Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. DNA Extraction and Metagenomic Sequencing
2.3. Statistical Analysis
3. Results
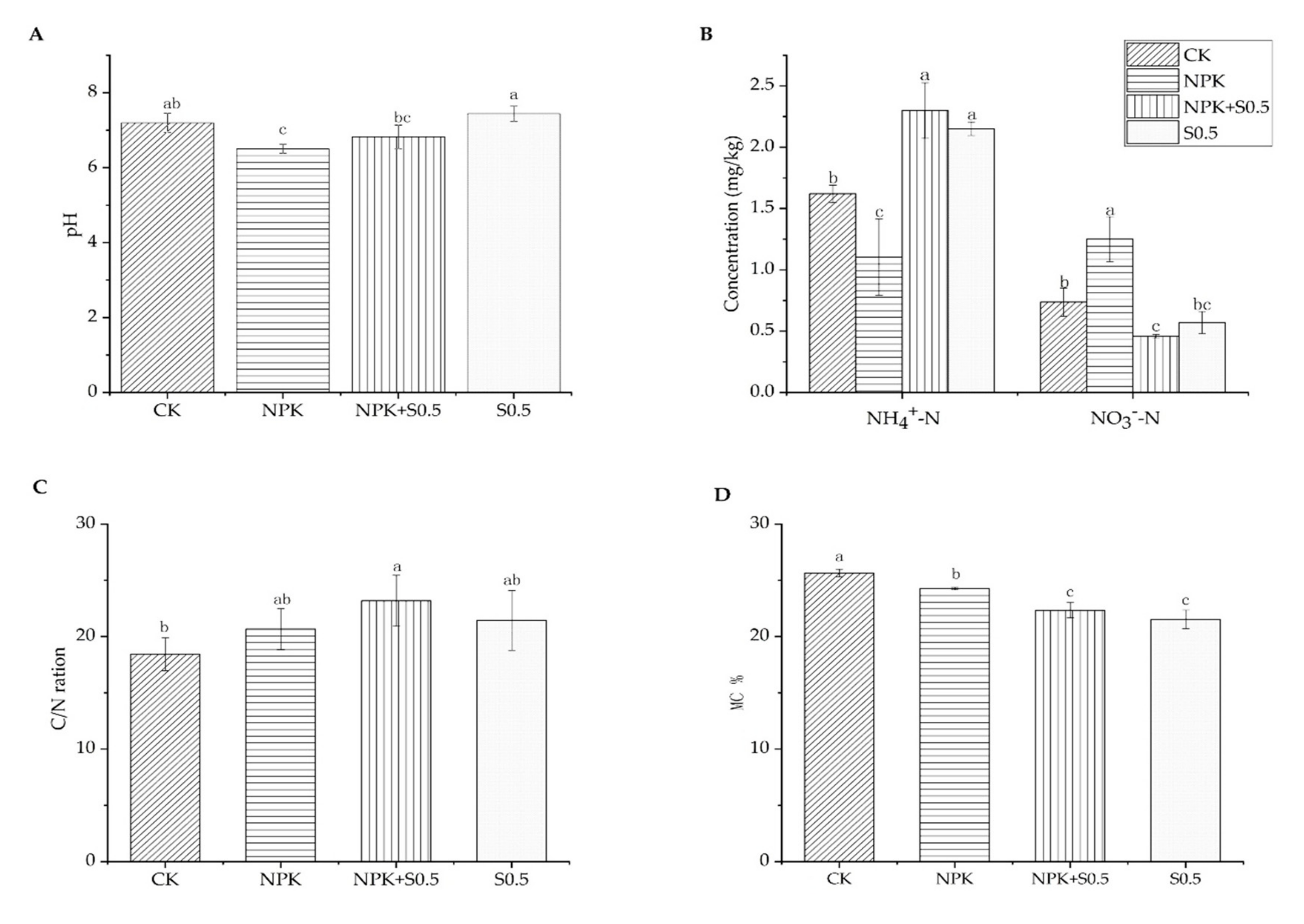
3.1. Soil Chemical Properties
3.2. Soil Microbiome Community
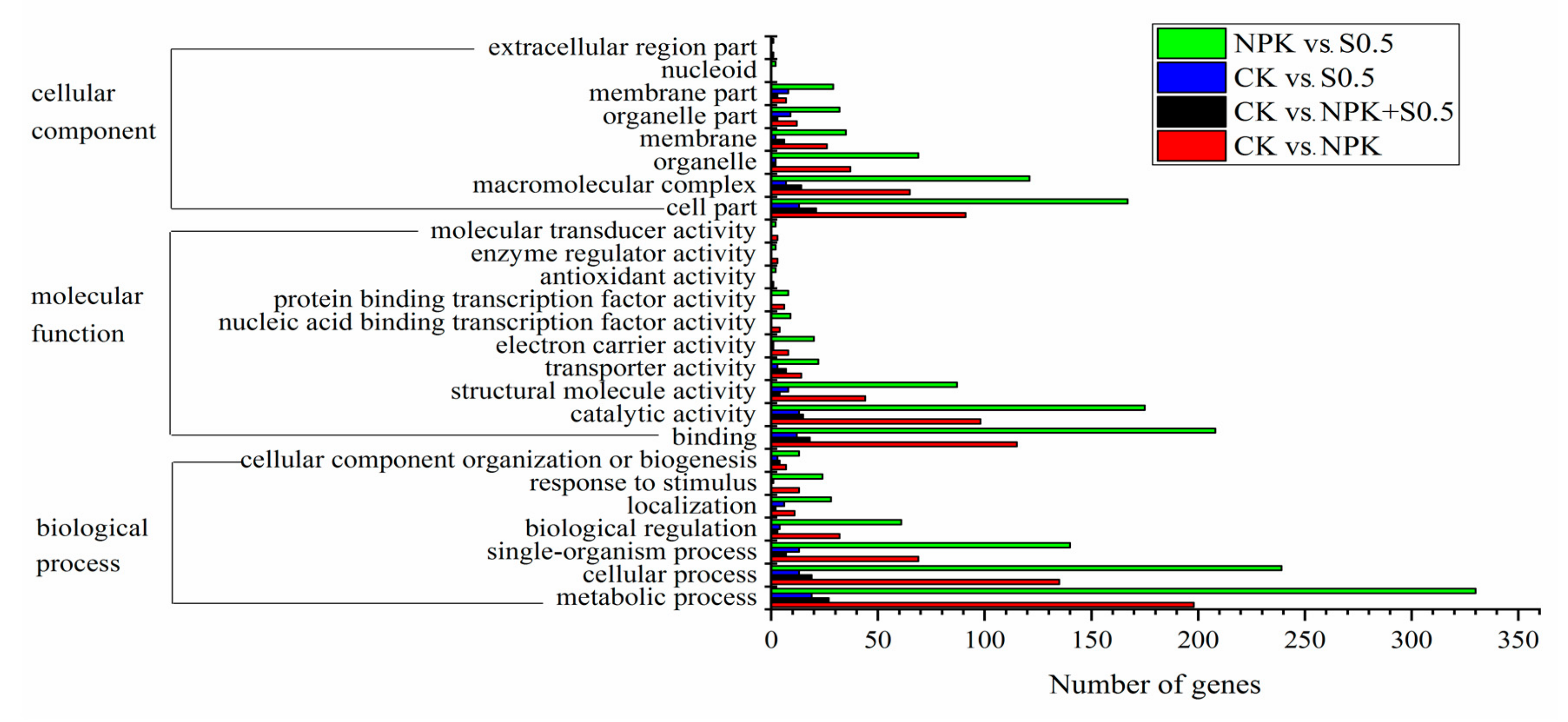
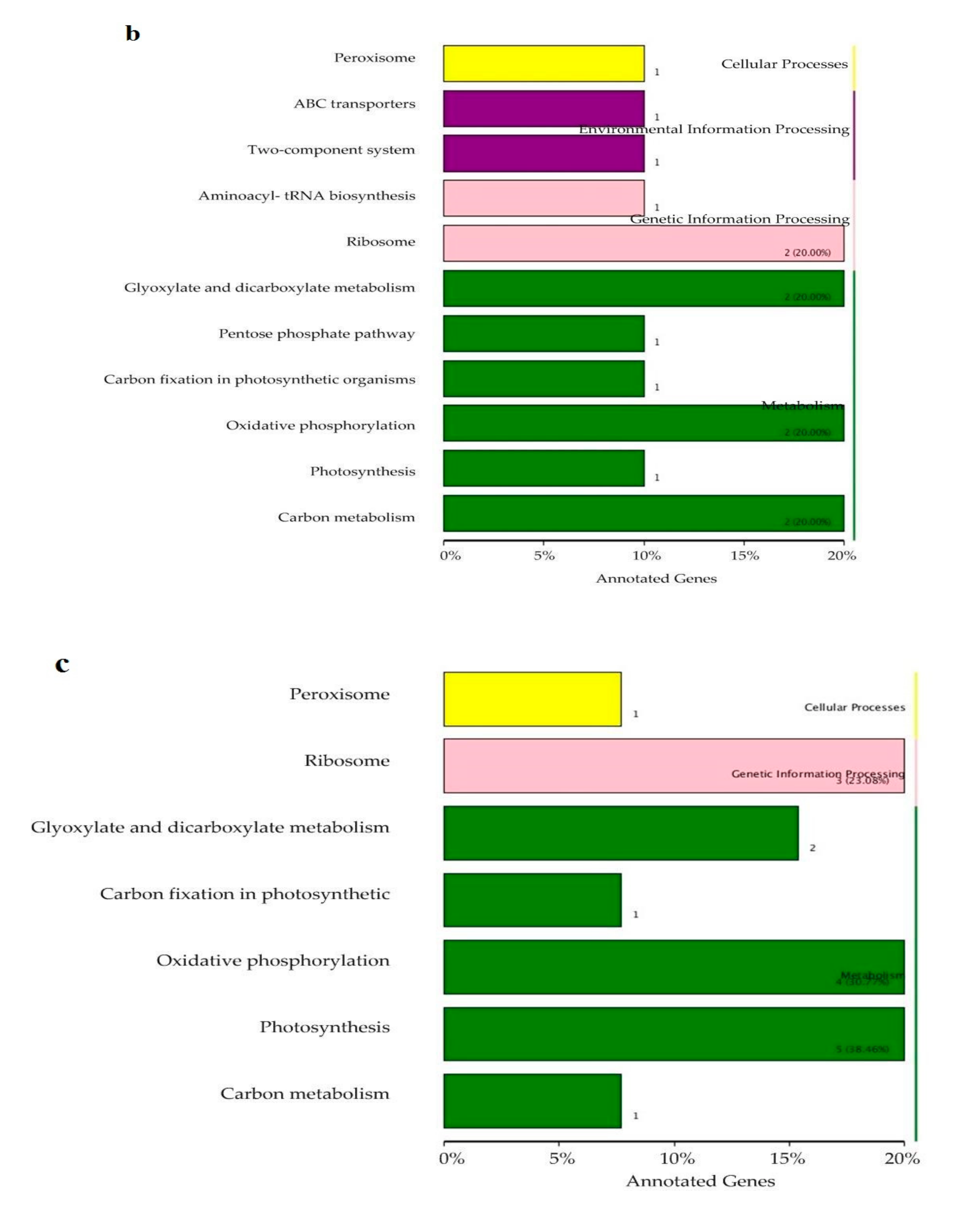
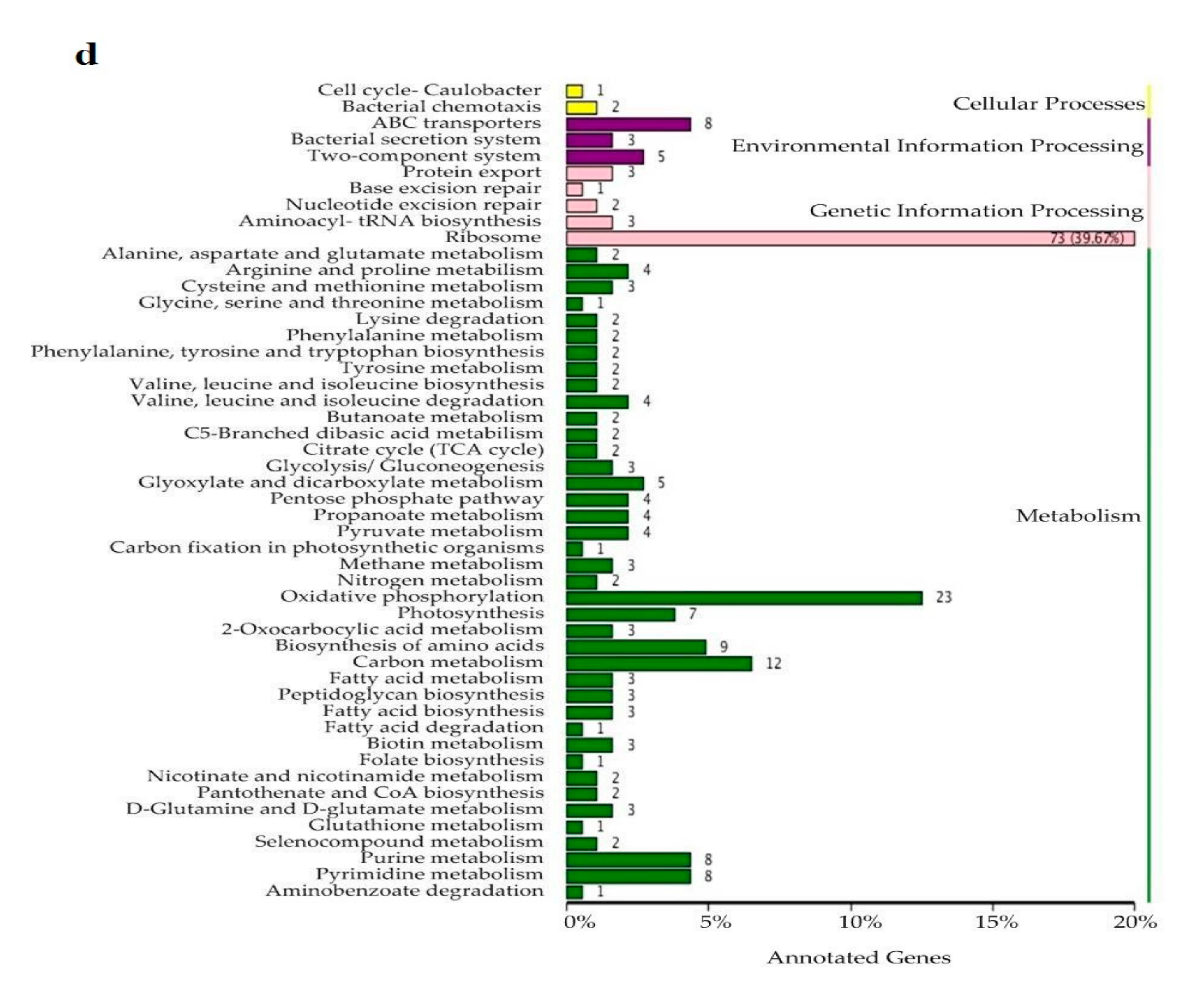
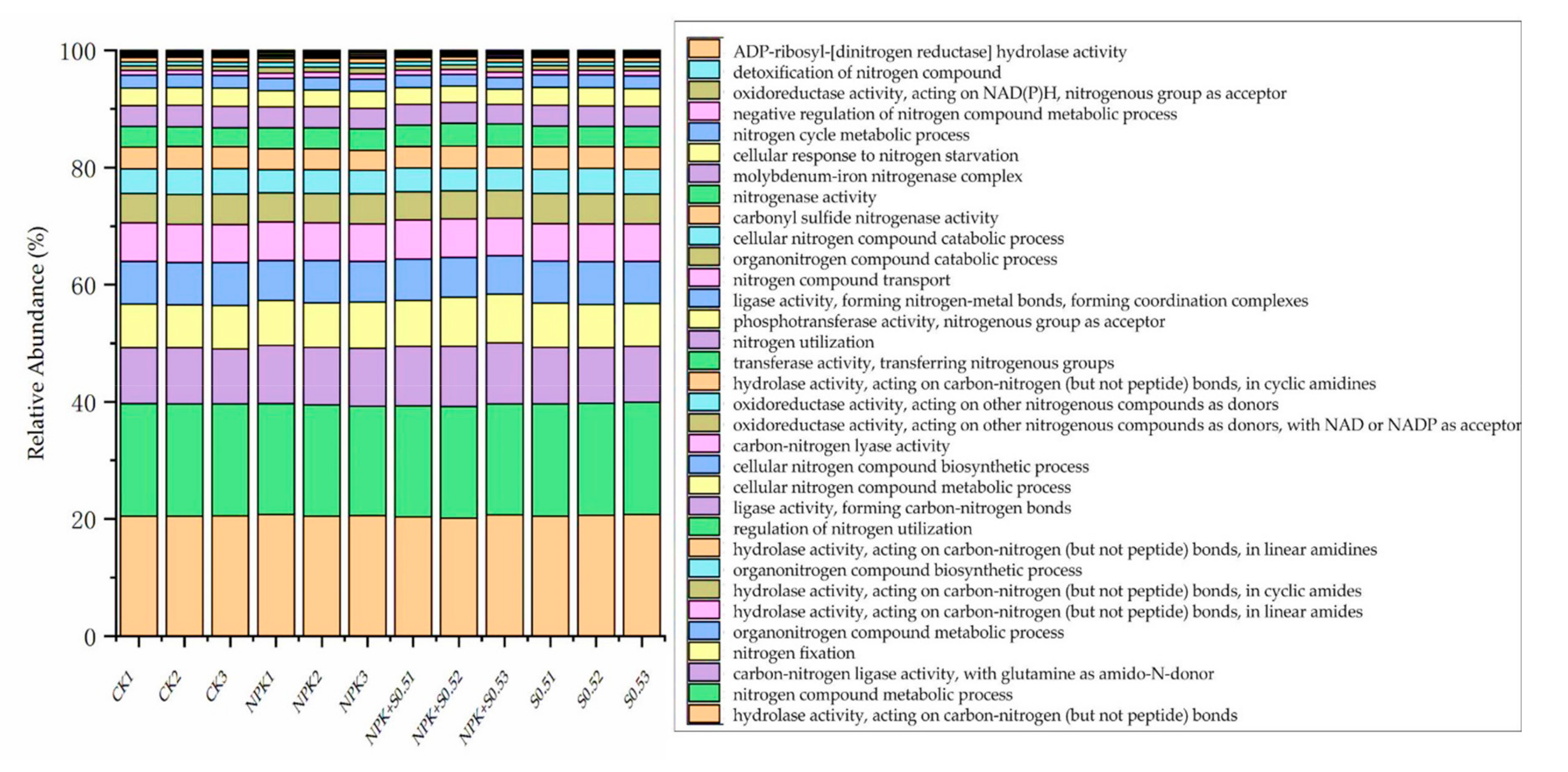
3.3. Soil Microbiome Functions
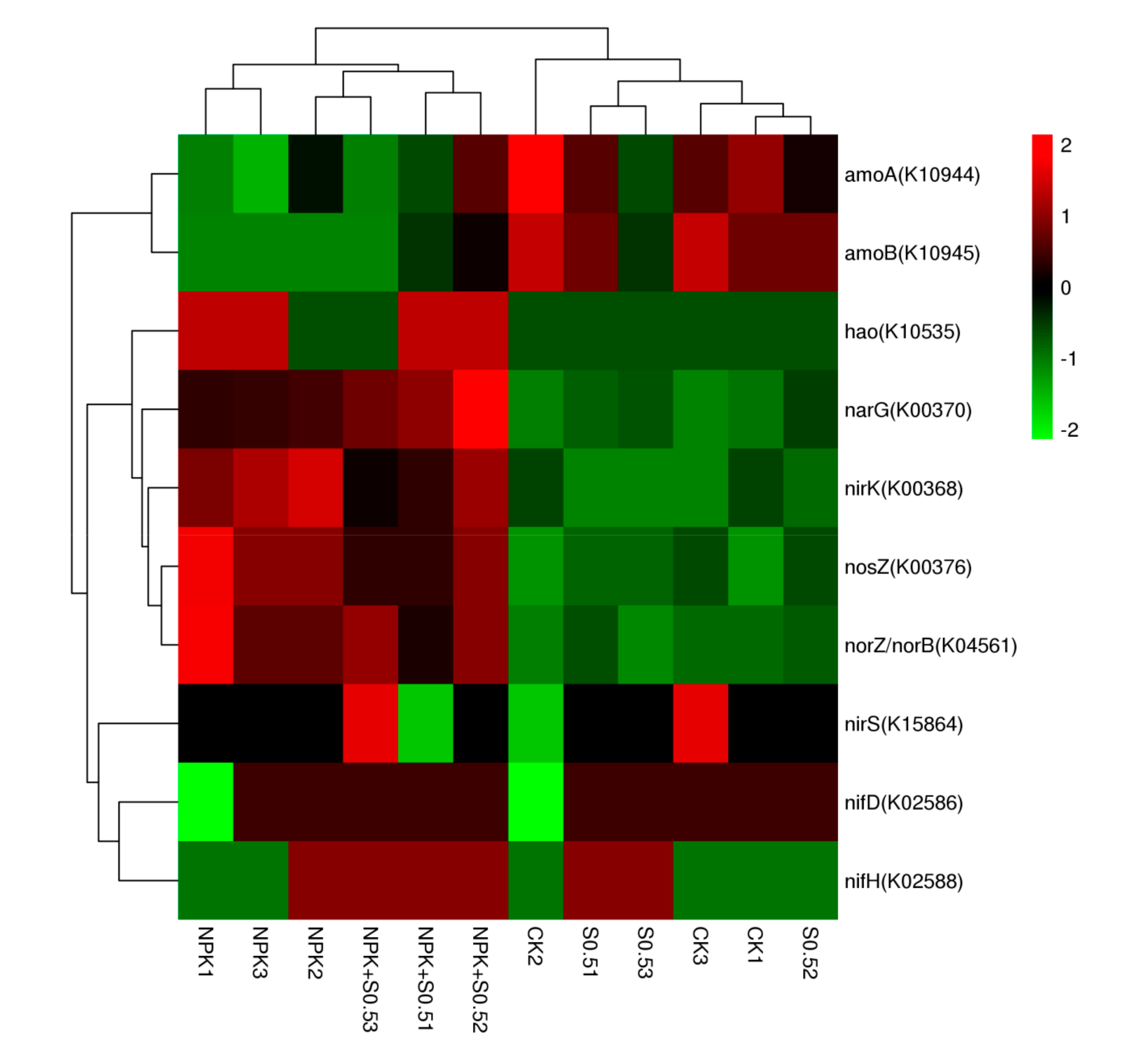
