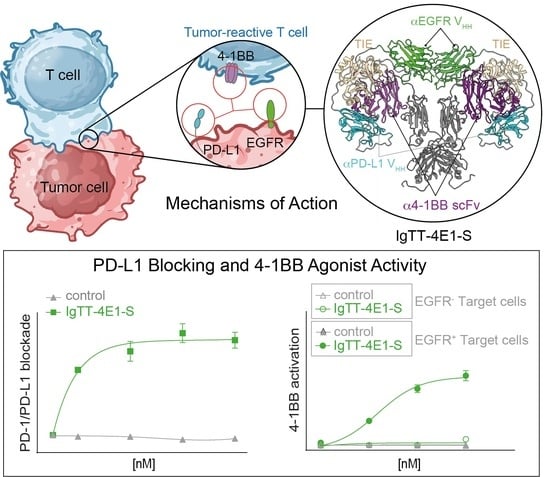
Characterization of a Trispecific PD-L1 Blocking Antibody That Exhibits EGFR-Conditional 4-1BB Agonist Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Construction of the Expression Vectors
2.3. Expression and Purification of the Recombinant Antibodies
2.4. Enzyme-Linked Immunosorbent Assay
2.5. Western Blotting
2.6. Size-Exclusion Chromatography
2.7. Molecular Modeling
2.8. Biolayer Interferometry
2.9. Flow Cytometry
2.10. ADCC Reporter Bioassay
2.11. PD-1/PD-L1 Blockade Bioassay
2.12. Antigen-Dependent Jurkat 4-1BB Activation Assay
2.13. IFNγ Secretion Analysis
2.14. Statistical Analysis
3. Results
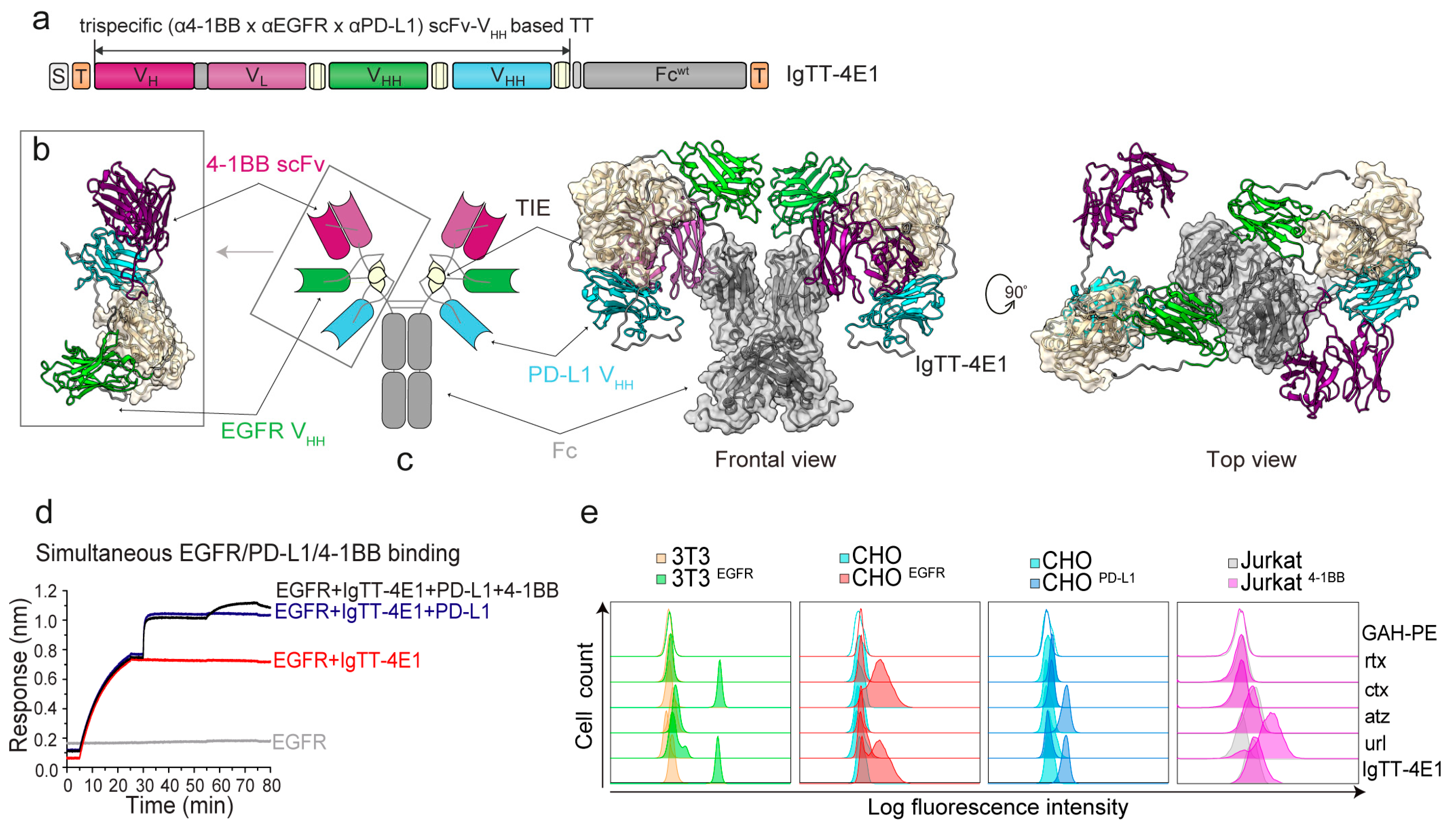
3.1. Generation of IgTT-4E1 and IgTT-4E1-S
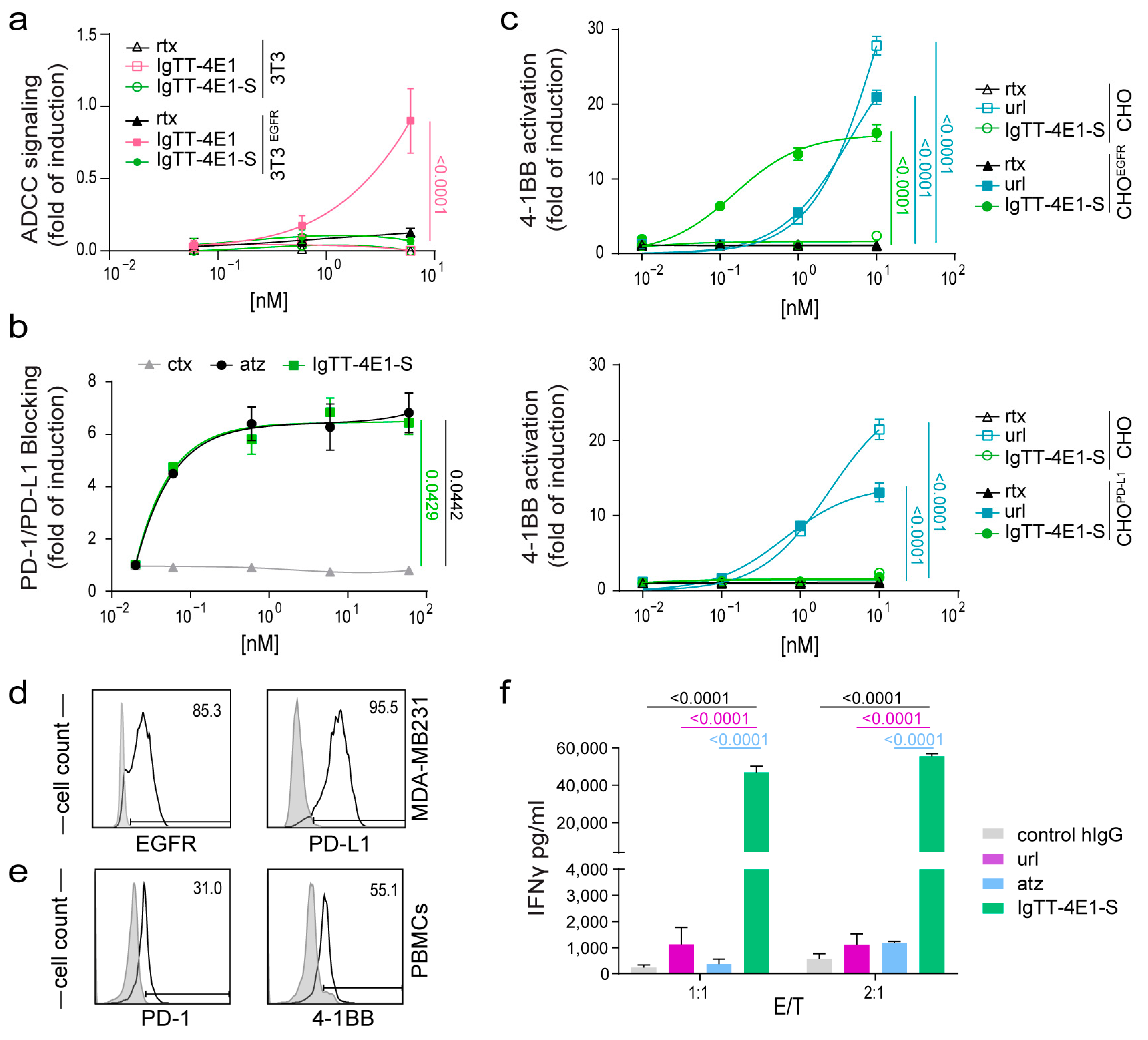
3.2. Determination of Fc-Mediated Effector Functions
3.3. Effect of IgTT-4E1-S on PD-1/PD-L1 Blockade
3.4. Costimulatory Activity of IgTT-4E1-S
3.5. IgTT-4E1-S Enhances the Activation of Primary Human T Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, P.; Goswami, S.; Raychaudhuri, D.; Siddiqui, B.A.; Singh, P.; Nagarajan, A.; Liu, J.; Subudhi, S.K.; Poon, C.; Gant, K.L.; et al. Immune checkpoint therapy-current perspectives and future directions. Cell 2023, 186, 1652–1669. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Antibody Therapeutics Approved or in Regulatory Review in the EU or US. The Antibody Society. Available online: https://www.antibodysociety.org/resources/approved-antibodies/ (accessed on 10 January 2024).
- Chester, C.; Sanmamed, M.F.; Wang, J.; Melero, I. Immunotherapy targeting 4-1BB: Mechanistic rationale, clinical results, and future strategies. Blood 2018, 131, 49–57. [Google Scholar] [CrossRef]
- Melero, I.; Shuford, W.W.; Newby, S.A.; Aruffo, A.; Ledbetter, J.A.; Hellström, K.E.; Mittler, R.S.; Chen, L. Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat. Med. 1997, 3, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ravetch, J.V. Antitumor activities of agonistic anti-TNFR antibodies require differential FcγRIIB coengagement in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 19501–19506. [Google Scholar] [CrossRef]
