Comparative Molecular Dynamics Study of 19 Bovine Antibodies with Ultralong CDR H3
Abstract
1. Introduction
2. Methods
3. Results
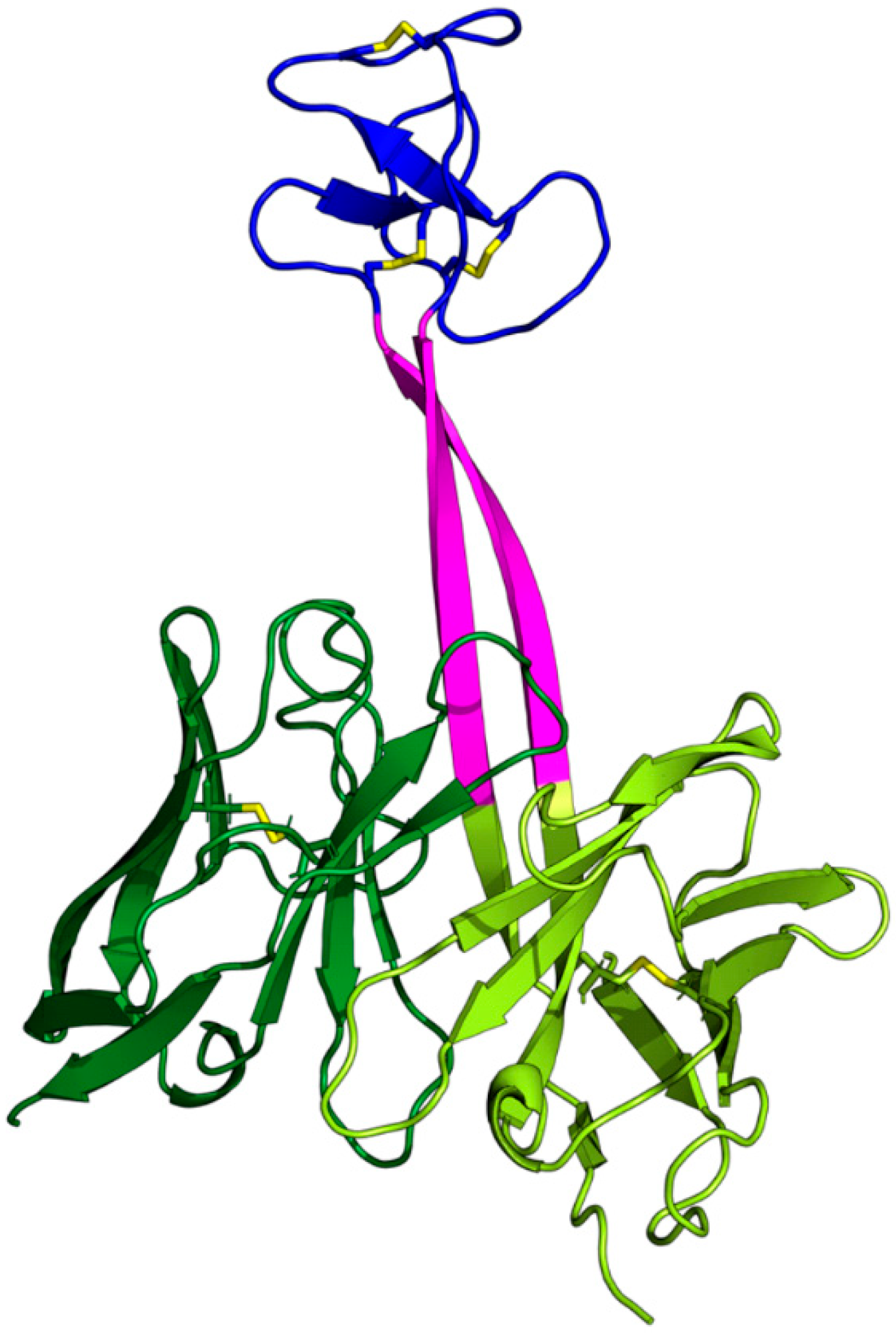