3.4. The Soil Microbiome Participating in N Cycling
4. Discussion
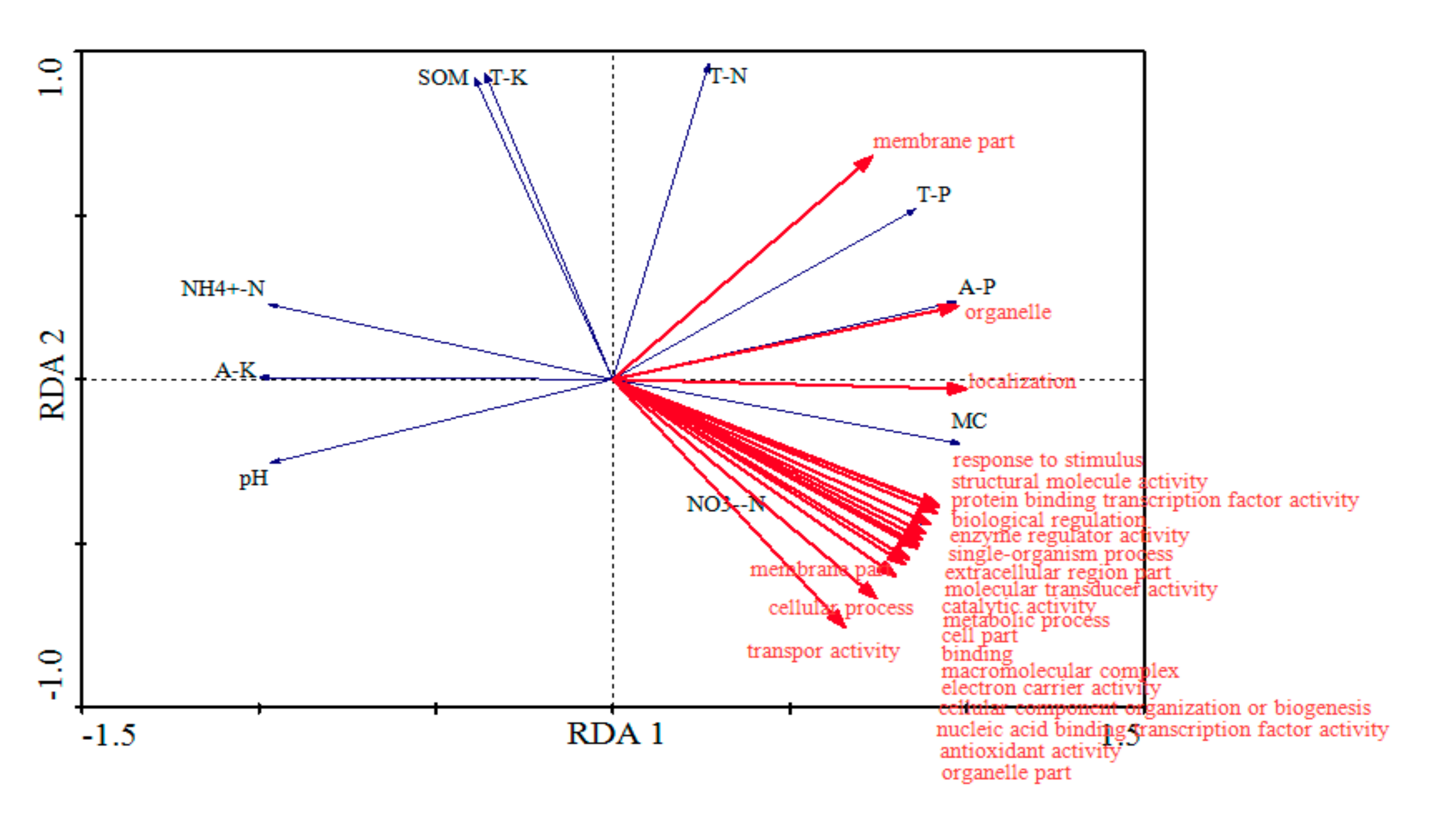
4.1. Soil Chemistry Properties
4.2. Soil Microbiome Community and Function
4.3. The Soil Microbiome Participating in the N Cycle
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Samples | Counting Numbers (×105) | Total Length (bp) (×108) | Largest Length (bp) (×104) | N50 (bp) | Mapped (%) |
|---|---|---|---|---|---|
| CK | 1.92 ± 0.23 a | 1.34 ± 0.26 b | 4.48 ± 0.17 b | 716.00 ± 29.87 b | 16.56 ± 1.98 b |
