New Anti-RSV Nucleoprotein Monoclonal Antibody Pairs Discovered Using Rabbit Phage Display Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Immunization
2.2. Library Construction
2.3. Library Validation
2.4. Phage Infection and Preparation
2.5. Phage Display
2.5.1. Panning Selection
2.5.2. Screening
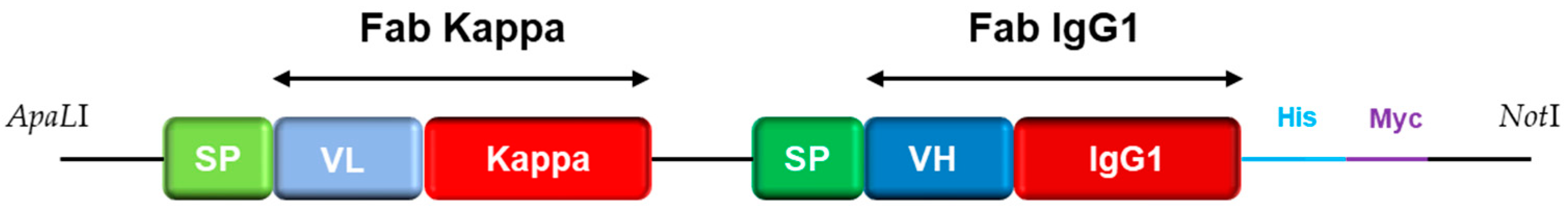
2.6. Recombinant Engineering
2.6.1. Identification of Candidates
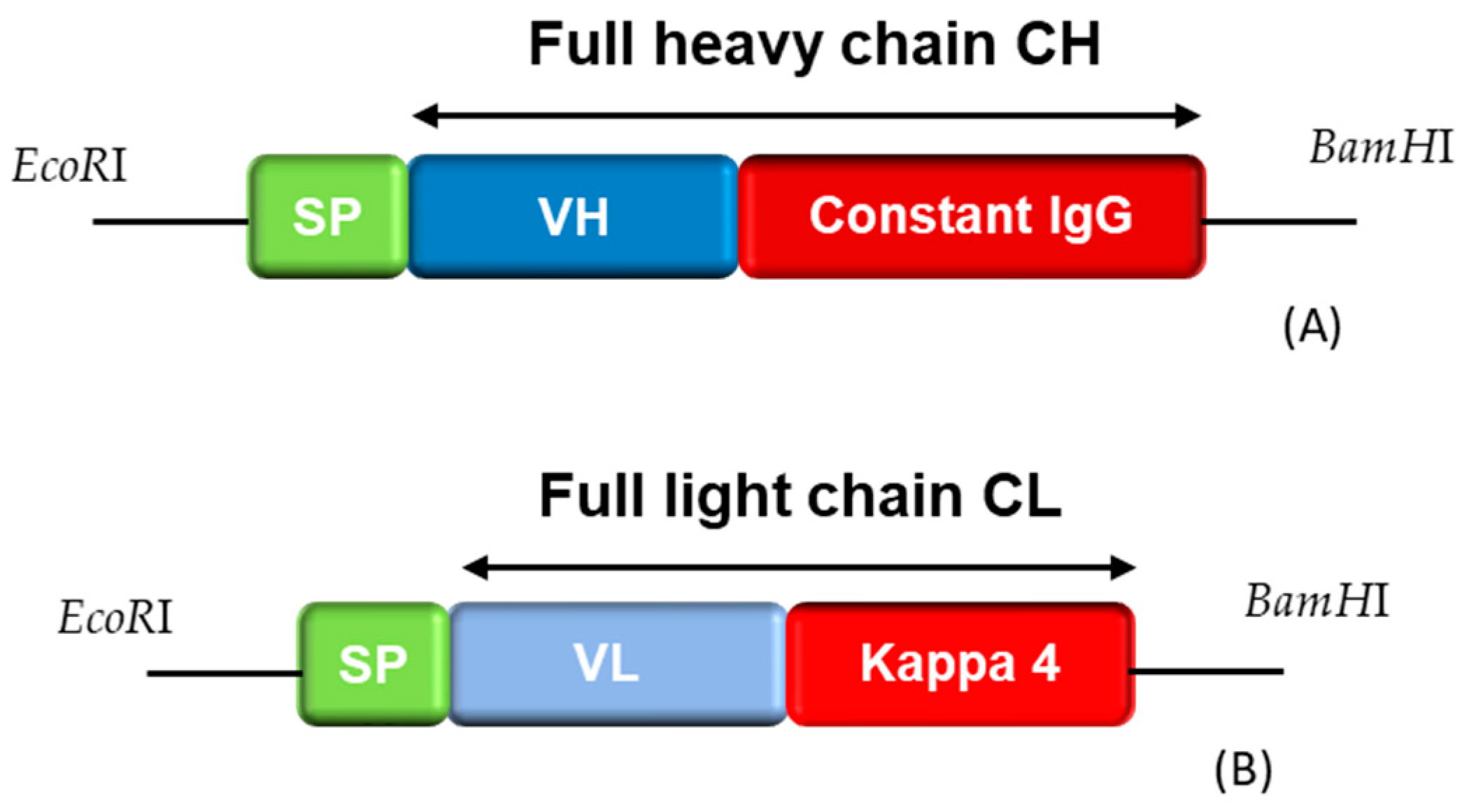
2.6.2. Reformatting and Recombinant Production of the Full IgG
2.7. Pairing Tests
2.7.1. Recombinant Strains
2.7.2. Native Lysates
3. Results
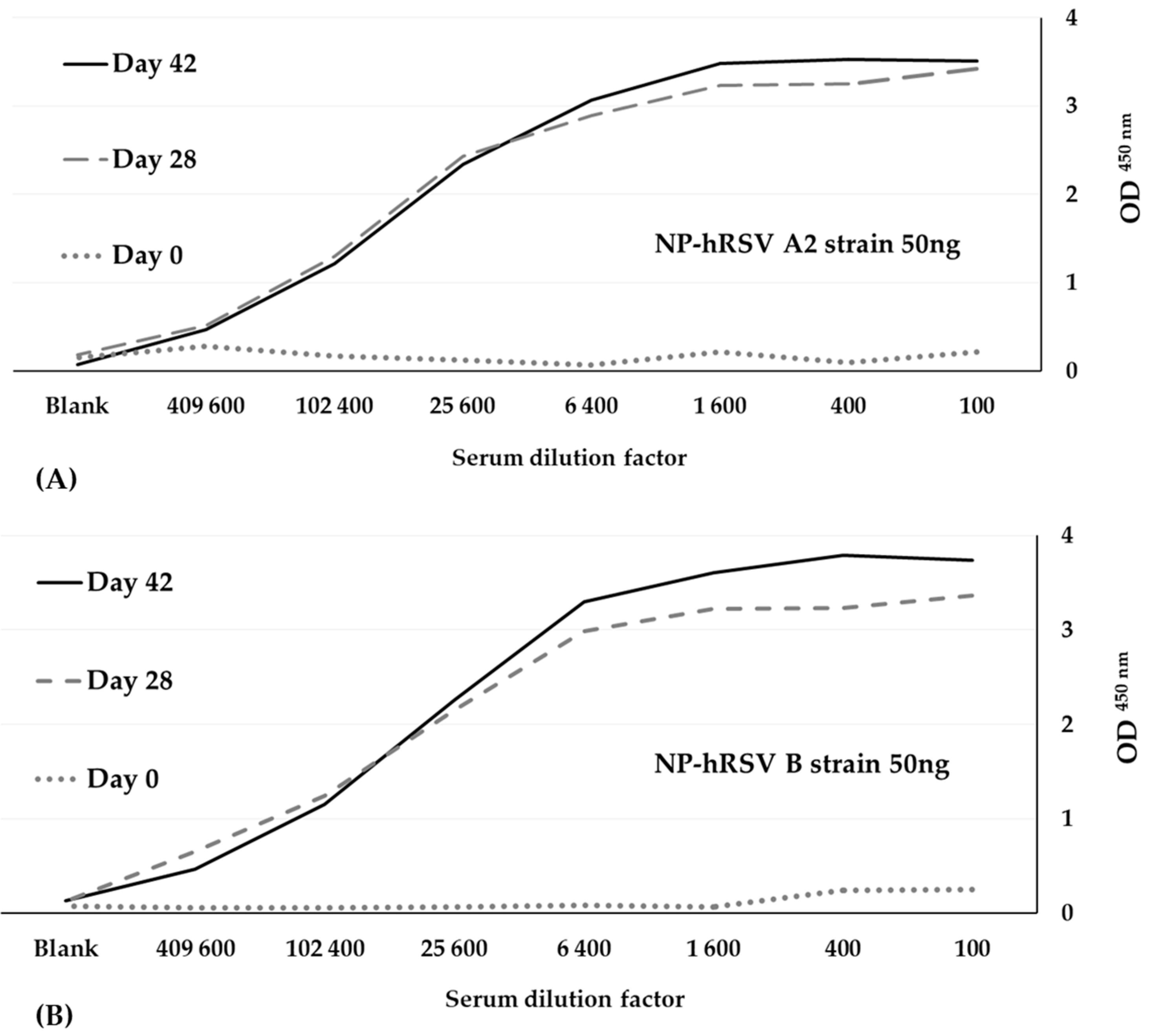
3.1. Immunization
3.2. Library Construction
- -
- The library size was to be >106 colony-forming units per mL (cfu/mL).
- -
- The insertion rate (IR) was determined after the colony PCRs on the randomly chosen clones from the bacterial spread of the electroporated library. A clone was qualified as positive when it showed a 1800 bp PCR product. A library with good quality should show >80% IR.
- -
- The quality of the PCR inserts cloned in our phagemid was assessed by sequencing eight randomly chosen positive PCR products to determine the percentage of plasmids with an ORF for the chimeric Fab and for the evaluation of the sequence variability. A library with good diversity should show <10% identical sequences.