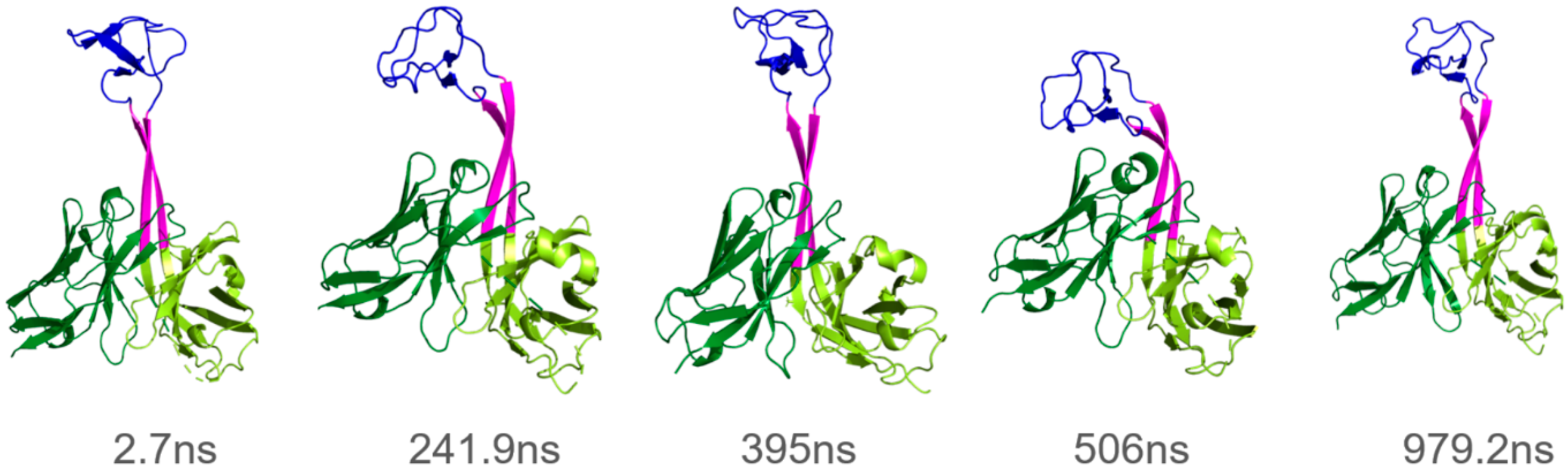
3.1. Overall Dynamics and Knob Motions
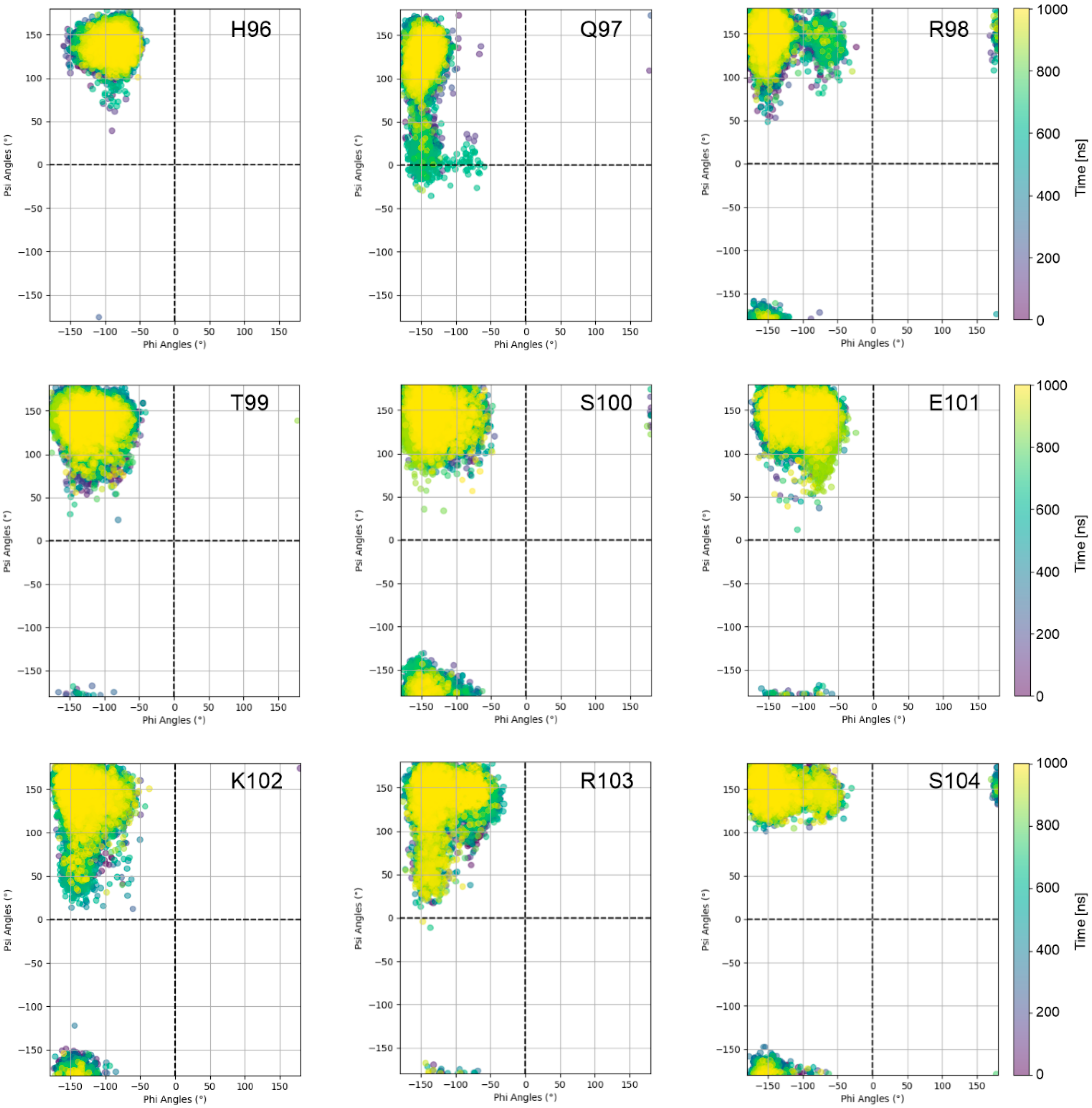
3.2. The Dynamics of the Stalk