| NPK | 2.39 ± 0.14 ab | 2.01 ± 0.11 a | 6.03 ± 0.26 a | 779.67 ± 1.53 a | 20.39 ± 1.00 a |
| NPK+S0.5 | 2.66 ± 0.10 a | 2.16 ± 0.98 a | 5.82 ± 0.12 a | 758.67 ± 25.42 a | 19.81 ± 0.60 a |
| S0.5 | 1.82 ± 0.50 a | 1.13 ± 0.13 c | 4.46 ± 0.11 b | 694.00 ± 14.00 b | 15.26 ± 2.37 b |
| DEG Set | DEG Numbers | High Abundance | Low Abundance | COG | GO | KEGG | NR | Pfam | SwissProt |
|---|---|---|---|---|---|---|---|---|---|
| CK vs. NPK | 1137 | 582 | 555 | 295 | 198 | 173 | 626 | 423 | 263 |
| CK vs. NPK+S0.5 | 304 | 153 | 151 | 38 | 27 | 16 | 84 | 64 | 39 |
| CK vs. S0.5 | 234 | 161 | 73 | 18 | 19 | 16 | 64 | 46 | 29 |
| NPK vs. S0.5 | 1581 | 702 | 879 | 464 | 330 | 310 | 885 | 649 | 444 |
| pH | MC | SOM | AP | A-K | NH4+-N | NO3−-N | T-P | T-K | T-N | |
|---|---|---|---|---|---|---|---|---|---|---|
| Proteobacteria | 0.74 | −0.66 | −0.02 | −0.53 | 0.85 | 0.72 | −0.41 | −0.65 | −0.23 | −0.19 |