3.3. Panning Selection
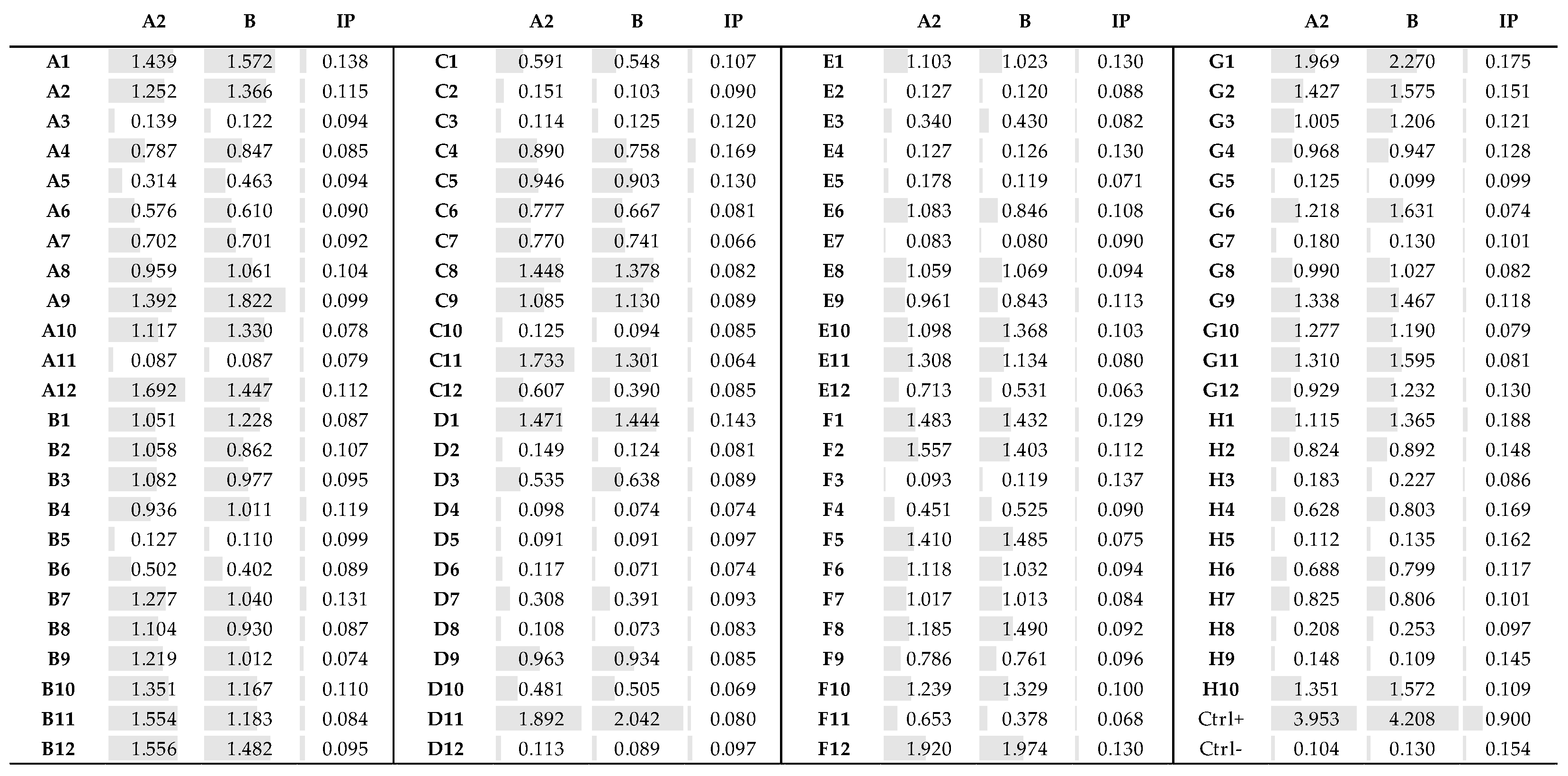
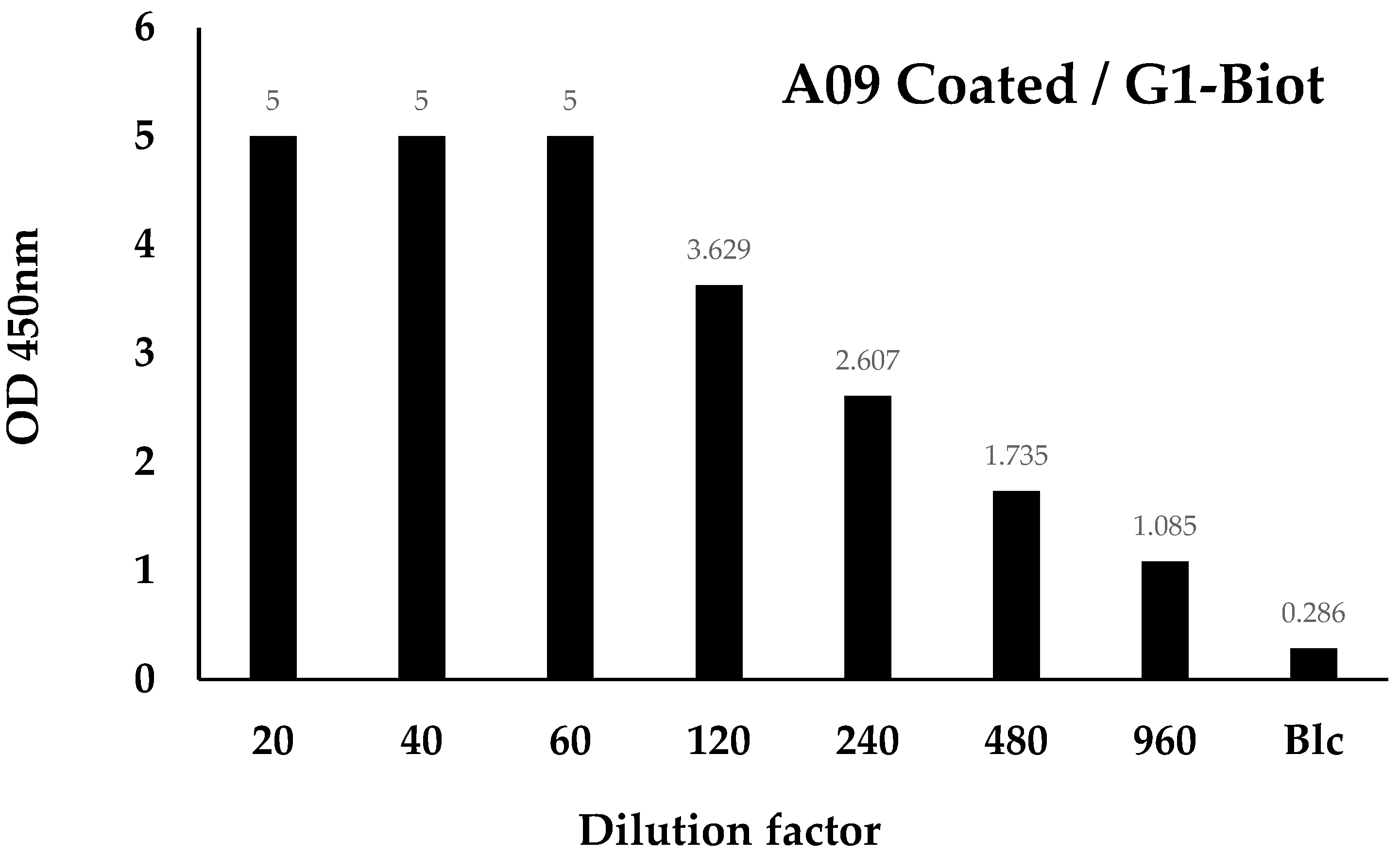
3.4. Fab Screening by ELISA
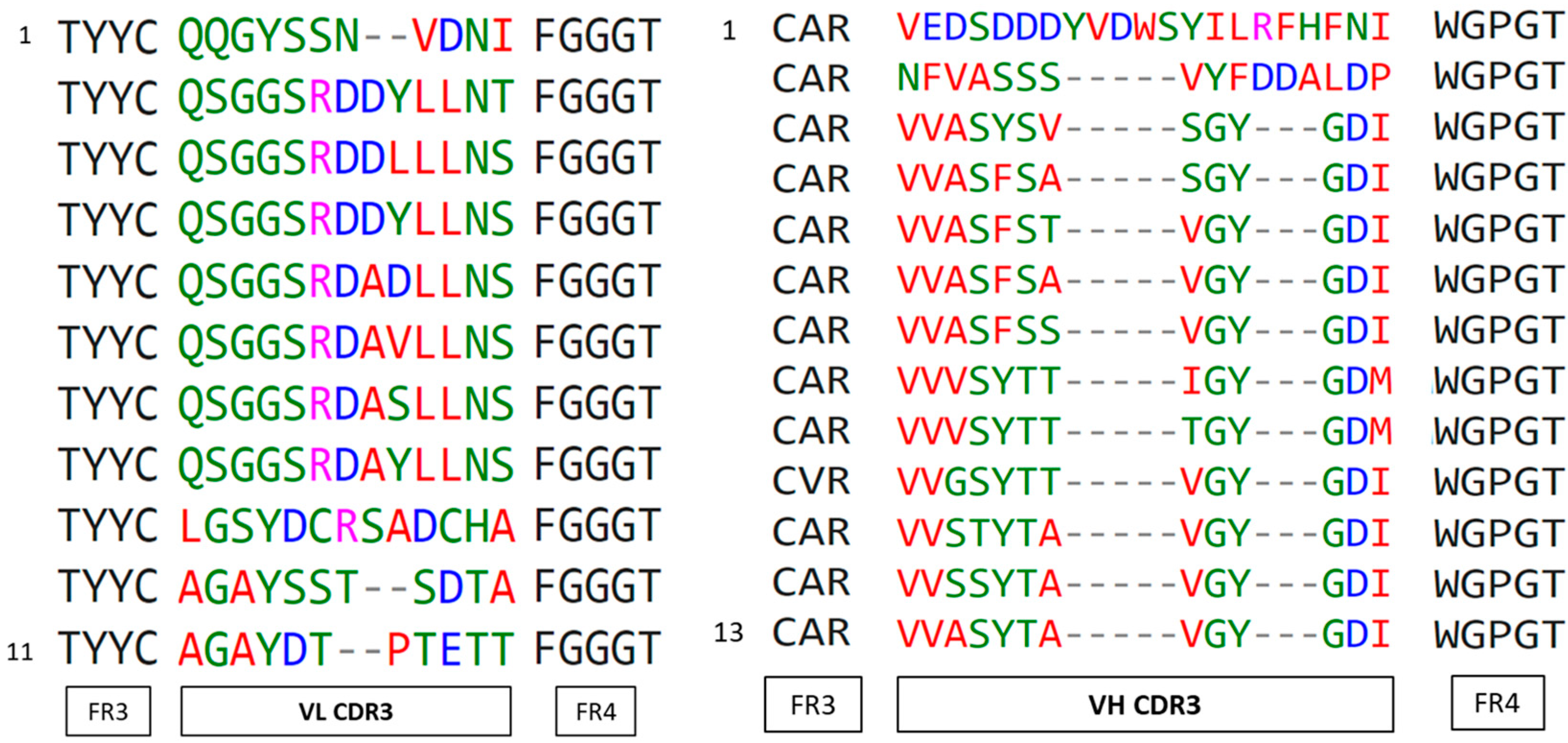
3.5. Identification of the Fab Clones
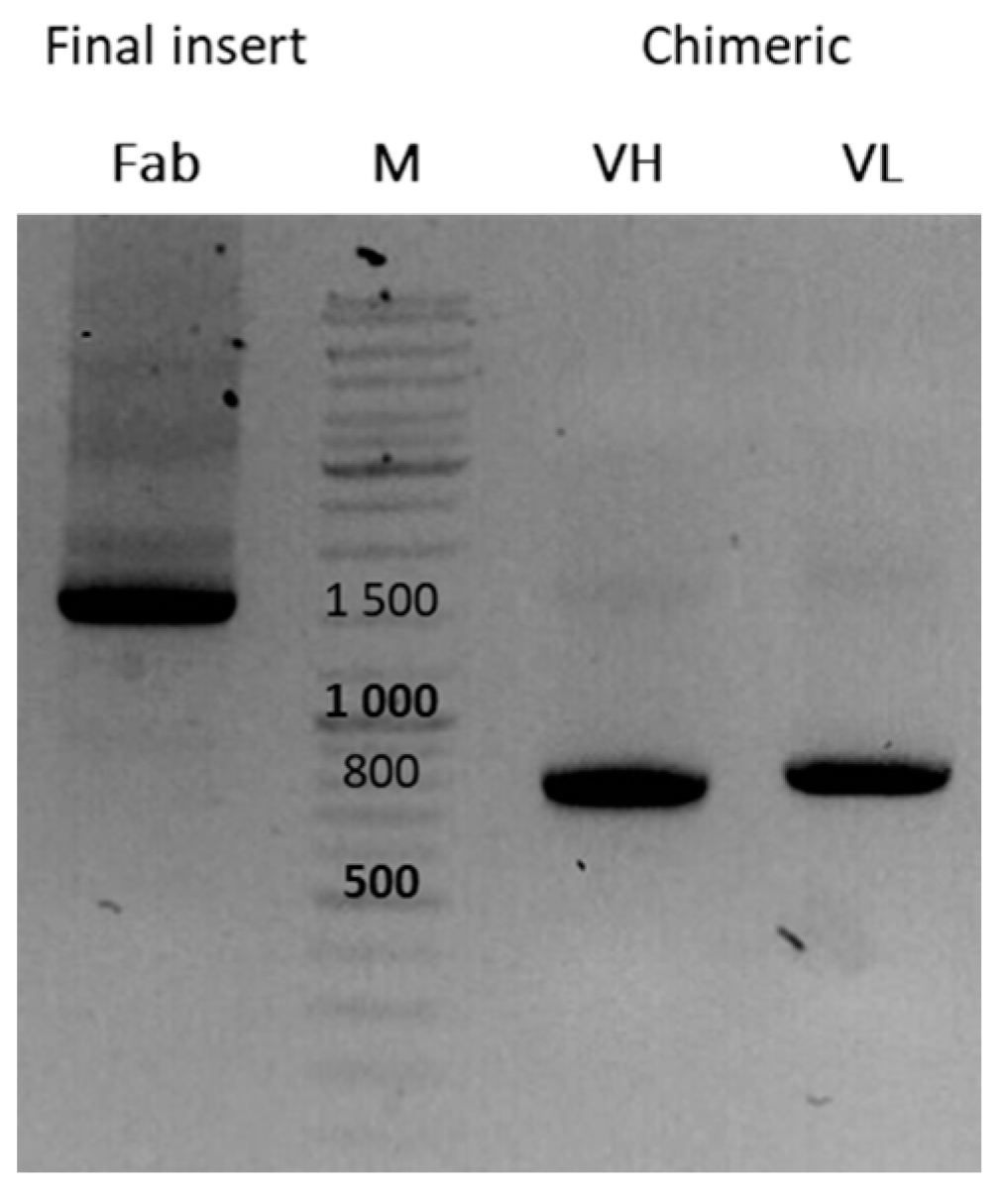
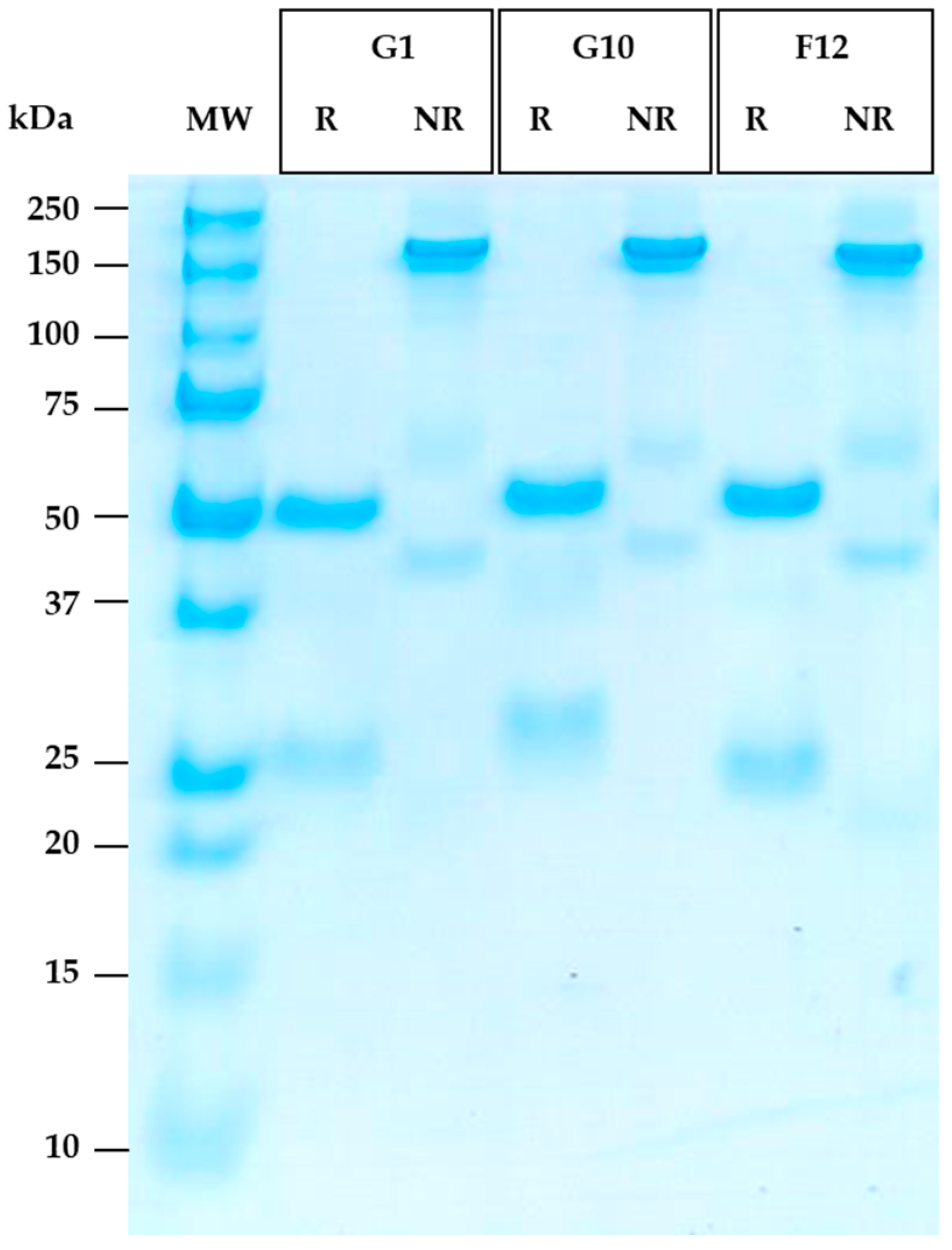
3.6. Reformatting and Production of the Full Recombinant Rabbit IgG
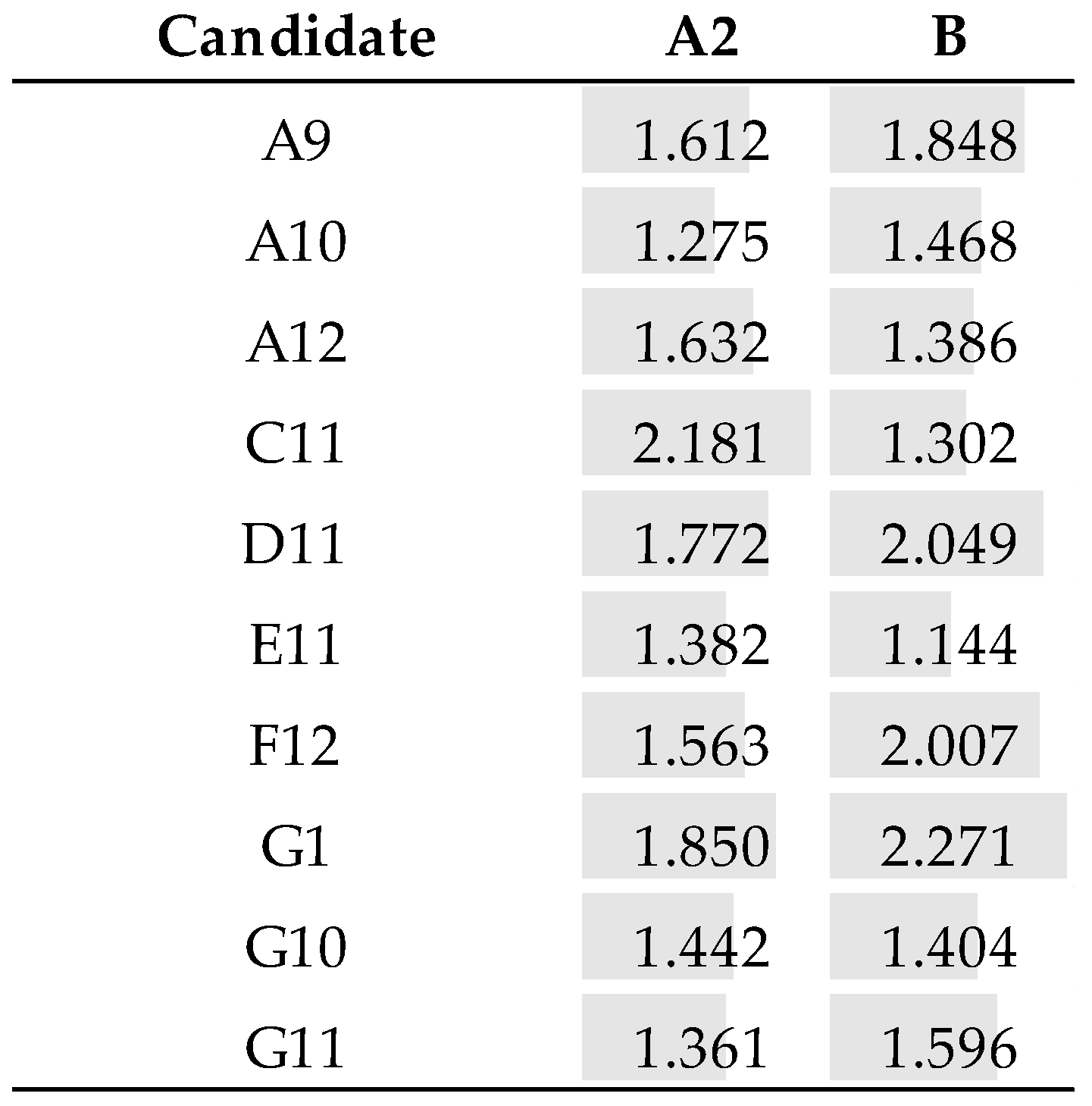
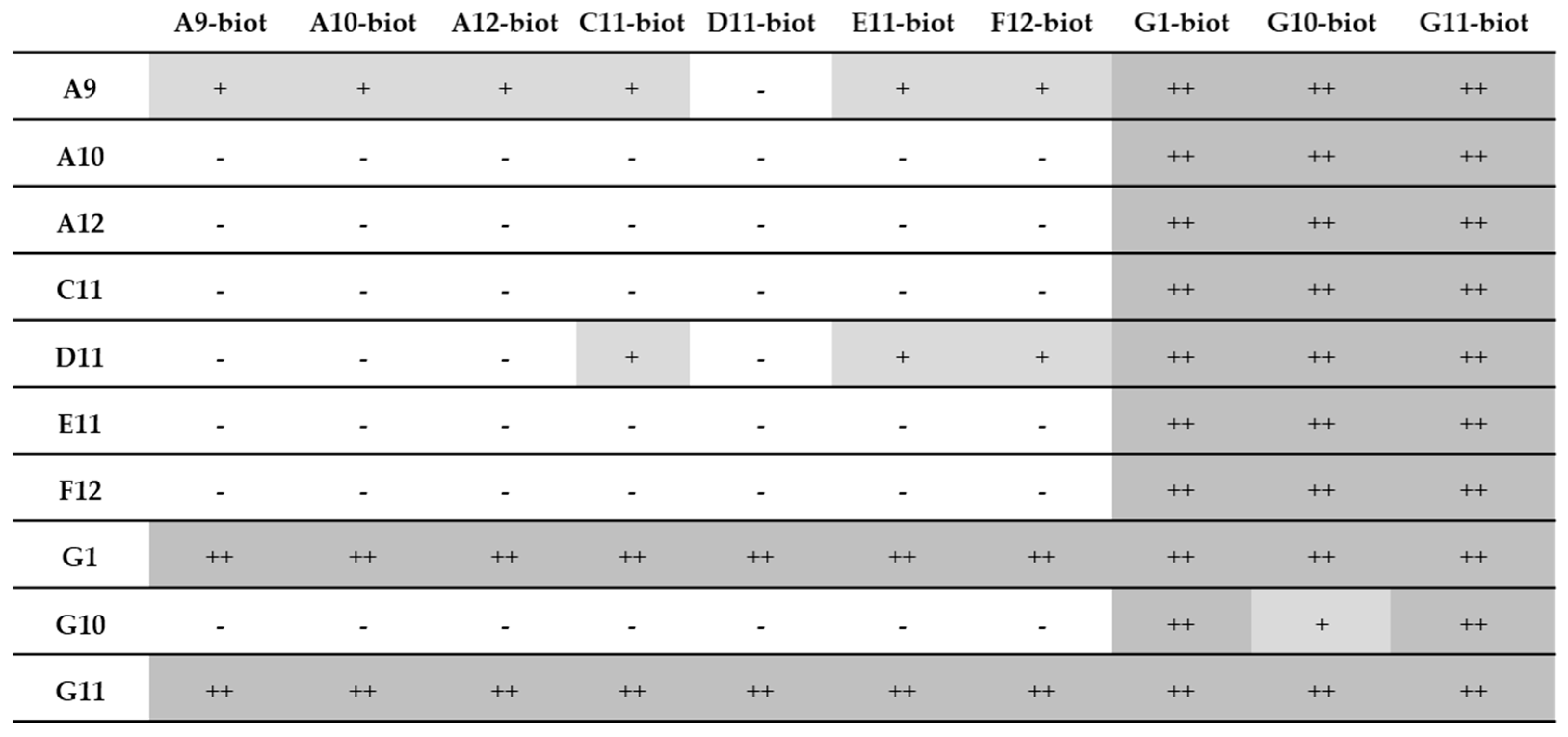
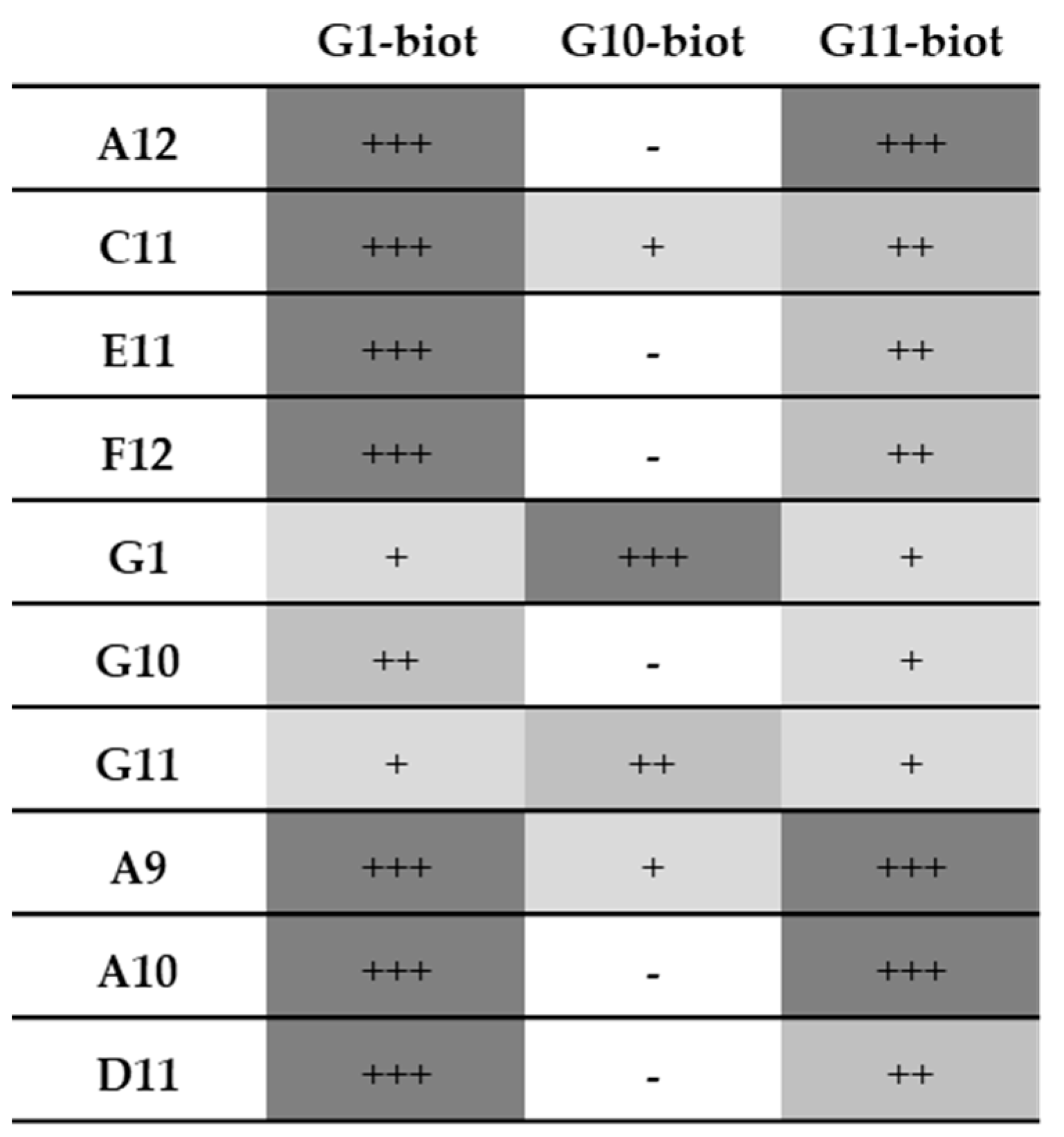
3.7. Pairing Tests
3.7.1. Recombinant Samples
3.7.2. Native Lysates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Respiratory Syncytial Virus (RSV) Disease. Available online: https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccine-standardization/respiratory-syncytial-virus-disease (accessed on 27 April 2023).
- Centre for Disease Control and Prevention. Respiratory Syncytial Virus Infection (RSV)|CDC. Available online: https://www.cdc.gov/rsv/index.html (accessed on 27 April 2023).
- Nair, H.; Nokes, D.J.; Gessner, B.D.; Dherani, M.; Madhi, S.A.; Singleton, R.J.; O’Brien, K.L.; Roca, A.; Wright, P.F.; Bruce, N.; et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet 2010, 375, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- González, L.A.; Vázquez, Y.; Mora, J.E.; Palavecino, C.E.; Bertrand, P.; Ferrés, M.; Contreras, A.M.; Beckhaus, A.A.; Riedel, C.A.; Bueno, S.M. Evaluation