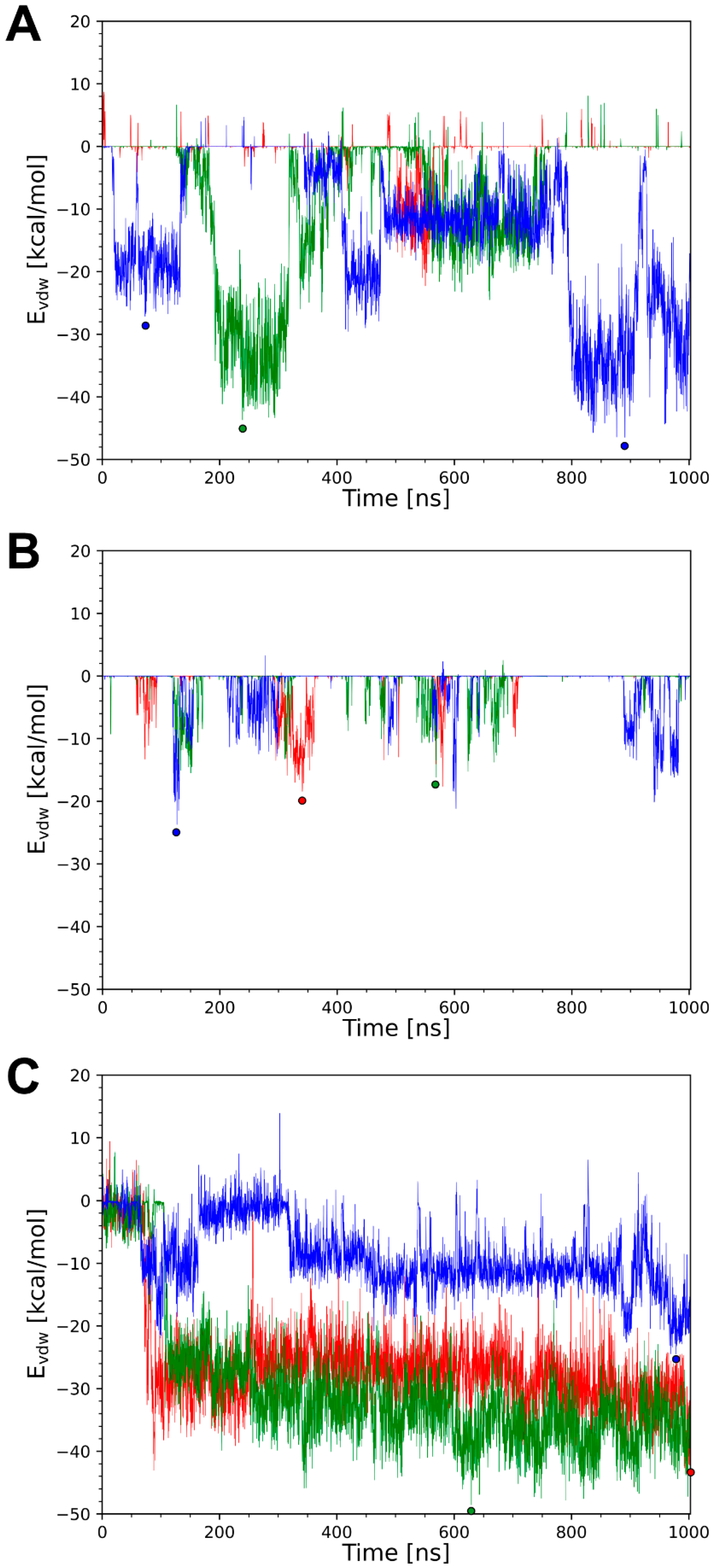
3.3. Interactions Between the Knob and the Light Chain
4. Discussion
4.1. Knobs Are Generally Highly Dynamic
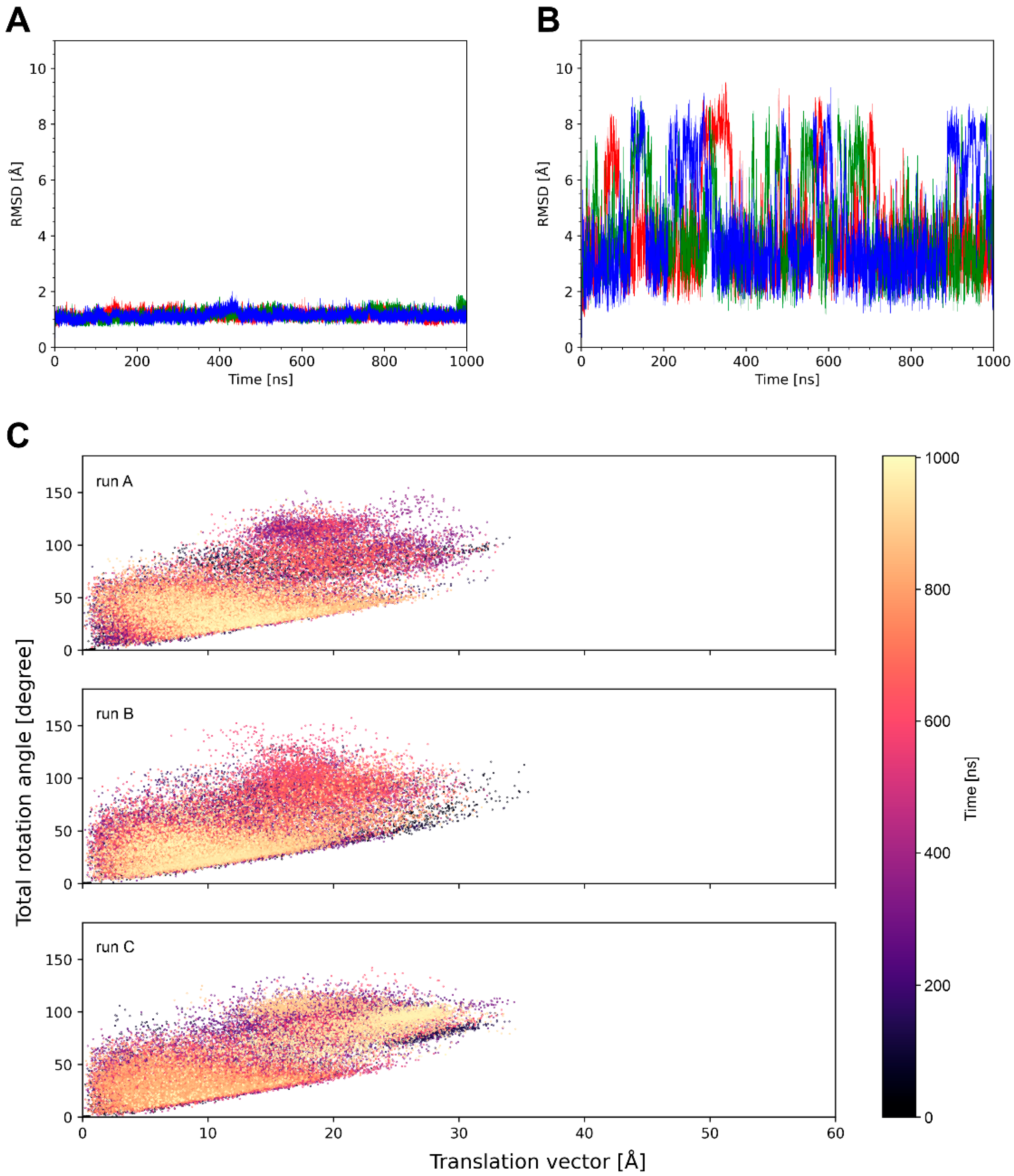