| Acidobacteria | −0.82 | 0.61 | 0.03 | 0.63 | −0.86 | −0.70 | 0.48 | 0.73 | 0.18 | 0.27 |
| Firmicutes | −0.07 | −0.06 | −0.67 | 0.19 | −0.06 | −0.14 | 0.63 | −0.08 | −0.95 * | −0.23 |
| Ascomycota | −0.90 | −0.03 | −0.04 | 0.970 * | −0.49 | −0.29 | 0.81 | 0.87 | −0.39 | 0.60 |
| Basidiomycota | 0.95 | −0.22 | 0.19 | −0.94 | 0.70 | 0.52 | −0.90 | −0.85 | 0.43 | −0.44 |
| Mucoromycota | −0.76 | 0.29 | −0.52 | 0.76 | −0.66 | −0.58 | 0.99 * | 0.58 | −0.74 | 0.14 |
| Crenarchaeota | −0.09 | 0.63 | 0.14 | −0.16 | −0.45 | −0.45 | −0.24 | 0.06 | 0.60 | −0.16 |
| Euryarchaeota | −0.07 | 0.32 | 0.48 | −0.11 | −0.20 | −0.14 | −0.44 | 0.14 | 0.84 | 0.11 |
| Thaumarchaeota | −0.88 | 0.69 | −0.34 | 0.71 | −0.97 * | −0.86 | 0.82 | 0.69 | −0.27 | 0.11 |
| Bradyrhizobium | −0.14 | −0.03 | −0.67 | 0.25 | −0.12 | −0.18 | 0.68 | −0.03 | −0.96 * | −0.20 |
| Streptomyces | −0.93 | −0.09 | 0.37 | 0.97 * | −0.44 | −0.17 | 0.52 | 0.99 ** | 0.09 | 0.83 |
