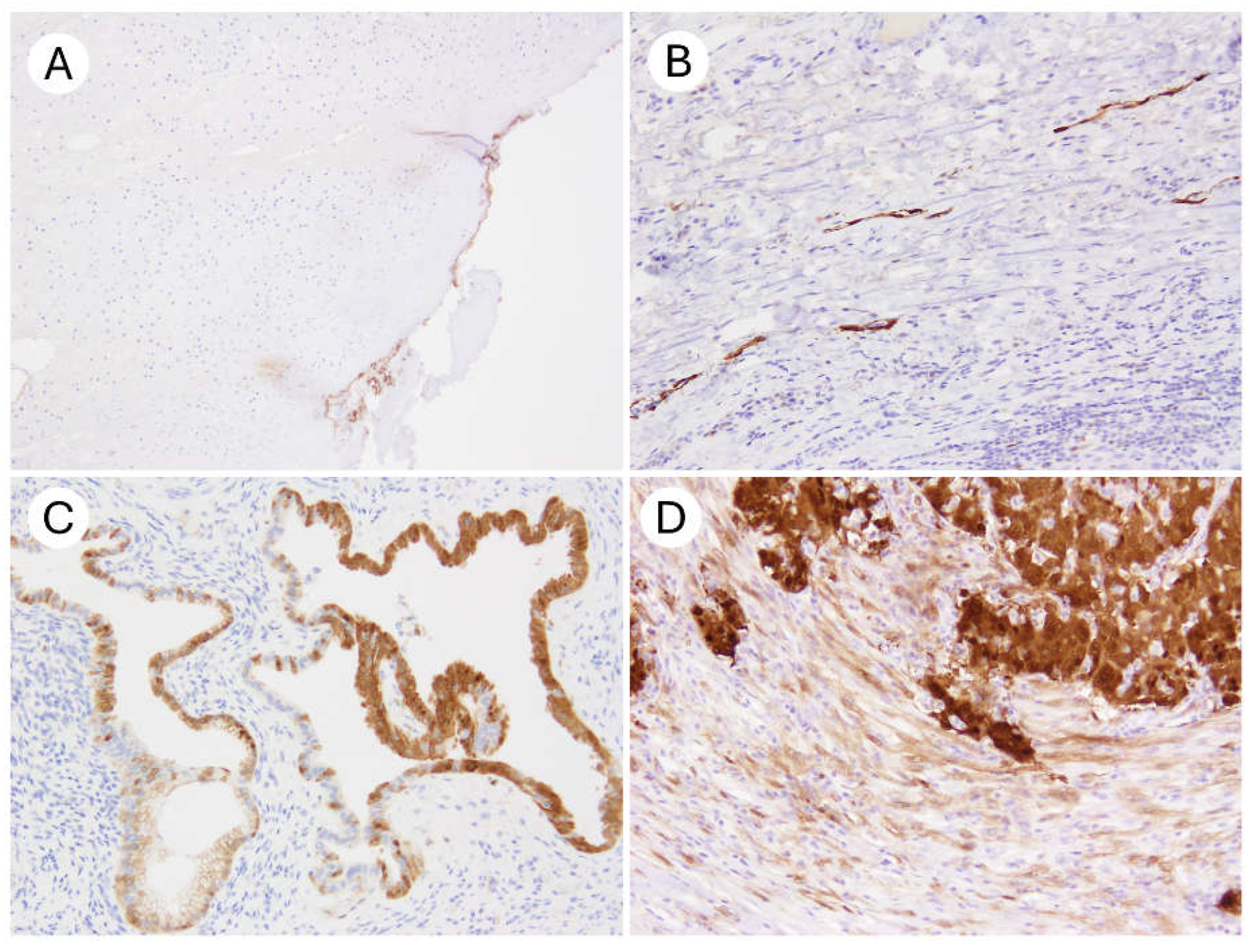
| Characteristics | N (%) |
|---|---|
| Total | 42 (100) |
| Organ | |
| Uterine cervix | 30 (71) |
| Uterine corpus | 4 (9.5) |
| Vulva | 6 (14) |
| Ovary | 2 (4.8) |
| Specimen type | |
| Surgical specimen | 26 (62) |
| Biopsy specimen | 16 (38) |
| p16 (E6H4) immunohistochemistry | |
| Positive | 21 (50) |
| Negative | 21 (50) |
| Diagnosis | |
| Squamous cell carcinoma, HPV-associated | 12 (29) |
| Cervicitis | 6 (14) |
| High-grade squamous intraepithelial neoplasia/CIN3 | 4 (9.5) |
| Squamous cell carcinoma, HPV-independent | 4 (9.5) |
| Adenocarcinoma in situ, HPV-associated | 2 (4.8) |
| Adenocarcinoma in situ, HPV-independent, gastric-type | 2 (4.8) |
| Adenocarcinoma, HPV-associated | 2 (4.8) |
| Adenocarcinoma, HPV-independent, gastric type | 2 (4.8) |
| High-grade serous carcinoma | 2 (4.8) |
| Neuroendocrine carcinoma | 2 (4.8) |
| Serous carcinoma | 2 (4.8) |
| Clear cell carcinoma | 1 (2.4) |
| Endometrioid carcinoma | 1 (2.4) |
| Characteristics | N (%) |
|---|---|
| Endometrial cancer | 44 (100) |
| Endometrioid carcinoma, grade 1/2 | 17 (39) |
| Endometrioid carcinoma, grade 3 | 16 (36) |
| Serous carcinoma | 5 (11) |
| Clear cell carcinoma | 3 (6.8) |
| Carcinosarcoma | 3 (6.8) |
| Ovarian cancer | 90 (100) |
| High-grade serous carcinoma | 35 (39) |
| Low-grade serous carcinoma | 3 (3.3) |
| Clear cell carcinoma | 27 (30) |
| Endometrioid carcinoma | 20 (22) |
| Mucinous carcinoma | 5 (5.6) |
| Characteristics | E6H4 | JC8 | 6H12 |
|---|---|---|---|
| Vendor | Ventana/Roche | Agilent/Dako | Leica |
| Product No. | 705-4713 | GA783 | PA0016 |
| Type of antibody | Mouse monoclonal | Mouse monoclonal | Mouse monoclonal |
| Target epitope | Human p16INK4a, amino acids 134-156 | Human p16INK4a, recombinant full-length human p16 | Human p16INK4a epitope not fully disclosed |
| Isotype | IgG2a | IgG2a | IgG2b |
| Dilution | Prediluted | Prediluted | Prediluted |
| Antigen retrieval | HIER using CC1 64 min. at 95 °C | HIER using EnVision FLEX High pH 30 min. at 97 °C | HIER using BERS2 20 min. at 100 °C |
| Primary antibody incubation time | 16 min | 20 min | 15 min |
| Autostainer | BenchMark Ultra | Dako Omnis | Leica BOND III |
