Elucidating Novel Targets for Ovarian Cancer Antibody–Drug Conjugate Development: Integrating In Silico Prediction and Surface Plasmon Resonance to Identify Targets with Enhanced Antibody Internalization Capacity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Online Tool/Public Data Source
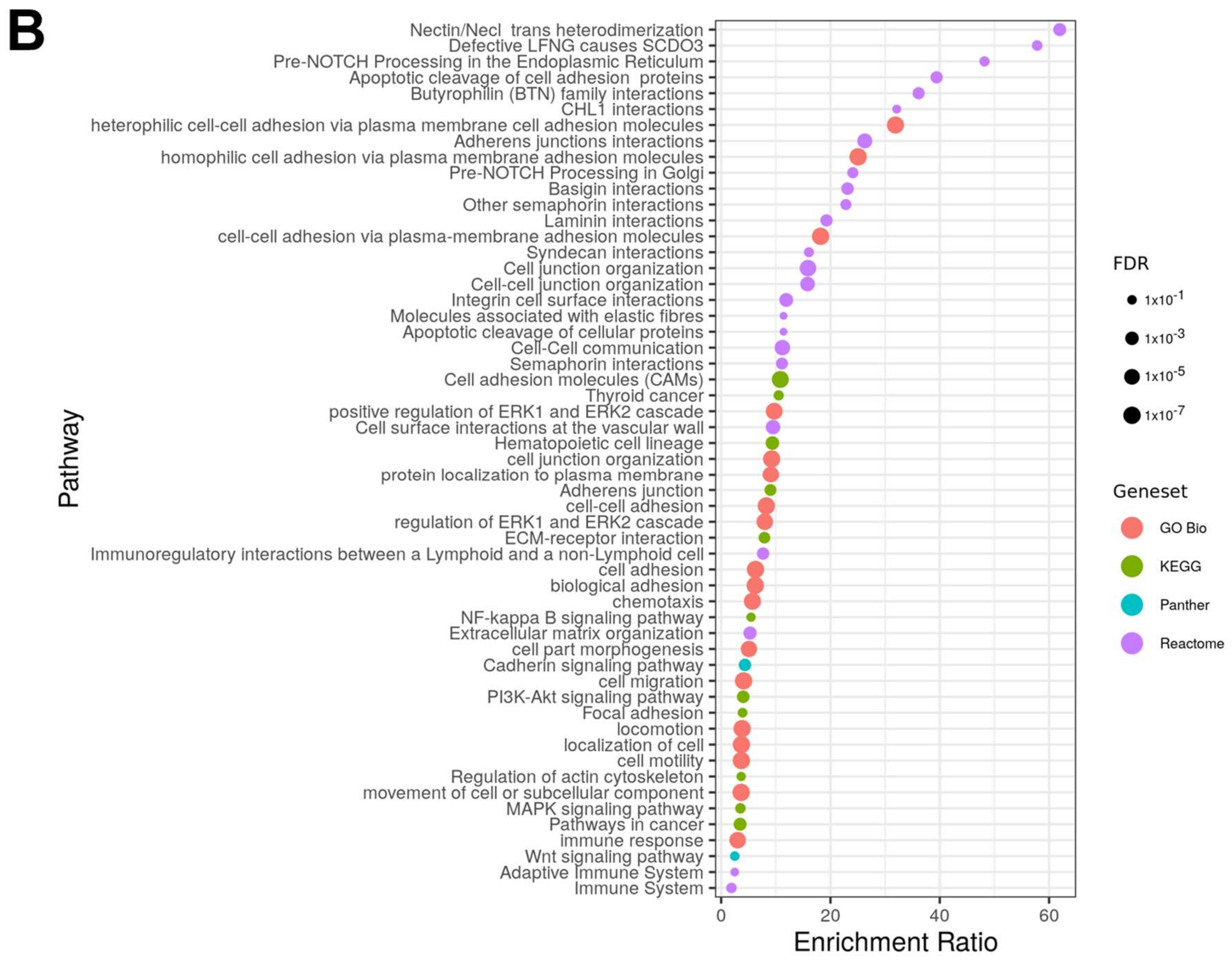
2.2. Pathway Analysis
2.3. Cell Culture
2.4. Western Blot
2.5. Gene Expression Analysis
2.6. Healthy Tissue Lysates
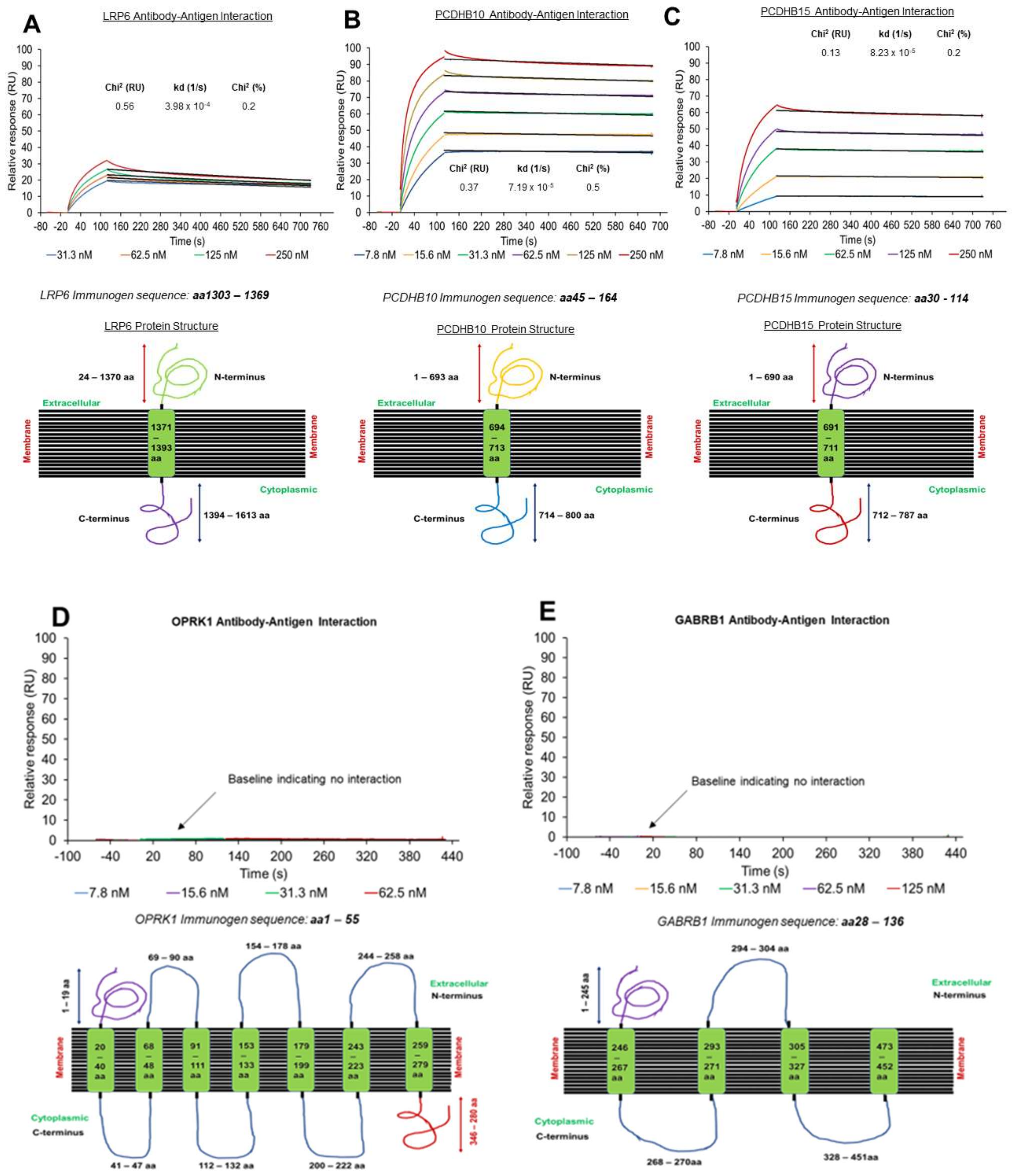
2.7. Antibodies and Recombinant Proteins
2.8. Off-Rate Screening
2.9. Immunofluorescence Assay
2.10. pHAb Amine Antibody Internalization Assay
2.11. Cell Viability
2.12. Statistics
3. Results
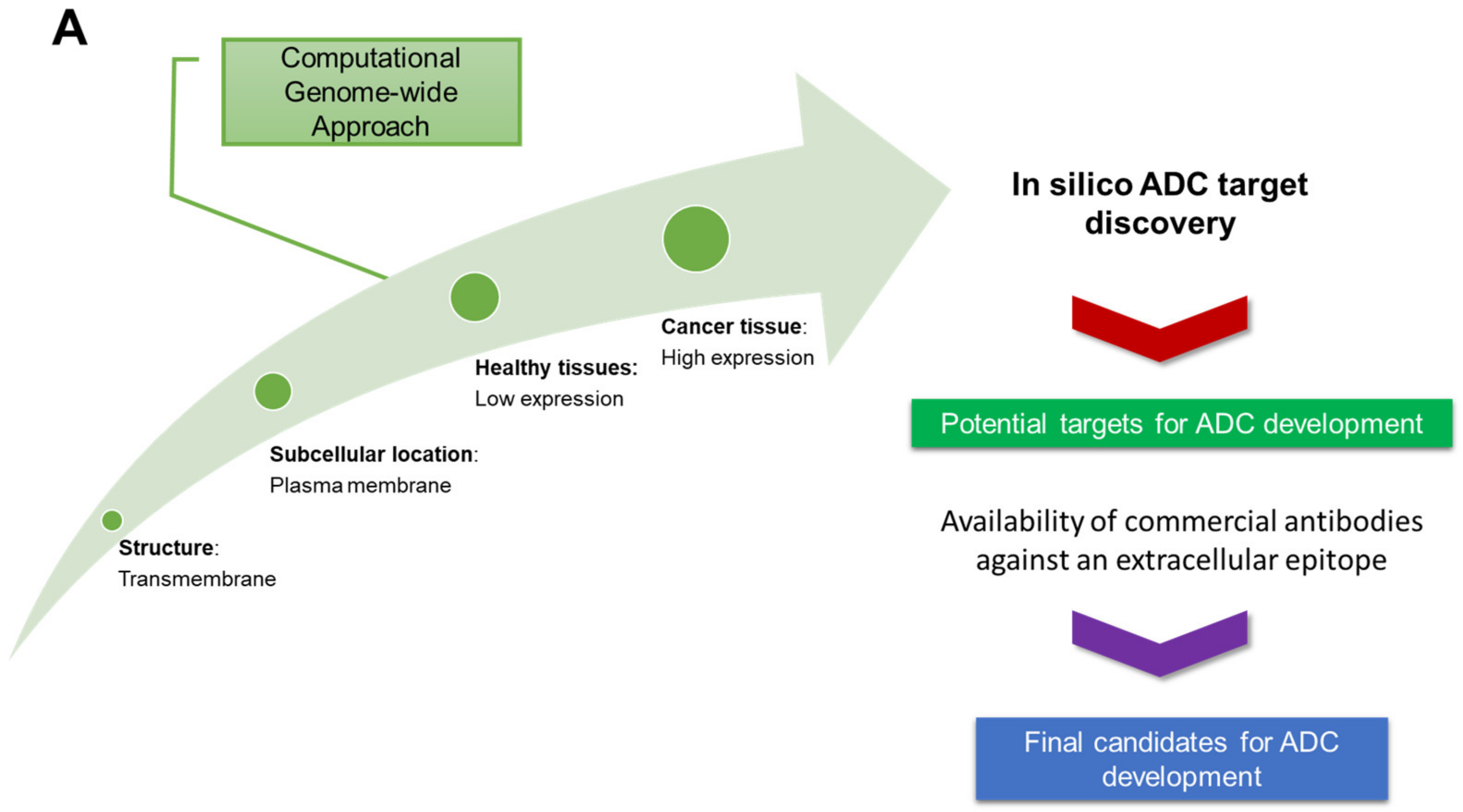
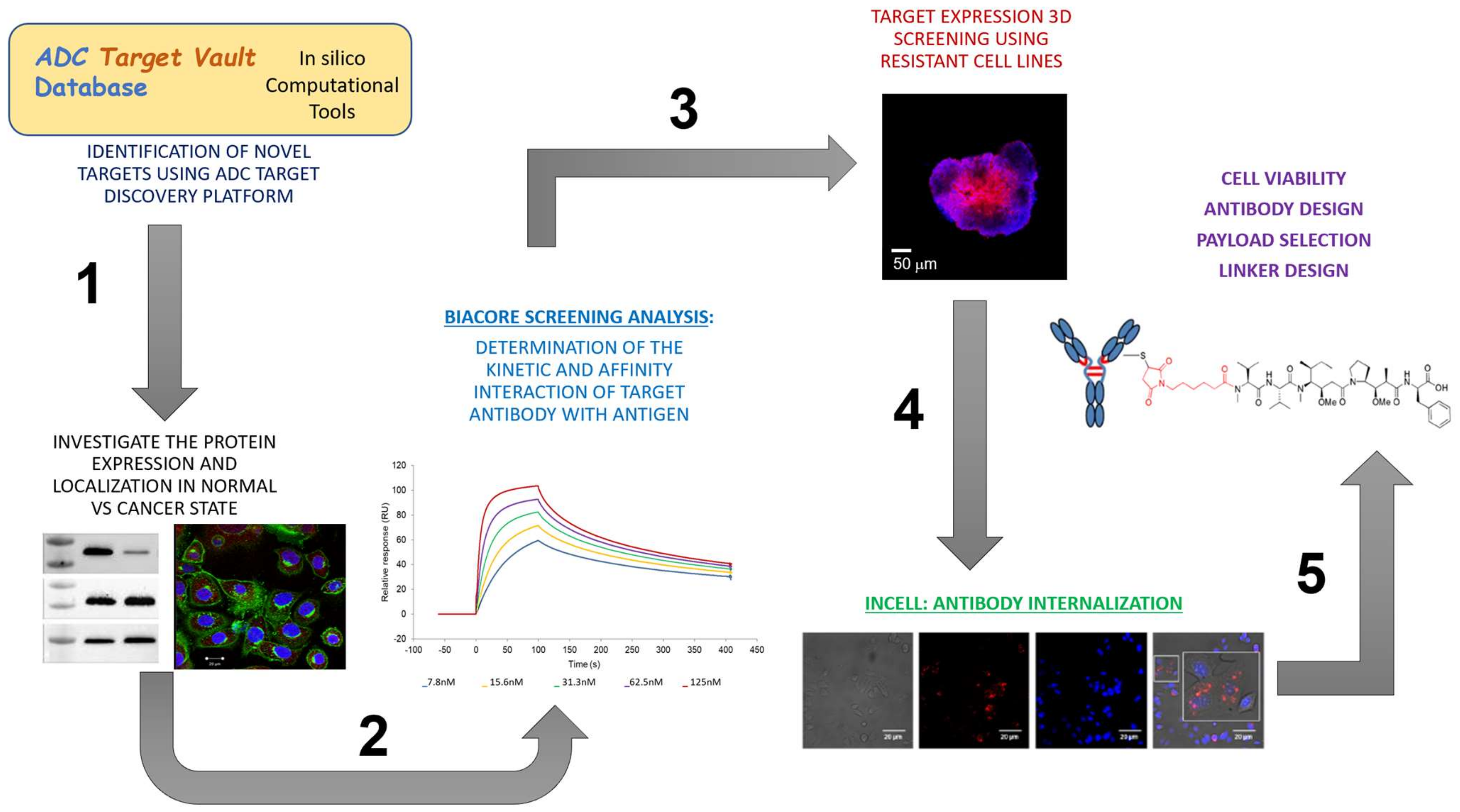
3.1. Identification of Candidate Targets via ADC Database
3.1.1. In Silico Database Development
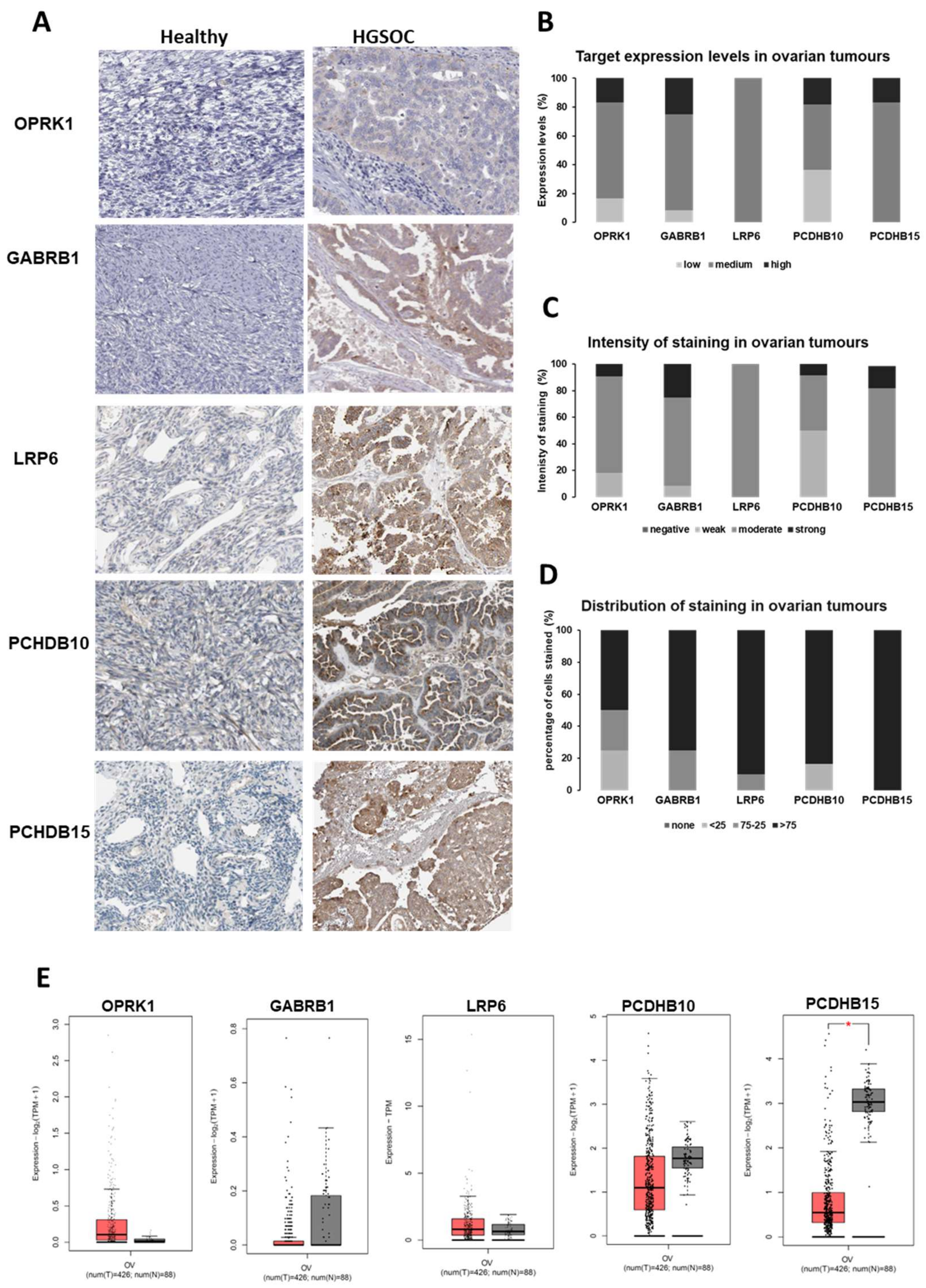
3.1.2. Target Expression in Healthy and Ovarian Cancer Tissues
3.2. Off-Rate Screening via SPR Platform as a Strategy to Enhance Antibody Selection
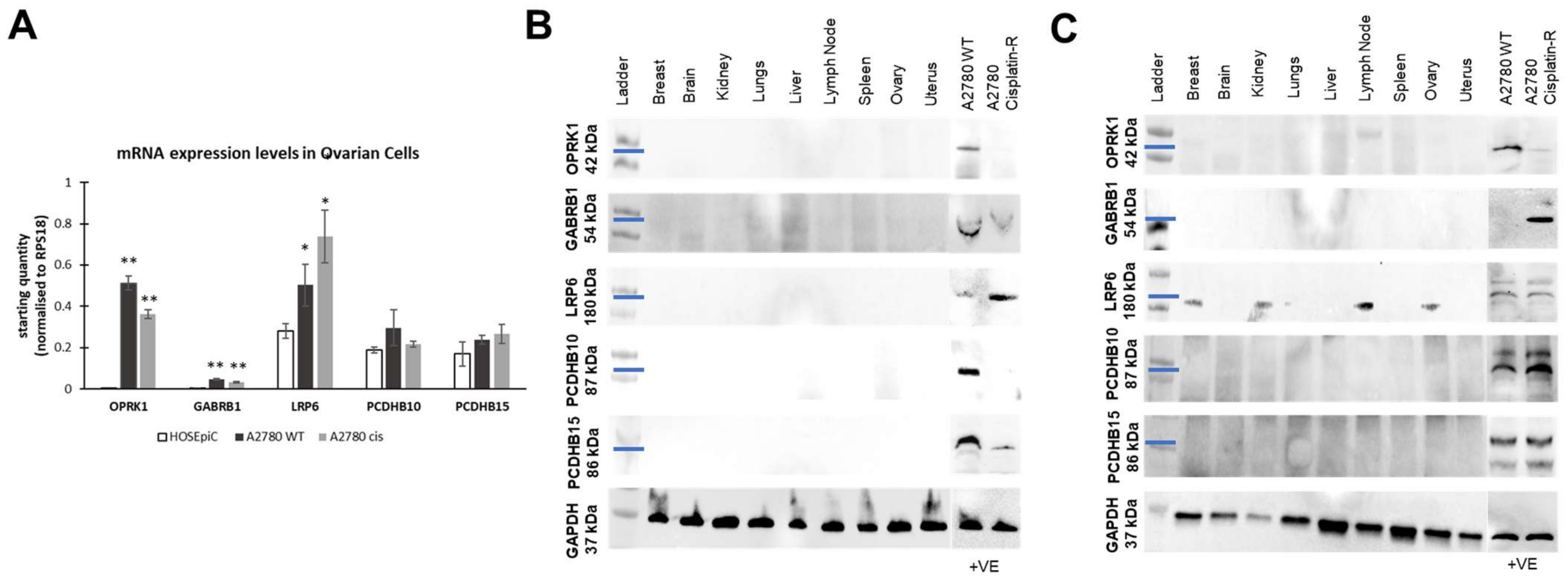
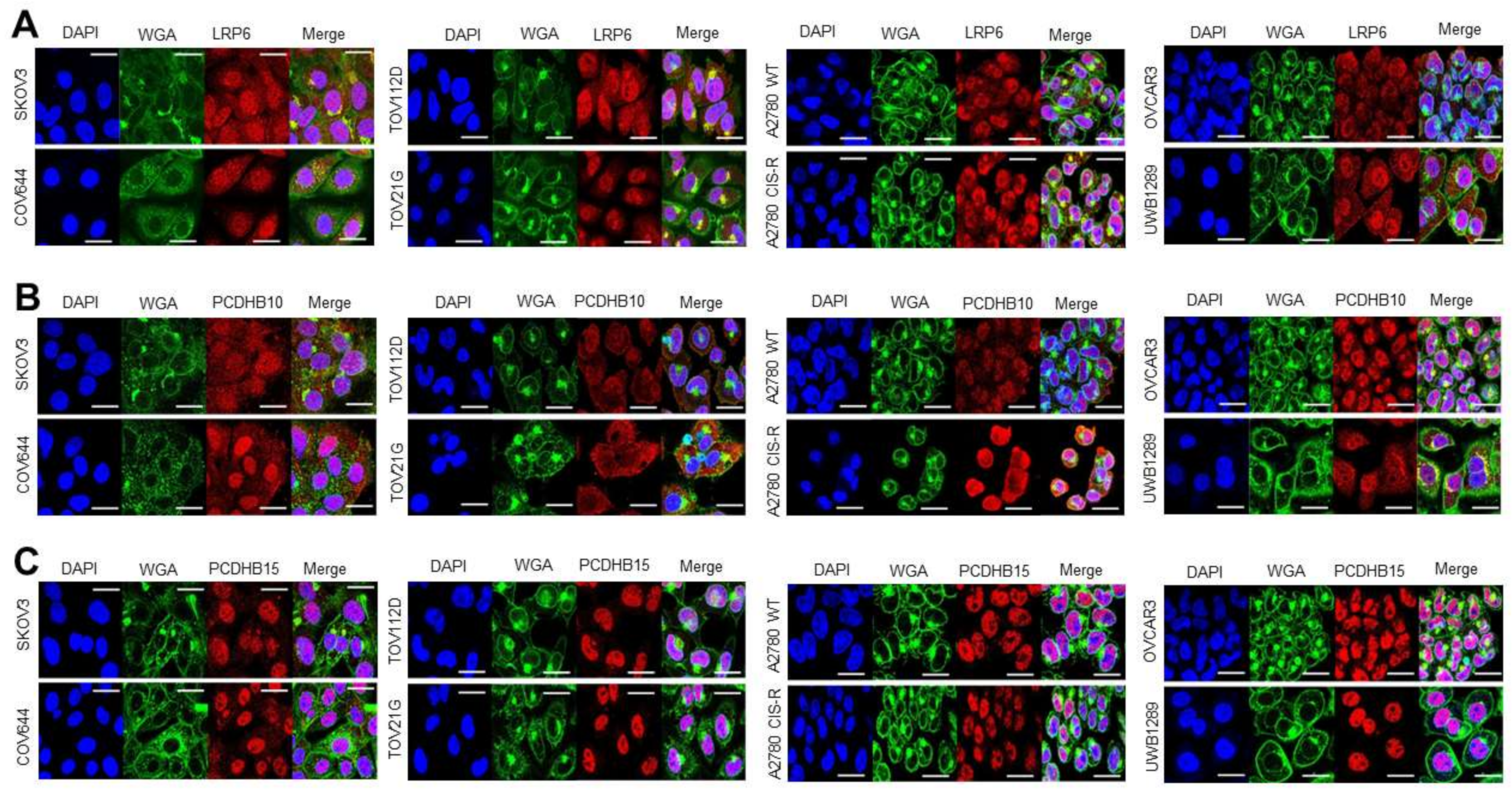
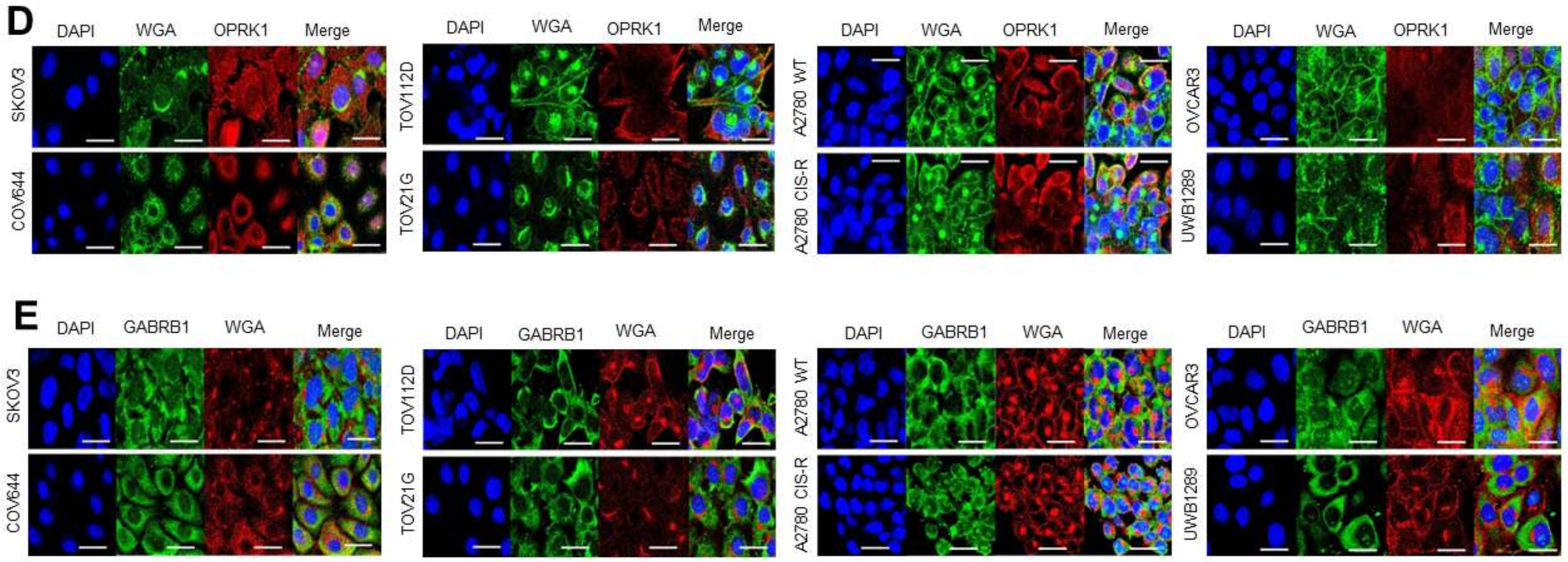
3.3. Target Localization and Basal Expression Patterns in Ovarian Cancer Cell Lines
3.3.1. Target Localisation and Basal Expression in Confluent 2D Monolayers
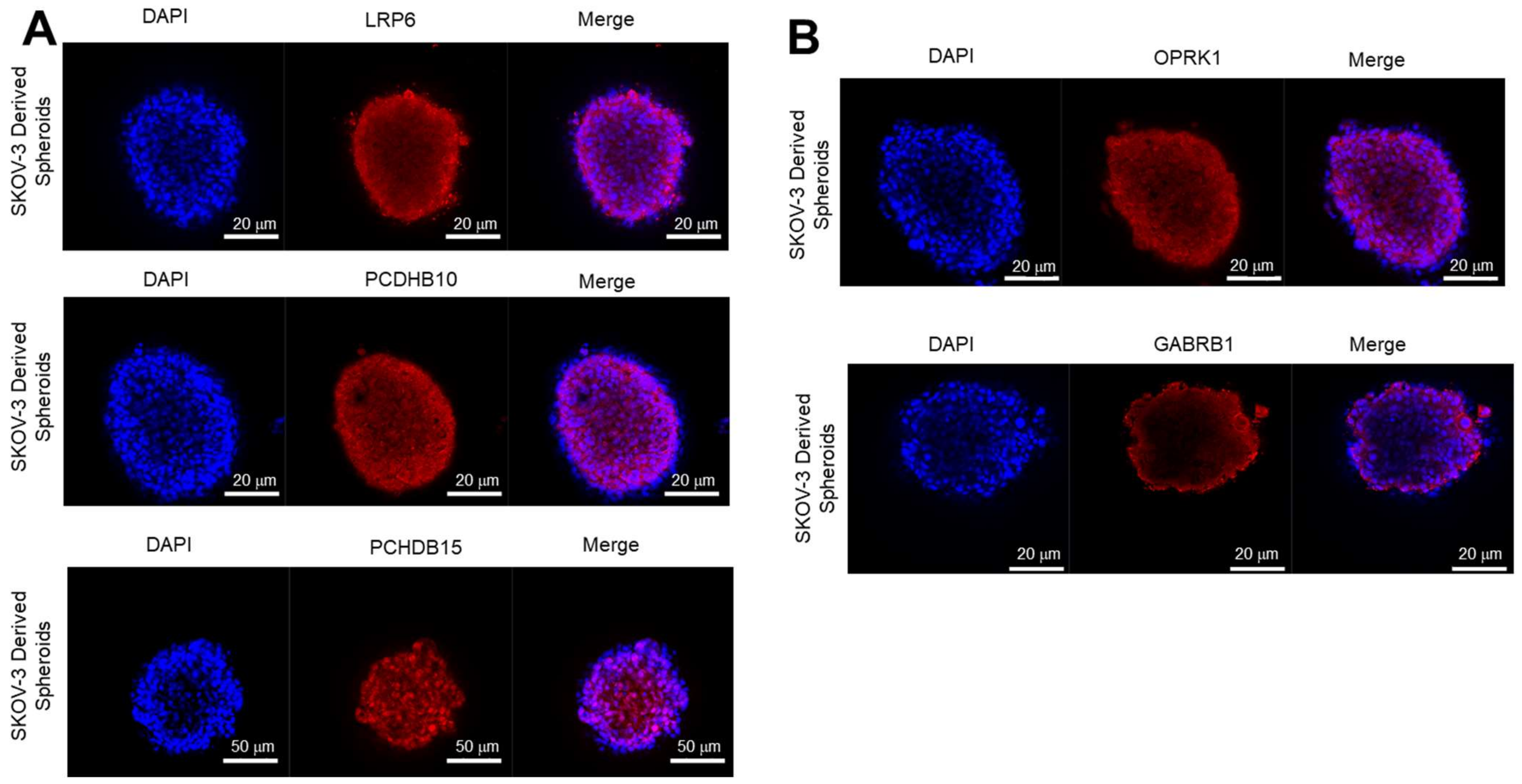
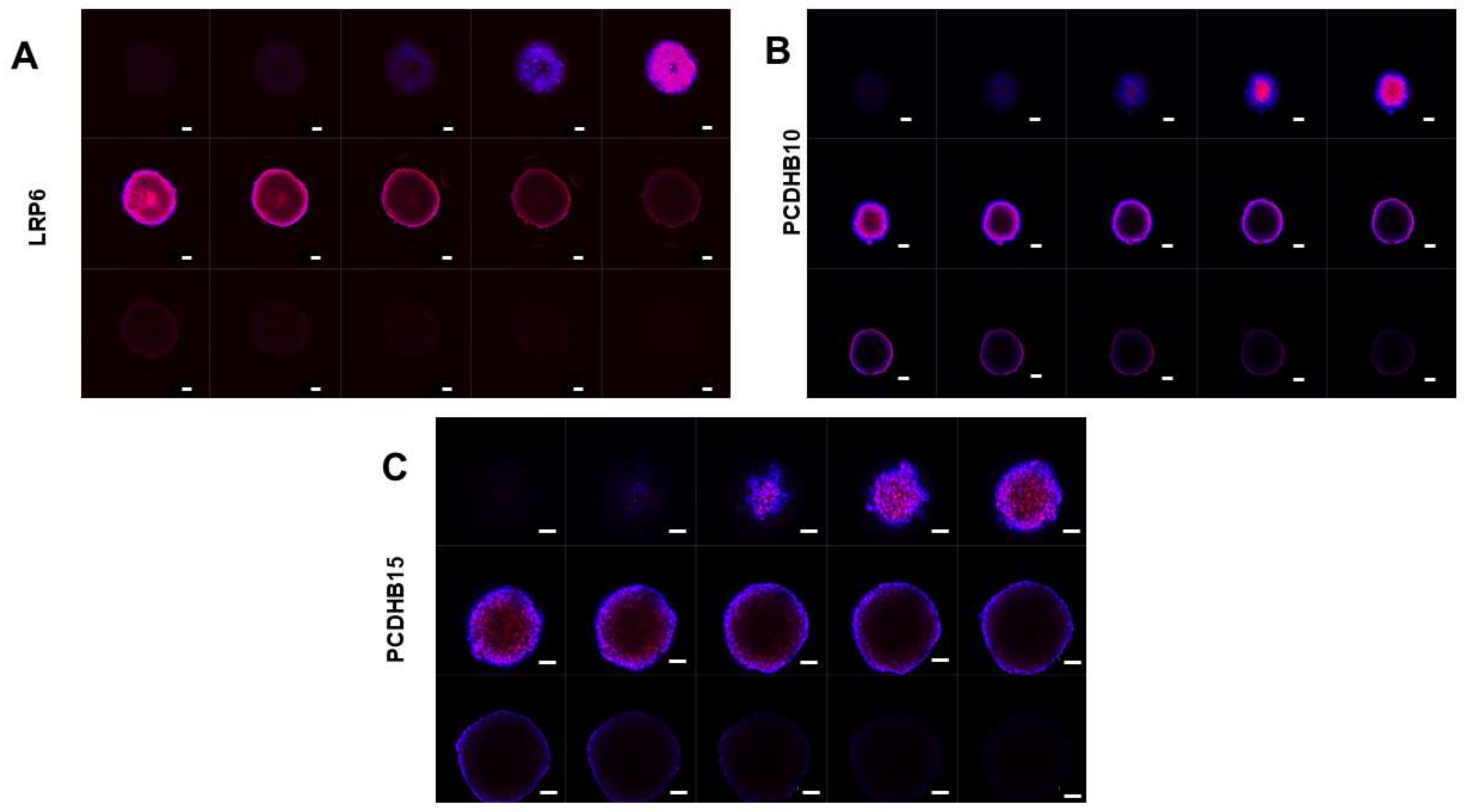
3.3.2. Expression of Target Proteins in 3D Spheroids
3.3.3. Antibody Penetration in 3D Spheroids
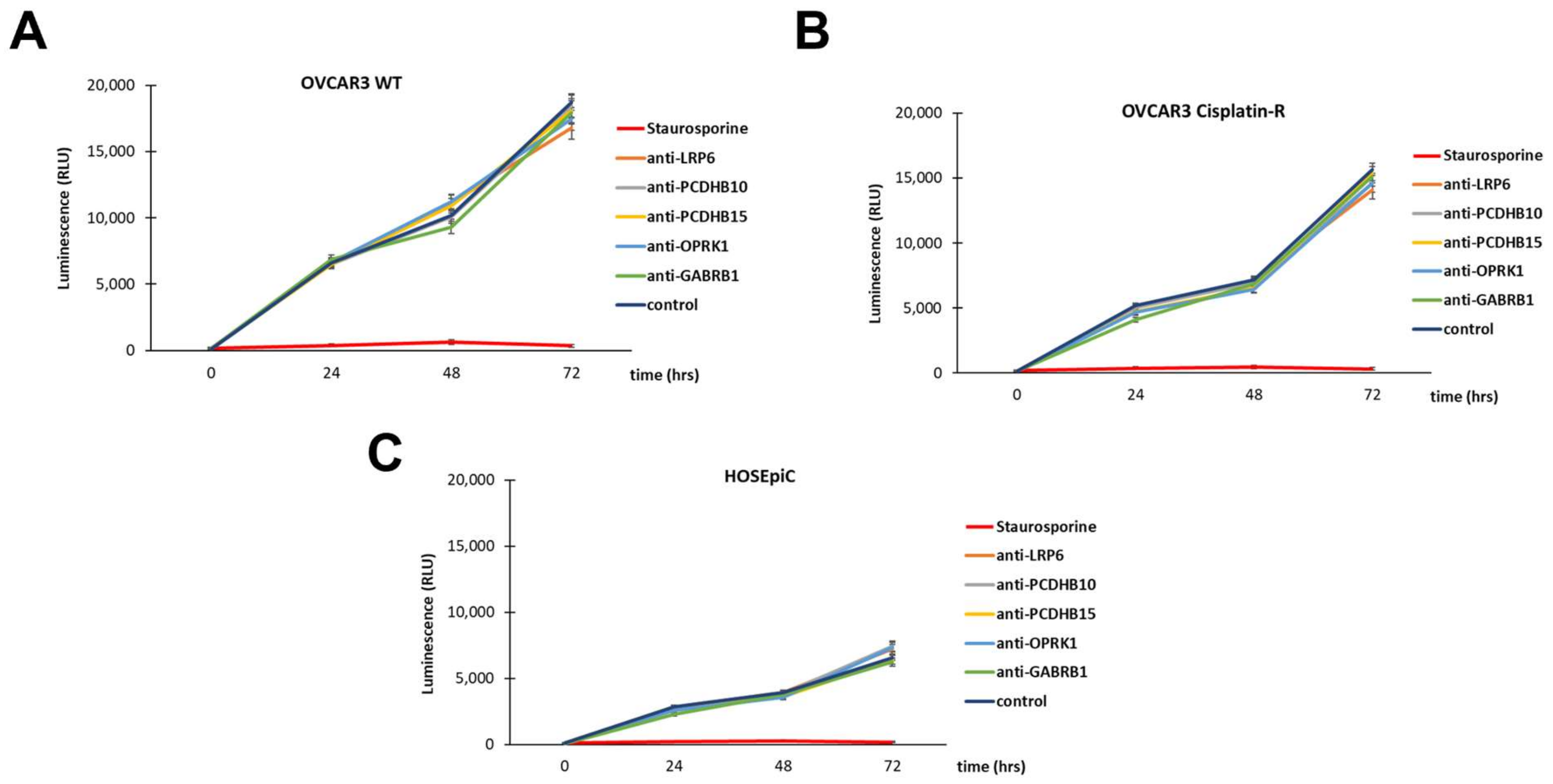
3.4. Antibody Internalization in Ovarian Cancer Cells
3.5. In Vitro Antibody-Mediated Ovarian Cancer Cell Toxicity
4. Discussion
4.1. Target Selection Significance
4.2. Challenges in ADC Development
4.3. Clinical Implications
4.4. Limitations of the Study and Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deb, B.; Uddin, A.; Chakraborty, S. MiRNAs and Ovarian Cancer: An Overview. J. Cell. Physiol. 2018, 233, 3846–3854. [Google Scholar] [CrossRef] [PubMed]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in Ovarian Cancer Therapy. Cancer Chemother. Pharmacol. 2017, 81, 17–38. [Google Scholar] [CrossRef]