of monoclonal antibodies that detect conserved proteins from Respiratory Syncytial Virus, Metapneumovirus and Adenovirus in human samples. J. Virol. Methods 2018, 254, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, L.; Tang, A.; Callahan, C.; Pristatsky, P.; Swoyer, R.; Cejas, P.; Nahas, D.; Galli, J.; Cosmi, S.; et al. Discovery and characterization of phage display-derived human monoclonal antibodies against RSV F glycoprotein. PLoS ONE 2016, 11, e0156798. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, Z.; Ni, F.; Chen, X.; Ma, J.; Patel, N.; Lu, H.; Liu, Y.; Tian, J.; Flyer, D. Structure basis of neutralization by a novel site II/IV antibody against respiratory syncytial virus fusion protein. PLoS ONE 2019, 14, e0210749. [Google Scholar] [CrossRef] [PubMed]
- Leirs, K.; Tewari Kumar, P.; Decrop, D.; Pérez-Ruiz, E.; Leblebici, P.; Van Kelst, B.; Compernolle, G.; Meeuws, H.; Van Wesenbeeck, L.; Lagatie, O.; et al. Bioassay Development for Ultrasensitive Detection of Influenza A Nucleoprotein Using Digital ELISA. Anal. Chem. 2016, 88, 8450–8458. [Google Scholar] [CrossRef] [PubMed]