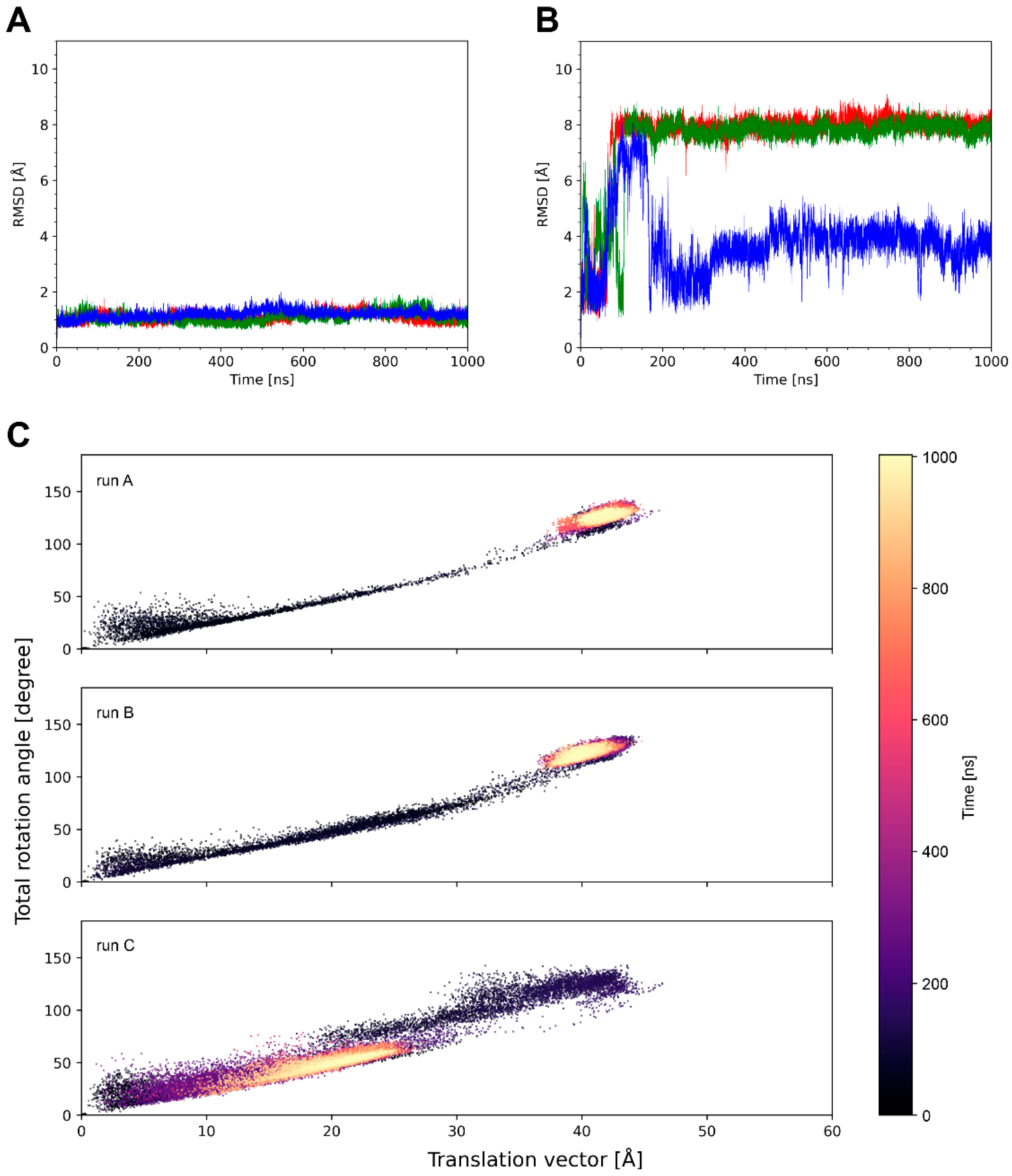
4.2. The Magnitude of Knob Motions Is System-Dependent
- (i)
- The average translational offset of the knobs ranges from 9 to 32 Å (Table 2).
- (ii)
- (iii)
- The frequency of the knob motions differs significantly (Figure 4B, Figure 5B and Figure S1; Supplementary Videos S1–S3).