- Van der Bilt, A.R.; van der Zee, A.G.; de Vries, E.G.; de Jong, S.; Bosscha, H.T.; Hoor, K.A.T.; Dunnen, W.F.D.; Hollema, H.; Reyners, A.K. Multiple VEGF Family Members Are Simultaneously Expressed in Ovarian Cancer: A Proposed Model for Bevacizumab Resistance. Curr. Pharm. Des. 2012, 18, 3784–3792. [Google Scholar] [CrossRef] [PubMed]
- Mitamura, T.; Pradeep, S.; McGuire, M.; Wu, S.Y.; Ma, S.; Hatakeyama, H.; Lyons, Y.A.; Hisamatsu, T.; Noh, K.; Villar-Prados, A.; et al. Induction of Anti-VEGF Therapy Resistance by Upregulated Expression of Microseminoprotein (MSMP). Oncogene 2018, 37, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Matulonis, U.A. PARP Inhibitor Resistance Mechanisms and Implications for Post-Progression Combination Therapies. Cancers 2020, 12, 2054. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Karakasis, K.; Madariaga, A.; Oza, A.M. Overcoming Platinum and PARP-Inhibitor Resistance in Ovarian Cancer. Cancers 2020, 12, 1607. [Google Scholar] [CrossRef] [PubMed]
- Bunn, P.A.; Minna, J.D.; Augustyn, A.; Gazdar, A.F.; Ouadah, Y.; Krasnow, M.A.; Berns, A.; Brambilla, E.; Rekhtman, N.; Massion, P.P.; et al. Small Cell Lung Cancer: Can Recent Advances in Biology and Molecular Biology Be Translated into Improved Outcomes? J. Thorac. Oncol. 2016, 11, 453–474. [Google Scholar] [CrossRef]
- Ou, J.; Si, Y.; Goh, K.; Yasui, N.; Guo, Y.; Song, J.; Wang, L.; Jaskula-Sztul, R.; Fan, J.; Zhou, L.; et al. Bioprocess Development of Antibody-Drug Conjugate Production for Cancer Treatment. PLoS ONE 2018, 13, e0206246. [Google Scholar] [CrossRef]
- Hafeez, U.; Parakh, S.; Gan, H.K.; Scott, A.M. Antibody—Drug Conjugates for Cancer Therapy. Molecules 2020, 25, 4764. [Google Scholar] [CrossRef]
- Moore, K.N.; Oza, A.M.; Colombo, N.; Oaknin, A.; Scambia, G.; Lorusso, D.; Konecny, G.E.; Banerjee, S.; Murphy, C.G.; Tanyi, J.L.; et al. Phase III, Randomized Trial of Mirvetuximab Soravtansine versus Chemotherapy in Patients with Platinum-Resistant Ovarian Cancer: Primary Analysis of FORWARD I. Ann. Oncol. 2021, 32, 757–765. [Google Scholar] [CrossRef]
- Luke, J.J.; Barlesi, F.; Chung, K.; Tolcher, A.W.; Kelly, K.; Hollebecque, A.; le Tourneau, C.; Subbiah, V.; Tsai, F.; Kao, S.; et al. Phase I Study of ABBV-428, a Mesothelin-CD40 Bispecific, in Patients with Advanced Solid Tumors. J. Immunother. Cancer 2021, 9, e002015. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.S.; Wesolowski, R.; Baffa, R.; Liao, K.H.; Hua, S.Y.; Gibson, B.L.; Pirie-Shepherd, S.; Tolcher, A.W. A Phase I, Dose-Escalation Study of PF-06650808, an Anti-Notch3 Antibody-Drug Conjugate, in Patients with Breast Cancer and Other Advanced Solid Tumors. Investig. New Drugs 2020, 38, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Christie, E.L.; Bowtell, D.D.L. Acquired Chemotherapy Resistance in Ovarian Cancer. Ann. Oncol. 2017, 28, viii13–viii15. [Google Scholar] [CrossRef]
- Liu, J.; Westin, S.N. Rational Selection of Biomarker Driven Therapies for Gynecologic Cancers: The More We Know, the More We Know We Don’t Know. Gynecol. Oncol. 2016, 141, 65. [Google Scholar] [CrossRef]
- Lakayan, D.; Haselberg, R.; Gahoual, R.; Somsen, G.W.; Kool, J. Affinity Profiling of Monoclonal Antibody and Antibody-Drug-Conjugate Preparations by Coupled Liquid Chromatography-Surface Plasmon Resonance Biosensing. Anal. Bioanal. Chem. 2018, 410, 7837–7848. [Google Scholar] [CrossRef]
- Healey, G.D.; Frostell, A.; Fagge, T.; Gonzalez, D.; Conlan, R.S. A RAGE-Targeted Antibody-Drug Conjugate: Surface Plasmon Resonance as a Platform for Accelerating Effective ADC Design and Development. Antibodies 2019, 8, 7. [Google Scholar] [CrossRef]
- Douzi, B. Protein–Protein Interactions: Surface Plasmon Resonance. Methods Mol. Biol. 2017, 1615, 257–275. [Google Scholar] [CrossRef]
- Healey, G.D.; Pan-Castillo, B.; Garcia-Parra, J.; Davies, J.; Roberts, S.; Jones, E.; Dhar, K.; Nandanan, S.; Tofazzal, N.; Piggott, L.; et al. Antibody Drug Conjugates against the Receptor for Advanced Glycation End Products (RAGE), a Novel Therapeutic Target in Endometrial Cancer. J. ImmunoTherapy Cancer 2019, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Beeg, M.; Nobili, A.; Orsini, B.; Rogai, F.; Gilardi, D.; Fiorino, G.; Danese, S.; Salmona, M.; Garattini, S.; Gobbi, M. A Surface Plasmon Resonance-Based Assay to Measure Serum Concentrations of Therapeutic Antibodies and Anti-Drug Antibodies. Sci. Rep. 2019, 9, 2064. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [PubMed]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS Web Server for Consensus Prediction of Membrane Protein Topology and Signal Peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Saethang, T.; Hodge, K.; Kimkong, I.; Payne, D.M.; Knepper, M.A.; Pisitkun, T. AbDesigner3D: A Structure-Guided Tool for Peptide-Based Antibody Production. Bioinformatics 2018, 34, 2158–2160. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Gillespie, M.; Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Weiswald, L.B.; Guinebretière, J.M.; Richon, S.; Bellet, D.; Saubaméa, B.; Dangles-Marie, V. In Situ Protein Expression in Tumour Spheres: Development of an Immunostaining Protocol for Confocal Microscopy. BMC Cancer 2010, 10, 106. [Google Scholar] [CrossRef]
- Reynolds, S.M.; Kall, L.; Riffle, M.E.; Bilmes, J.A.; Noble, W.S. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput. Biol. 2008, 4, e1000213. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 2005, 21, i251–i257. [Google Scholar] [CrossRef] [PubMed]
- Viklund, H.; Bernsel, A.; Skwark, M.; Elofsson, A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics 2008, 24, 2928–2929. [Google Scholar] [CrossRef] [PubMed]
- Viklund, H.; Elofsson, A. OCTOPUS: Improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 2008, 24, 1662–1668. [Google Scholar] [CrossRef]
- Bernsel, A.; Viklund, H.; Falk, J.; Lindahl, E.; von Heijne, G.; Elofsson, A. Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. USA 2008, 105, 7177–7181. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef]
- Ruan, X.; Liu, A.; Zhong, M.; Wei, J.; Zhang, W.; Rong, Y.; Liu, W.; Li, M.; Qing, X.; Chen, G.; et al. Silencing LGR6 Attenuates Stemness and Chemoresistance via Inhibiting Wnt/β-Catenin Signaling in Ovarian Cancer. Mol. Ther. Oncolytics 2019, 14, 94–106. [Google Scholar] [CrossRef]
- Moradi, Z.; Barati, M.; Azad, M.; Safari, A.; Chahardouli, B.; Rezvany, M.R. DNA Methylation and the Expression of the Tumor-Suppressor Genes Protocadherin-10 and Reprimo in Pediatric Acute Lymphoblastic Leukemia. Iran J. Pediatr. Hematol. Oncol. 2023, 13, 132–143. [Google Scholar] [CrossRef]
- Carrier, A.; Desjobert, C.; Lobjois, V.; Rigal, L.; Busato, F.; Tost, J.; Ensenyat-Mendez, M.; Marzese, D.M.; Pradines, A.; Favre, G.; et al. Epigenetically Regulated PCDHB15 Impairs Aggressiveness of Metastatic Melanoma Cells. Clin. Epigenetics 2022, 14, 156. [Google Scholar] [CrossRef]