- Compte, M.; Harwood, S.L.; Muñoz, I.G.; Navarro, R.; Zonca, M.; Perez-Chacon, G.; Erce-Llamazares, A.; Merino, N.; Tapia-Galisteo, A.; Cuesta, A.M.; et al. A tumor-targeted trimeric 4-1BB-agonistic antibody induces potent anti-tumor immunity without systemic toxicity. Nat. Commun. 2018, 9, 4809. [Google Scholar] [CrossRef] [PubMed]
- Claus, C.; Ferrara, C.; Xu, W.; Sam, J.; Lang, S.; Uhlenbrock, F.; Albrecht, R.; Herter, S.; Schlenker, R.; Hüsser, T.; et al. Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy. Sci. Transl. Med. 2019, 11, eaav5989. [Google Scholar] [CrossRef]
- Hinner, M.J.; Aiba, R.S.B.; Jaquin, T.J.; Berger, S.; Dürr, M.C.; Schlosser, C.; Allersdorfer, A.; Wiedenmann, A.; Matschiner, G.; Schüler, J.; et al. Tumor-Localized Costimulatory T-Cell Engagement by the 4-1BB/HER2 Bispecific Antibody-Anticalin Fusion PRS-343. Clin. Cancer Res. 2019, 25, 5878–5889. [Google Scholar] [CrossRef]
- Compte, M.; Harwood, S.L.; Martínez-Torrecuadrada, J.; Perez-Chacon, G.; González-García, P.; Tapia-Galisteo, A.; Van Bergen En Henegouwen, P.M.P.; Sánchez, A.; Fabregat, I.; Sanz, L.; et al. Case Report: An EGFR-Targeted 4-1BB-agonistic Trimerbody Does Not Induce Hepatotoxicity in Transgenic Mice With Liver Expression of Human EGFR. Front. Immunol. 2020, 11, 614363. [Google Scholar] [CrossRef]
- Hangiu, O.; Compte, M.; Dinesen, A.; Navarro, R.; Tapia-Galisteo, A.; Mandrup, O.A.; Erce-Llamazares, A.; Lázaro-Gorines, R.; Nehme-Álvarez, D.; Domínguez-Alonso, C.; et al. Tumor targeted 4-1BB agonist antibody-albumin fusions with high affinity to FcRn induce anti-tumor immunity without toxicity. iScience 2022, 25, 104958. [Google Scholar] [CrossRef]
- Compte, M.; Harwood, S.L.; Erce-Llamazares, A.; Tapia-Galisteo, A.; Romero, E.; Ferrer, I.; Garrido-Martin, E.M.; Enguita, A.B.; Ochoa, M.C.; Blanco, B.; et al. An Fc-free EGFR-specific 4-1BB-agonistic Trimerbody Displays Broad Antitumor Activity in Humanized Murine Cancer Models without Toxicity. Clin. Cancer Res. 2021, 27, 3167–3177. [Google Scholar] [CrossRef]