| Detection system | UltraView | EnVision Flex+ | Bond Polymer Refine Detection (DS9800) |
| Clinical Validation | FDA-approved as part of CINtec® Histology kit | RUO | RUO |
| JC8 | |||
| Positive | Negative | ||
| E6H4 | Positive | 21 | 0 |
| Negative | 0 | 21 | |
| 6H12 | |||
| Positive | Negative | ||
| E6H4 | Positive | 21 | 0 |
| Negative | 0 | 21 | |
| 6H12 | |||
| Positive | Negative | ||
| JC8 | Positive | 21 | 0 |
| Negative | 0 | 21 | |
| JC8 | ||||
| Positive Block staining | Negative Mosaic staining | Negative Absence | ||
| E6H4 | Positive Block staining | 40 | 0 | 0 |
| Negative Mosaic staining | 0 | 68 | 0 | |
| Negative Absence | 0 | 0 | 26 | |
| 6H12 | ||||
| Positive | ||||
| E6H4 | Positive Block staining | 40 | 0 | 0 |
| Negative Mosaic staining | 0 | 68 | 0 | |
| Negative Absence | 0 | 0 | 26 | |
| 6H12 | ||||
| Positive | ||||
| JC8 | Positive Block staining | 40 | 0 | 0 |
| Negative Mosaic staining | 0 | 68 | 0 | |
| Negative Absence | 0 | 0 | 26 | |
| Pathologist B | |||
|---|---|---|---|
| Positive | Negative | ||
| Pathologist A | Positive | 30 | 0 |
| Negative | 0 | 30 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, H.; Sugitani, A.; Kobayashi-Kato, M.; Uno, M.; Ishikawa, M. Comparative Evaluation of Three Primary Antibody Clones for p16 Immunohistochemistry in Gynecologic Tumors. Antibodies 2025, 14, 77. https://doi.org/10.3390/antib14030077
Yoshida H, Sugitani A, Kobayashi-Kato M, Uno M, Ishikawa M. Comparative Evaluation of Three Primary Antibody Clones for p16 Immunohistochemistry in Gynecologic Tumors. Antibodies. 2025; 14(3):77. https://doi.org/10.3390/antib14030077
Chicago/Turabian StyleYoshida, Hiroshi, Ayumi Sugitani, Mayumi Kobayashi-Kato, Masaya Uno, and Mitsuya Ishikawa. 2025. "Comparative Evaluation of Three Primary Antibody Clones for p16 Immunohistochemistry in Gynecologic Tumors" Antibodies 14, no. 3: 77. https://doi.org/10.3390/antib14030077
APA StyleYoshida, H., Sugitani, A., Kobayashi-Kato, M., Uno, M., & Ishikawa, M. (2025). Comparative Evaluation of Three Primary Antibody Clones for p16 Immunohistochemistry in Gynecologic Tumors. Antibodies, 14(3), 77. https://doi.org/10.3390/antib14030077