- (iv)
- About half of the systems show energetically favorable knob–LC interactions over the simulation time with significant differences in the interaction energies (Table 3).
- (v)
- The sequence position and number of stalk residues with large torsion-angle fluctuations, which contribute to knob bending, differ significantly (Figure 7).
4.3. Implications of Knob Dynamics for Biological Function and Antibody Design
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Chi, X.; Li, Y.; Qiu, X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: Mechanism and regulation. Immunology 2020, 160, 233–247. [Google Scholar] [CrossRef]
- Collis, A.V.J.; Brouwer, A.P.; Martin, A.C.R. Analysis of the antigen combining site: Correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 2003, 325, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Haakenson, J.K.; Huang, R.; Smider, V.V. Diversity in the Cow Ultralong CDR H3 Antibody Repertoire. Front. Immunol. 2018, 9, 1262. [Google Scholar] [CrossRef]
- Passon, M.; de Smedt, S.; Svilenov, H.L. Principles of antibodies with ultralong complementarity-determining regions and picobodies. Biotechnol. Adv. 2023, 64, 108120. [Google Scholar] [CrossRef]
- Stanfield, R.L.; Wilson, I.A.; Smider, V.V. Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies. Sci. Immunol. 2016, 1, aaf7962. [Google Scholar] [CrossRef]
- Wang, F.; Ekiert, D.C.; Ahmad, I.; Yu, W.; Zhang, Y.; Bazirgan, O.; Torkamani, A.; Raudsepp, T.; Mwangi, W.; Criscitiello, M.F.; et al. Reshaping antibody diversity. Cell 2013, 153, 1379–1393. [Google Scholar] [CrossRef]
- Dong, J.; Finn, J.A.; Larsen, P.A.; Smith, T.P.L.; Crowe, J.E. Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains. Front. Immunol. 2019, 10, 558. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Berndsen, Z.T.; Huang, R.; Sok, D.; Warner, G.; Torres, J.L.; Burton, D.R.; Ward, A.B.; Wilson, I.A.; Smider, V.V. Structural basis of broad HIV neutralization by a vaccine-induced cow antibody. Sci. Adv. 2020, 6, eaba0468. [Google Scholar] [CrossRef]
- Huang, R.; Warner Jenkins, G.; Kim, Y.; Stanfield, R.L.; Singh, A.; Martinez-Yamout, M.; Kroon, G.J.; Torres, J.L.; Jackson, A.M.; Kelley, A.; et al. The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2023, 120, e2303455120. [Google Scholar] [CrossRef]
- Clarke, J.D.; Douangamath, A.; Mikolajek, H.; Bonnet-Di Placido, M.; Ren, J.; Fry, E.E.; Stuart, D.I.; Hammond, J.A.; Owens, R.J. The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies. Acta Crystallogr. F Struct. Biol. Commun. 2024, 80, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Altman, P.X.; Ozorowski, G.; Stanfield, R.L.; Haakenson, J.; Appel, M.; Parren, M.; Lee, W.-H.; Sang, H.; Woehl, J.; Saye-Francisco, K.; et al. Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire. PLoS Pathog. 2024, 20, e1012042. [Google Scholar] [CrossRef] [PubMed]
- Svilenov, H.L.; Sacherl, J.; Protzer, U.; Zacharias, M.; Buchner, J. Mechanistic principles of an ultra-long bovine CDR reveal strategies for antibody design. Nat. Commun. 2021, 12, 6737. [Google Scholar] [CrossRef]
- Pei, J.; Tang, M.; Grishin, N.V. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, W30–W34. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cisneros, G.A.; Cruzeiro, V.; et al. Amber 2022; University of California: San Francisco, CA, USA, 2022. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Cheatham, T.E., III; Miller, J.L.; Fox, T.; Darden, T.A.; Kollman, P.A. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. J. Am. Chem. Soc. 1995, 117, 4193–4194. [Google Scholar] [CrossRef]
- Götz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Le Grand, S.; Götz, A.W.; Walker, R.C. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013, 184, 374–380. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E. Parallelization of CPPTRAJ enables large scale analysis of molecular dynamics trajectory data. J. Comput. Chem. 2018, 39, 2110–2117. [Google Scholar] [CrossRef]
- Zhang, T.; Faraggi, E.; Zhou, Y. Fluctuations of backbone torsion angles obtained from NMR-determined structures and their prediction. Proteins 2010, 78, 3353–3362. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Sayle, R.A.; Milner-White, E.J. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 1995, 20, 374. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. The PyMOL Molecular Graphics System, Version 3.0; Schrödinger, LLC: New York, NY, USA, 2020; Available online: http://www.pymol.org/pymol (accessed on 10 July 2024).