- Mikkelsen, K.; Harwood, S.L.; Compte, M.; Merino, N.; Mølgaard, K.; Lykkemark, S.; Alvarez-Mendez, A.; Blanco, F.J.; Álvarez-Vallina, L. Carcinoembryonic Antigen (CEA)-Specific 4-1BB-Costimulation Induced by CEA-Targeted 4-1BB-Agonistic Trimerbodies. Front. Immunol. 2019, 10, 1791. [Google Scholar] [CrossRef]
- Shen, A.; Liu, W.; Wang, H.; Zeng, X.; Wang, M.; Zhang, D.; Zhao, Q.; Fang, Q.; Wang, F.; Cheng, L.; et al. A novel 4-1BB/HER2 bispecific antibody shows potent antitumor activities by increasing and activating tumor-infiltrating T cells. Am. J. Cancer Res. 2023, 13, 3246–3256. [Google Scholar]
- Tapia-Galisteo, A.; Compte, M.; Álvarez-Vallina, L.; Sanz, L. When three is not a crowd: Trispecific antibodies for enhanced cancer immunotherapy. Theranostics 2023, 13, 1028–1041. [Google Scholar] [CrossRef]
- Geuijen, C.; Tacken, P.; Wang, L.-C.; Klooster, R.; van Loo, P.F.; Zhou, J.; Mondal, A.; Liu, Y.; Kramer, A.; Condamine, T.; et al. A human CD137×PD-L1 bispecific antibody promotes anti-tumor immunity via context-dependent T cell costimulation and checkpoint blockade. Nat. Commun. 2021, 12, 4445. [Google Scholar] [CrossRef]
- Cheng, L.-S.; Zhu, M.; Gao, Y.; Liu, W.-T.; Yin, W.; Zhou, P.; Zhu, Z.; Niu, L.; Zeng, X.; Zhang, D.; et al. An Fc-muted bispecific antibody targeting PD-L1 and 4-1BB induces antitumor immune activity in colorectal cancer without systemic toxicity. Cell Mol. Biol. Lett. 2023, 28, 47. [Google Scholar] [CrossRef]
- Jeong, S.; Park, E.; Kim, H.-D.; Sung, E.; Kim, H.; Jeon, J.; Kim, Y.; Jung, U.; Son, Y.-G.; Hong, Y.; et al. Novel anti-4-1BB×PD-L1 bispecific antibody augments anti-tumor immunity through tumor-directed T-cell activation and checkpoint blockade. J. Immunother. Cancer 2021, 9, e002428. [Google Scholar] [CrossRef]