- Hillig, T.; Kristensen, J.; Brasen, C.; Brandslund, I.; Olsen, D.; Davidsen, C.; Madsen, J.; Jensen, C.; Hansen, Y.; Friis-Hansen, L. Sensitivity and performance of three novel quantitative assays of SARS-CoV-2 nucleoprotein in blood. Sci. Rep. 2023, 13, 2868. [Google Scholar] [CrossRef] [PubMed]
- Gómez, R.S.; Mora, J.E.; Cortés, C.M.; Riedel, C.A.; Ferrés, M.; Bueno, S.M.; Kalergis, A.M. Respiratory syncytial virus detection in cells and clinical samples by using three new monoclonal antibodies. J. Med. Virol. 2014, 86, 1256–1266. [Google Scholar] [CrossRef] [PubMed]
- Thuy Tien, T.T.; Park, H.; Tuong, H.T.; Yu, S.-T.; Choi, D.-Y.; Yeo, S.-J. Development of a Rapid Fluorescent Immunochromatographic Test to Detect Respiratory Syncytial Virus. Int. J. Mol. Sci. 2018, 19, 3013. [Google Scholar] [CrossRef]
- Weber, J.; Peng, H.; Rader, C. From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 2017, 49, e305. [Google Scholar] [CrossRef] [PubMed]
- Seeber, S.; Ros, F.; Thorey, I.; Tiefenthaler, G.; Kaluza, K.; Lifke, V.; Fischer, J.A.A.; Klostermann, S.; Endl, J.; Kopetzki, E.; et al. A robust high throughput platform to generate functional recombinant monoclonal antibodies using rabbit B cells from peripheral blood. PLoS ONE 2014, 9, e86184. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.; Nunes, C.; Rocha, G.; Oliveira, F.; Sanches, F.; Gobbi, H. Rabbit monoclonal antibodies show higher sensitivity than mouse monoclonals for estrogen and progesterone receptor evaluation in breast cancer by immunohistochemistry. Pathol. Res. Pract. 