- Henzler-Wildman, K.; Kern, D. Dynamic personalities of proteins. Nature 2007, 450, 964–972. [Google Scholar] [CrossRef]
- Wang, C.; Palmer, A.G. Solution NMR methods for quantitative identification of chemical exchange in 15 N-labeled proteins. Magn. Reson. Chem. 2003, 41, 866–876. [Google Scholar] [CrossRef]
- Trommer, J.; Lesniowski, F.; Buchner, J.; Svilenov, H.L. Specific features of a scaffolding antibody light chain. Protein Sci. 2024, 33, e4990. [Google Scholar] [CrossRef]
- Ramsland, P.A.; Kaushik, A.; Marchalonis, J.J.; Edmundson, A.B. Incorporation of long CDR3s into V domains: Implications for the structural evolution of the antibody-combining site. Exp. Clin. Immunogenet. 2001, 18, 176–198. [Google Scholar] [CrossRef]
- Macpherson, A.; Scott-Tucker, A.; Spiliotopoulos, A.; Simpson, C.; Staniforth, J.; Hold, A.; Snowden, J.; Manning, L.; van den Elsen, J.; Lawson, A.D.G. Isolation of antigen-specific, disulphide-rich knob domain peptides from bovine antibodies. PLoS Biol. 2020, 18, e3000821. [Google Scholar] [CrossRef] [PubMed]
- Kuravsky, M.; Gibbons, G.F.; Joyce, C.; Scott-Tucker, A.; Macpherson, A.; Lawson, A.D.G. Modular design of bi- and multi-specific knob domain fusions. Front. Immunol. 2024, 15, 1384467. [Google Scholar] [CrossRef] [PubMed]




| No. | System (PDB-ID) | Light Chain | Heavy Chain | Protein Residues | Residues Modeled | Water Molecules | Total Atoms |
|---|---|---|---|---|---|---|---|
| 1 | 4k3d [7] | V3-G107 | V2-S113 | 283 | - | 29,230 | 91,722 |
| 2 | 4k3e [7] | V3-G107 | V2-S113 | 273 | 3 | 26,599 | 83,732 |
| 3 | 5e99 [6] | V3-G107 | V2-S168 | 287 | - | 25,759 | 81,274 |
| 4 | 5ijv [6] | V3-G107 | V2-S149 | 271 | - | 27,250 | 85,539 |
| 5 | 5ilt [6] | V3-G107 | V2-S166 | 288 | - | 24,564 | 77,732 |
| 6 | 6e8v [8] | V3-G112 | V28-S195 | 284 | - | 24,491 | 77,426 |
| 7 | 6e9g [8] | V3-G111 | L4-S167 | 279 | 10 | 26,918 | 84,737 |
| 8 | 6e9h [8] | V3-G112 | Q29-S197 | 282 | 7 | 28,469 | 89,436 |
| 9 | 6e9i [8] | V3-L111 | V28-S194 | 283 | - | 30,014 | 93,998 |
| 10 | 6e9k [8] | V3-G112 | Q3-S162 | 275 | 5 | 22,086 | 70,105 |
| 11 | 6e9q [8] | V3-L111 | V2-S158 | 271 | 18 | 21,871 | 69,458 |
| 12 | 6e9u [8] | V3-G112 | V2-S171 | 288 | - | 26,203 | 82,603 |
| 13 | 6oo0 [9] | E3-G107 | V2-S165 | 283 | 4 | 27,374 | 86,178 |
| 14 | 8bs8 [11] | V31-G139 | V30-S197 | 288 | 1 | 27,379 | 86,142 |
| 15 | 8ecq [10] | V3-G107 | V2-S166 | 285 | - | 22,681 | 71,984 |
| 16 | 8ecv [10] | V3-G107 | V2-S174 | 280 | - | 22,415 | 71,212 |
| 17 | 8ecz [10] | V3-G107 | V2-S176 | 286 | 11 | 29,643 | 92,975 |
| 18 | 8ed1 [10] | V3-G107 | V2-S166 | 286 | - | 28,656 | 90,015 |
| 19 | 8edf [10] | V3-G107 | V2-S166 | 281 | - | 29,327 | 91,962 |