- Peper-Gabriel, J.K.; Pavlidou, M.; Pattarini, L.; Morales-Kastresana, A.; Jaquin, T.J.; Gallou, C.; Hansbauer, E.-M.; Richter, M.; Lelievre, H.; Scholer-Dahirel, A.; et al. The PD-L1/4-1BB Bispecific Antibody-Anticalin Fusion Protein PRS-344/S095012 Elicits Strong T-Cell Stimulation in a Tumor-Localized Manner. Clin. Cancer Res. 2022, 28, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Altintas, I.; Gieseke, F.; Schoedel, K.B.; Burm, S.M.; Toker, A.; Salcedo, T.W.; Verzijl, D.; Eisel, D.; Grunwitz, C.; et al. An Fc-inert PD-L1×4-1BB bispecific antibody mediates potent anti-tumor immunity in mice by combining checkpoint inhibition and conditional 4-1BB co-stimulation. Oncoimmunology 2022, 11, 2030135. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Garralda, E.; Altintas, I.; Gieseke, F.; Geva, R.; Ben-Ami, E.; Maurice-Dror, C.; Calvo, E.; LoRusso, P.M.; Alonso, G.; et al. Preclinical Characterization and Phase I Trial Results of a Bispecific Antibody Targeting PD-L1 and 4-1BB (GEN1046) in Patients with Advanced Refractory Solid Tumors. Cancer Discov. 2022, 12, 1248–1265. [Google Scholar] [CrossRef]
- Alvarez-Cienfuegos, A.; Nuñez-Prado, N.; Compte, M.; Cuesta, A.M.; Blanco-Toribio, A.; Harwood, S.L.; Villate, M.; Merino, N.; Bonet, J.; Navarro, R.; et al. Intramolecular trimerization, a novel strategy for making multispecific antibodies with controlled orientation of the antigen binding domains. Sci. Rep. 2016, 6, 28643. [Google Scholar] [CrossRef]
- Rubio-Pérez, L.; Lázaro-Gorines, R.; Harwood, S.L.; Compte, M.; Navarro, R.; Tapia-Galisteo, A.; Bonet, J.; Blanco, B.; Lykkemark, S.; Ramírez-Fernández, Á.; et al. A PD-L1/EGFR bispecific antibody combines immune checkpoint blockade and direct anti-cancer action for an enhanced anti-tumor response. Oncoimmunology 2023, 12, 2205336. [Google Scholar] [CrossRef]
- Delidakis, G.; Kim, J.E.; George, K.; Georgiou, G. Improving Antibody Therapeutics by Manipulating the Fc Domain: Immunological and Structural Considerations. Annu. Rev. Biomed. Eng. 2022, 24, 249–274. [Google Scholar] [CrossRef]