- Chiang, C.-C.; Lin, G.-L.; Yang, S.-Y.; Tu, C.-W.; Huang, W.-L.; Wei, C.-F.; Wang, F.-C.; Lin, P.-J.; Huang, W.-H.; Chuang, Y.-M.; et al. PCDHB15 as a potential tumor suppressor and epigenetic biomarker for breast cancer. Oncol. Lett. 2022, 23, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, Z.; Lei, Y. The expression of kappa-opioid receptor promotes the migration of breast cancer cells in vitro. BMC Anesthesiol. 2021, 21, 210. [Google Scholar] [CrossRef]
- Yan, L.; Gong, Y.; Shao, M.; Ruan, G.; Xie, H.; Liao, X.; Wang, X.; Han, Q.; Zhou, X.; Zhu, L.; et al. Distinct diagnostic and prognostic values of γ-aminobutyric acid type A receptor family genes in patients with colon adenocarcinoma. Oncol. Lett. 2020, 20, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Enayatkhani, M.; Salimi, M.; Azadmanesh, K.; Teimoori-Toolabi, L. In-Silico Identification of New Inhibitors for Low-Density Lipoprotein Receptor-Related Protein6 (LRP6). J. Biomol. Struct. Dyn. 2022, 40, 4440–4450. [Google Scholar] [CrossRef] [PubMed]
- Rismani, E.; Rahimi, H.; Arab, S.S.; Azadmanesh, K.; Karimipoor, M.; Teimoori-Toolabi, L. Computationally Design of Inhibitory Peptides against Wnt Signaling Pathway: In Silico Insight on Complex of DKK1 and LRP6. Int. J. Pept. Res. Ther. 2018, 24, 49–60. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, Y.; Li, X.; Chen, L.; Wang, H.; Wu, J.; Zheng, J.; Wu, D. Characterization of the Kremen-binding Site on Dkk1 and Elucidation of the Role of Kremen in Dkk-mediated Wnt Antagonism. J. Biol. Chem. 2008, 283, 23371–23375. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.H.L.; Hough, R.; Bernaudo, S.; Peng, C. Wnt/β-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019, 12, 122. [Google Scholar] [CrossRef]
- Zhen, Y.; Pavez, M.; Li, X. The role of Pcdh10 in neurological disease and cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 8153–8164. [Google Scholar] [CrossRef]
- Echizen, K.; Nakada, M.; Hayashi, T.; Sabit, H.; Furuta, T.; Nakai, M.; Koyama-Nasu, R.; Nishimura, Y.; Taniue, K.; Morishita, Y.; et al. PCDH10 is required for the tumorigenicity of glioblastoma cells. Biochem. Biophys. Res. Commun. 2014, 444, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Moh, M.C.; Shen, S. The Roles of Cell Adhesion Molecules in Tumor Suppression and Cell Migration: A New Paradox. Cell Adh. Migr. 2009, 3, 334–336. [Google Scholar] [CrossRef]
- Schilling, V.; Beyerlein, P.; Chien, J. A Bioinformatics Analysis of Ovarian Cancer Data Using Machine Learning. Algorithms 2023, 16, 330. [Google Scholar] [CrossRef]