| System | RMSD [Å] (Core Domain) | RMSD [Å] (Entire ulCAB) | Translational Offset [Å] (Knob Domain) | Rotational Offset [deg] (Knob Domain) |
|---|---|---|---|---|
| 4k3d | 1.52 | 5.76 | 21.36 | 69.11 |
| 4k3e | 1.15 | 3.11 | 15.18 | 37.20 |
| 5e99 | 1.24 | 5.91 | 25.23 | 67.94 |
| 5ijv | 1.50 | 3.68 | 14.35 | 43.73 |
| 5ilt | 1.11 | 6.25 | 32.58 | 95.71 |
| 6e8v | 1.41 | 5.66 | 22.35 | 79.28 |
| 6e9g | 1.92 | 6.28 | 20.13 | 70.66 |
| 6e9h | 1.40 | 6.27 | 26.97 | 93.45 |
| 6e9i | 1.43 | 3.42 | 10.82 | 28.74 |
| 6e9k | 1.36 | 3.62 | 10.85 | 37.01 |
| 6e9q | 1.35 | 3.30 | 10.79 | 48.31 |
| 6e9u | 1.44 | 1.81 | 3.10 | 13.53 |
| 6oo0 | 1.38 | 2.92 | 9.05 | 33.37 |
| 8bs8 | 1.40 | 4.36 | 14.36 | 43.12 |
| 8ecq | 1.44 | 4.66 | 19.99 | 65.15 |
| 8ecv | 1.33 | 4.02 | 15.94 | 62.00 |
| 8ecz | 1.18 | 5.23 | 23.25 | 59.69 |
| 8ed1 | 1.31 | 4.78 | 15.69 | 54.05 |
| 8edf | 1.14 | 4.20 | 12.03 | 48.25 |
| System | EvdW (runA) | EvdW (runB) | EvdW (runC) | Average EvdW |
|---|---|---|---|---|
| 4k3d | −0.59 | −7.39 | −13.28 | −7.08 |
| 4k3e | −0.09 | −0.59 | −3.00 | −1.23 |
| 5e99 | −0.49 | −2.15 | −16.61 | −6.41 |
| 5ijv | 1.27 | 7.25 | 4.96 | 4.49 |
| 5ilt | −25.88 | −29.66 | −8.82 | −21.45 |
| 6e8v | −6.88 | −4.75 | −6.76 | −6.13 |
| 6e9g | −11.56 | −10.84 | −2.28 | −8.22 |
| 6e9h | −4.32 | −2.56 | −10.77 | −5.88 |
| 6e9i | −2.71 | −3.81 | 0.17 | −2.12 |
| 6e9k | −4.16 | −11.43 | −3.26 | −6.28 |
| 6e9q | −0.10 | −0.05 | −0.24 | −0.13 |
| 6e9u | −12.11 | −13.75 | −11.83 | −12.56 |
| 6oo0 | 0.13 | 0.09 | −0.03 | 0.06 |
| 8bs8 | 0.20 | −0.43 | −0.46 | −0.23 |
| 8ecq | −5.91 | −5.53 | −4.23 | −5.22 |
| 8ecv | −0.85 | −0.51 | −2.88 | −1.41 |
| 8ecz | −0.92 | −7.63 | −0.63 | −3.06 |
| 8ed1 | −0.47 | −1.30 | −0.48 | −0.75 |
| 8edf | −0.81 | −0.84 | −1.44 | −1.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denysenko, O.; Horn, A.H.C.; Sticht, H. Comparative Molecular Dynamics Study of 19 Bovine Antibodies with Ultralong CDR H3. Antibodies 2025, 14, 70. https://doi.org/10.3390/antib14030070
Denysenko O, Horn AHC, Sticht H. Comparative Molecular Dynamics Study of 19 Bovine Antibodies with Ultralong CDR H3. Antibodies. 2025; 14(3):70. https://doi.org/10.3390/antib14030070
Chicago/Turabian StyleDenysenko, Olena, Anselm H. C. Horn, and Heinrich Sticht. 2025. "Comparative Molecular Dynamics Study of 19 Bovine Antibodies with Ultralong CDR H3" Antibodies 14, no. 3: 70. https://doi.org/10.3390/antib14030070
APA StyleDenysenko, O., Horn, A. H. C., & Sticht, H. (2025). Comparative Molecular Dynamics Study of 19 Bovine Antibodies with Ultralong CDR H3. Antibodies, 14(3), 70. https://doi.org/10.3390/antib14030070