2008, 204, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Laurino, L.; Furlanetto, A.; Chinellato, S.; Orvieto, E.; Canal, F.; Facchetti, F.; Dei Tos, A.P. Rabbit monoclonal antibodies: A comparative study between a novel category of immunoreagents and the corresponding mouse monoclonal antibodies. Am. J. Clin. Pathol. 2005, 124, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Stahl, S.J.; Watts, N.R.; Rader, C.; DiMattia, M.A.; Mage, R.G.; Palmer, I.; Kaufman, J.D.; Grimes, J.M.; Stuart, D.I.; Steven, A.C.; et al. Generation and Characterization of a Chimeric Rabbit/Human Fab for Co-Crystallization of HIV-1 Rev. J. Mol. Biol. 2010, 397, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Makdasi, E.; Levy, Y.; Alcalay, R.; Noy-Porat, T.; Zahavy, E.; Mechaly, A.; Epstein, E.; Peretz, E.; Cohen, H.; Bar-On, L.; et al. Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope. Viruses 2021, 13, 566. [Google Scholar] [CrossRef] [PubMed]
- Baurand, P.-E.; Balland, J.; Reynas, C.; Ramseyer, M.; Vivier, D.; Bellaye, P.-S.; Collin, B.; Paul, C.; Denat, F.; Asgarov, K.; et al. Development of Anti-LRRC15 Small Fragments for Imaging Purposes Using a Phage-Display ScFv Approach. Int. J. Mol. Sci. 2022, 23, 12677. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Lee, J.S.; Shim, H. Construction of Rabbit Immune Antibody Libraries. In Phage Display: Methods and Protocols, Methods in Molecular Biology; Hust, M., Lim, T., Eds.; Humana Press: New York, NY, USA, 2018; pp. 133–146. [Google Scholar] [CrossRef]
- Russo, G.; Meier, D.; Helmsing, S.; Wenzel, E.; Oberle, F.; Frenzel, A.; Hust, M. Parallelized Antibody Selection in Microtiter Plates. In Phage Display: Methods and Protocols, Methods in Molecular Biology; Hust, M., Lim, T., Eds.; Humana Press: New York, NY, USA, 2018; Volume 1701, pp. 273–284. [Google Scholar] [CrossRef]
- Rader, C.; Ritter, G.; Nathan, S.; Elia, M.; Gout, I.; Jungbluth, A.A.; Cohen, L.S.; Welt, S.; Old, L.J.; Barbas, C.F. The Rabbit Antibody Repertoire as a Novel Source for the Generation of Therapeutic Human Antibodies. J. Biol. Chem. 2000, 275, 13668–13676. [Google Scholar] [CrossRef] [PubMed]
- Rader, C. Generation and Selection of Rabbit Antibody Libraries by Phage Display. In Therapeutic Antibodies, Protocol, Methods in Molecular Biology; Dimitrov, A., Ed.; Humana Press: New York, NY, USA, 2009; Volume 525, pp. 101–128. [Google Scholar] [CrossRef]
- Ridder, R.; Gram, H. Generation of Rabbit Immune Libraries. In Antibody Engineering Vol.1, Springer Protocols Handbooks; Kontermann, R., Dübel, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 115–123. [Google Scholar] [CrossRef]