- Roskoski, R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- Silva-Pilipich, N.; Blanco, E.; Lozano, T.; Martisova, E.; Igea, A.; Herrador-Cañete, G.; Ballesteros-Briones, M.C.; Gorraiz, M.; Sarrión, P.; González-Sapienza, G.; et al. Local delivery of optimized nanobodies targeting the PD-1/PD-L1 axis with a self-amplifying RNA viral vector induces potent antitumor responses. Cancer Lett. 2023, 561, 216139. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Saphire, E.O.; Parren, P.W.; Pantophlet, R.; Zwick, M.B.; Morris, G.M.; Rudd, P.M.; Dwek, R.A.; Stanfield, R.L.; Burton, D.R.; Wilson, I.A. Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science 2001, 293, 1155–1159. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Liu, S.; Miersch, S.; Li, P.; Bai, B.; Liu, C.; Qin, W.; Su, J.; Huang, H.; Pan, J.; Sidhu, S.S.; et al. A Synthetic Human Antibody Antagonizes IL-18Rβ Signaling Through an Allosteric Mechanism. J. Mol. Biol. 2020, 432, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.; Kim, H.S.; Tong, R.K.; Bainbridge, T.W.; Vernes, J.-M.; Zhang, Y.; Lin, Y.L.; Chung, S.; Dennis, M.S.; Zuchero, Y.J.Y.; et al. Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice. J. Biol. Chem. 2017, 292, 3900–3908. [Google Scholar] [CrossRef] [PubMed]
- Zhai, T.; Wang, C.; Xu, Y.; Huang, W.; Yuan, Z.; Wang, T.; Dai, S.; Peng, S.; Pang, T.; Jiang, W.; et al. Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer. J. Immunother. Cancer 2021, 9, e002131. [Google Scholar] [CrossRef] [PubMed]
- Warmuth, S.; Gunde, T.; Snell, D.; Brock, M.; Weinert, C.; Simonin, A.; Hess, C.; Tietz, J.; Johansson, M.; Spiga, F.M.; et al. Engineering of a trispecific tumor-targeted immunotherapy incorporating 4-1BB co-stimulation and PD-L1 blockade. Oncoimmunology 2021, 10, 2004661. [Google Scholar] [CrossRef]
- Bitra, A.; Doukov, T.; Croft, M.; Zajonc, D.M. Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier. J. Biol. Chem. 2018, 293, 9958–9969. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.R.; Bagchi, A.; Roovers, R.C.; van Bergen en Henegouwen, P.M.P.; Ferguson, K.M. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure 2013, 21, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Bluemel, C.; Hausmann, S.; Fluhr, P.; Sriskandarajah, M.; Stallcup, W.B.; Baeuerle, P.A.; Kufer, P. Epitope distance to the target cell membrane and antigen size determine the potency of T cell-mediated lysis by BiTE antibodies specific for a large melanoma surface antigen. Cancer Immunol. Immunother. 2010, 59, 1197–1209. [Google Scholar] [CrossRef]
- Zhang, W.; Pang, Q.; Yan, C.; Wang, Q.; Yang, J.; Yu, S.; Liu, X.; Yuan, Z.; Wang, P.; Xiao, Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco. Targets Ther. 2017, 10, 763–771. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubio-Pérez, L.; Frago, S.; Compte, M.; Navarro, R.; Harwood, S.L.; Lázaro-Gorines, R.; Gómez-Rosel, M.; Hangiu, O.; Silva-Pilipich, N.; Vanrell, L.; et al. Characterization of a Trispecific PD-L1 Blocking Antibody That Exhibits EGFR-Conditional 4-1BB Agonist Activity. Antibodies 2024, 13, 34. https://doi.org/10.3390/antib13020034
Rubio-Pérez L, Frago S, Compte M, Navarro R, Harwood SL, Lázaro-Gorines R, Gómez-Rosel M, Hangiu O, Silva-Pilipich N, Vanrell L, et al. Characterization of a Trispecific PD-L1 Blocking Antibody That Exhibits EGFR-Conditional 4-1BB Agonist Activity. Antibodies. 2024; 13(2):34. https://doi.org/10.3390/antib13020034
Chicago/Turabian StyleRubio-Pérez, Laura, Susana Frago, Marta Compte, Rocío Navarro, Seandean L. Harwood, Rodrigo Lázaro-Gorines, Marina Gómez-Rosel, Oana Hangiu, Noelia Silva-Pilipich, Lucía Vanrell, and et al. 2024. "Characterization of a Trispecific PD-L1 Blocking Antibody That Exhibits EGFR-Conditional 4-1BB Agonist Activity" Antibodies 13, no. 2: 34. https://doi.org/10.3390/antib13020034
APA StyleRubio-Pérez, L., Frago, S., Compte, M., Navarro, R., Harwood, S. L., Lázaro-Gorines, R., Gómez-Rosel, M., Hangiu, O., Silva-Pilipich, N., Vanrell, L., Smerdou, C., & Álvarez-Vallina, L. (2024). Characterization of a Trispecific PD-L1 Blocking Antibody That Exhibits EGFR-Conditional 4-1BB Agonist Activity. Antibodies, 13(2), 34. https://doi.org/10.3390/antib13020034