- Huang, G.; Mao, L.; Hu, X. Circ_0011373 promotes papillary thyroid carcinoma progression by regulating miR-1271/LRP6 axis. Hormones 2023, 22, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, L.; Sun, Q.; Chen, R.; Zhang, C.; Yang, P.; Tan, Y.; Peng, C.; Wang, T.; Jin, C.; et al. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin. Transl. Med. 2021, 11, e565. [Google Scholar] [CrossRef]
- Tolcher, A.; Hamilton, E.; Coleman, R.L. The evolving landscape of antibody-drug conjugates in gynecologic cancers. Cancer Treat. Rev. 2023, 116, 102546. [Google Scholar] [CrossRef]
- Solorzano, S.R.; Imaz-Rosshandler, I.; Camacho-Arroyo, I.; García-Tobilla, P.; Morales-Montor, G.; Salazar, P.; Arena-Ortiz, M.L.; Rodríguez-Dorantes, M. GABA promotes gastrin-releasing peptide secretion in NE/NE-like cells: Contribution to prostate cancer progression. Sci. Rep. 2018, 8, 10272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-F.; Xu, Q.-X.; Liao, L.-D.; Xu, X.-E.; Wu, J.-Y.; Shen, J.; Wu, Z.-Y.; Shen, J.-H.; Li, E.-M.; Xu, L.-Y. κ-Opioid receptor in the nucleus is a novel prognostic factor of esophageal squamous cell carcinoma. Hum. Pathol. 2013, 44, 1756–1765. [Google Scholar] [CrossRef]
- Sherman, S.K.; Maxwell, J.E.; Carr, J.C.; Wang, D.; Bellizzi, A.M.; O’dorisio, M.S.; O’dorisio, T.M.; Howe, J.R. Gene expression accurately distinguishes liver metastases of small bowel and pancreas neuroendocrine tumors. Clin. Exp. Metastasis 2014, 31, 935–944. [Google Scholar] [CrossRef]
- Saxena, J.; Nicolini, F.; Lockley, M. Reversing chemoresistance in high grade serous ovarian cancer 2019. Cancer Res. 2019, 79, 3801. [Google Scholar] [CrossRef]
- Hurwitz, J.; Haggstrom, L.R.; Lim, E. Antibody–Drug Conjugates: Ushering in a New Era of Cancer Therapy. Pharmaceutics 2023, 15, 2017. [Google Scholar] [CrossRef]
- Nejadmoghaddam, M.-R.; Minai-Tehrani, A.; Ghahremanzadeh, R.; Mahmoudi, M.; Dinarvand, R.; Zarnani, A.-H. Antibody-Drug Conjugates: Possibilities and Challenges. Avicenna J. Med. Biotechnol. 2019, 11, 3–23. [Google Scholar]
- Nejadmoghaddam, M.R.; Zarnani, A.H.; Ghahremanzadeh, R.; Ghods, R.; Mahmoudian, J.; Yousefi, M.; Nazari, M.; Ghahremani, M.H.; Abolhasani, M.; Anissian, A.; et al. Placenta-Specific1 (PLAC1) Is a Potential Target for Antibody-Drug Conjugate-Based Prostate Cancer Immunotherapy. Sci. Rep. 2017, 7, 13373. [Google Scholar] [CrossRef]