- Pfeil, J.; Tabatabai, J.; Sander, A.; Ries, M.; Grulich-Henn, J.; Schnitzler, P. Screening for Respiratory Syncytial Virus and Isolation Strategies in Children Hospitalized With acute Respiratory Tract Infection. Medicine 2014, 93, e144. [Google Scholar] [CrossRef] [PubMed]
| Library | ||||
|---|---|---|---|---|
| Size (cfu/mL)/transformant number | 3.6 × 106/1.5 × 107 | |||
| Insertion rate (colony PCR) | >95% (46/48 positives) | |||
| Library | 100% coding sequences (8/8) 100% diversity (8/8) | |||
| Panning | Recombinant strain | Input (pfu) | Output (pfu) | Ratio (out/in) |
| First round | B | 8 × 1010 | 4 × 104 | 5 × 10−7 |
| Second round | A2 | 2 × 1010 | 2 × 105 | 10−5 |
| A1 | B1 | C4 | D1 | E1 | F1 | G1 | H1 | |
| A2 | B2 | C5 | D9 | E6 | F2 | G2 | H2 | |
| A4 | B3 | C8 | D11 | E8 | F5 | G3 | H6 | |
| A7 | B4 | C9 | E10 | F6 | G6 | H7 | ||
| A8 | B9 | C11 | E11 | F8 | G9 | H10 | ||
| A9 | B10 | F10 | G10 | |||||
| A10 | B11 | F12 | G11 | |||||
| A12 | B12 | Total | ||||||
| 8 | 8 | 5 | 3 | 5 | 7 | 7 | 5 | 48 |
| Sequence Number | Unique Clones | Family (CDR3) | |
|---|---|---|---|
| VH | 48 | 28 | 13 |
| VL | 48 | 24 | 11 |
| Concentration (mg/mL) | Quantity (mg) | Production Yield (µg/mL) | |
|---|---|---|---|
| A9 | 0.9 | 1.9 | 38 |