- Kovtun, Y.V.; Audette, C.A.; Ye, Y.; Xie, H.; Ruberti, M.F.; Phinney, S.J.; Leece, B.A.; Chittenden, T.; Blättler, W.A.; Goldmacher, V.S. Antibody-Drug Conjugates Designed to Eradicate Tumors with Homogeneous and Heterogeneous Expression of the Target Antigen. Cancer Res. 2006, 66, 3214–3221. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Panowski, S.; Bhakta, S.; Raab, H.; Polakis, P.; Junutula, J.R. Site-Specific Antibody Drug Conjugates for Cancer Therapy. mAbs 2014, 6, 34–45. [Google Scholar] [CrossRef]
- Bornstein, G.G. Antibody Drug Conjugates: Preclinical Considerations. AAPS J. 2015, 17, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Chu, R.; Reczek, D.; Brondyk, W. Capture-Stabilize Approach for Membrane Protein SPR Assays. Sci. Rep. 2014, 4, 7360. [Google Scholar] [CrossRef]
- Staudacher, A.H.; Brown, M.P. Antibody Drug Conjugates and Bystander Killing: Is Antigen-Dependent Internalisation Required? Br. J. Cancer 2017, 117, 1736. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.; Tchistiakova, L.; Scott, N. Implications of Receptor-Mediated Endocytosis and Intracellular Trafficking Dynamics in the Development of Antibody Drug Conjugates. mAbs 2013, 5, 13. [Google Scholar] [CrossRef]
- Rudnick, S.I.; Lou, J.; Shaller, C.C.; Tang, Y.; Klein-Szanto, A.J.; Weiner, L.M.; Marks, J.D.; Adams, G.P. Influence of affinity and antigen internalization on the uptake and penetration of Anti-HER2 antibodies in solid tumors. Cancer Res. 2011, 71, 2250–2259. [Google Scholar] [CrossRef] [PubMed]
- Goldmacher, V.S.; Kovtun, Y.V. Antibody-drug conjugates: Using monoclonal antibodies for delivery of cytotoxic payloads to cancer cells. Ther. Deliv. 2011, 2, 397–416. [Google Scholar] [CrossRef]
- Vainshtein, I.; Roskos, L.K.; Cheng, J.; Sleeman, M.A.; Wang, B.; Liang, M. Quantitative Measurement of the Target-Mediated Internalization Kinetics of Biopharmaceuticals. Pharm. Res. 2015, 32, 286. [Google Scholar] [CrossRef] [PubMed]
- Ab, O.; Whiteman, K.R.; Bartle, L.M.; Sun, X.; Singh, R.; Tavares, D.; LaBelle, A.; Payne, G.; Lutz, R.J.; Pinkas, J.; et al. IMGN853, a Folate Receptor-α (FRα)–Targeting Antibody–Drug Conjugate, Exhibits Potent Targeted Antitumor Activity against FRα-Expressing Tumors. Mol. Cancer Ther. 2015, 14, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.; Lorusso, D.; Oaknin, A.; Pignata, S.; Denys, H.; Colombo, N.; Van Gorp, T.; Konner, J.; Marin, M.R.; Harter, P.; et al. Efficacy and Safety of Mirvetuximab Soravtansine in Patients with Platinum-Resistant Ovarian Cancer with High Folate Receptor Alpha Expression: Results from the SORAYA Study (LBA 4). Gynecol. Oncol. 2022, 166, S50. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onyido, E.K.; James, D.; Garcia-Parra, J.; Sinfield, J.; Moberg, A.; Coombes, Z.; Worthington, J.; Williams, N.; Francis, L.W.; Conlan, R.S.; et al. Elucidating Novel Targets for Ovarian Cancer Antibody–Drug Conjugate Development: Integrating In Silico Prediction and Surface Plasmon Resonance to Identify Targets with Enhanced Antibody Internalization Capacity. Antibodies 2023, 12, 65. https://doi.org/10.3390/antib12040065
Onyido EK, James D, Garcia-Parra J, Sinfield J, Moberg A, Coombes Z, Worthington J, Williams N, Francis LW, Conlan RS, et al. Elucidating Novel Targets for Ovarian Cancer Antibody–Drug Conjugate Development: Integrating In Silico Prediction and Surface Plasmon Resonance to Identify Targets with Enhanced Antibody Internalization Capacity. Antibodies. 2023; 12(4):65. https://doi.org/10.3390/antib12040065
Chicago/Turabian StyleOnyido, Emenike Kenechi, David James, Jezabel Garcia-Parra, John Sinfield, Anna Moberg, Zoe Coombes, Jenny Worthington, Nicole Williams, Lewis Webb Francis, Robert Steven Conlan, and et al. 2023. "Elucidating Novel Targets for Ovarian Cancer Antibody–Drug Conjugate Development: Integrating In Silico Prediction and Surface Plasmon Resonance to Identify Targets with Enhanced Antibody Internalization Capacity" Antibodies 12, no. 4: 65. https://doi.org/10.3390/antib12040065
APA StyleOnyido, E. K., James, D., Garcia-Parra, J., Sinfield, J., Moberg, A., Coombes, Z., Worthington, J., Williams, N., Francis, L. W., Conlan, R. S., & Gonzalez, D. (2023). Elucidating Novel Targets for Ovarian Cancer Antibody–Drug Conjugate Development: Integrating In Silico Prediction and Surface Plasmon Resonance to Identify Targets with Enhanced Antibody Internalization Capacity. Antibodies, 12(4), 65. https://doi.org/10.3390/antib12040065