| A10 | 0.7 | 3.2 | 64 |
| A12 | 1.2 | 5.8 | 116 |
| C11 | 1.5 | 3.4 | 68 |
| D11 | 0.6 | 2.5 | 50 |
| E11 | 1.2 | 2.5 | 50 |
| F12 | 1.1 | 8.2 | 164 |
| G1 | 1.1 | 5.9 | 118 |
| G10 | 1.4 | 3.5 | 70 |
| G12 | 1.5 | 7.5 | 150 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baurand, P.-E.; Balland, J.; Galli, E.; Eklin, S.; Bruley, R.; Ringenbach, L. New Anti-RSV Nucleoprotein Monoclonal Antibody Pairs Discovered Using Rabbit Phage Display Technology. Antibodies 2023, 12, 73. https://doi.org/10.3390/antib12040073
Baurand P-E, Balland J, Galli E, Eklin S, Bruley R, Ringenbach L. New Anti-RSV Nucleoprotein Monoclonal Antibody Pairs Discovered Using Rabbit Phage Display Technology. Antibodies. 2023; 12(4):73. https://doi.org/10.3390/antib12040073
Chicago/Turabian StyleBaurand, Pierre-Emmanuel, Jérémy Balland, Emilia Galli, Suvi Eklin, Rémy Bruley, and Laurence Ringenbach. 2023. "New Anti-RSV Nucleoprotein Monoclonal Antibody Pairs Discovered Using Rabbit Phage Display Technology" Antibodies 12, no. 4: 73. https://doi.org/10.3390/antib12040073
APA StyleBaurand, P.-E., Balland, J., Galli, E., Eklin, S., Bruley, R., & Ringenbach, L. (2023). New Anti-RSV Nucleoprotein Monoclonal Antibody Pairs Discovered Using Rabbit Phage Display Technology. Antibodies, 12(4), 73. https://doi.org/10.3390/antib12040073