| Mycobacterium | −0.76 | 0.29 | −0.52 | 0.77 | −0.67 | −0.58 | 0.99 | 0.58 | −0.74 | 0.14 |
| Sordaria | 0.62 | −0.66 | −0.10 | −0.39 | 0.77 | 0.65 | −0.24 | −0.55 | −0.38 | −0.15 |
| Fusarium | −0.33 | −0.79 | 0.43 | 0.59 | 0.35 | 0.55 | 0.18 | 0.51 | −0.11 | 0.79 |
| Paracoccidioides | 0.33 | 0.79 | −0.43 | −0.59 | −0.35 | −0.55 | −0.18 | −0.51 | 0.11 | −0.79 |
| Halorubrum | −0.19 | −0.32 | −0.39 | 0.37 | 0.07 | 0.08 | 0.59 | 0.12 | −0.82 | 0.10 |
| Halococcus | −0.76 | 0.29 | −0.52 | 0.76 | −0.66 | −0.58 | 0.99 * | 0.58 | −0.74 | 0.14 |
| Haloferax | 0.43 | −0.98 | 0.70 | −0.18 | 0.93 | 0.99 | −0.63 | −0.14 | 0.38 | 0.48 |
| Alphabaculovirus | −0.67 | −0.30 | −0.05 | 0.83 | −0.19 | −0.02 | 0.72 | 0.67 | −0.52 | 0.57 |
| Jd18virus | 0.50 | −0.86 | 0.82 | −0.31 | 0.90 | 0.96 * | −0.83 | −0.19 | 0.64 | 0.44 |
| Betabaculovirus | 0.738 | −0.685 | 0.011 | −0.524 | 0.863 | 0.738 | −0.424 | −0.639 | −0.213 | −0.17 |
References
- Zhao, J.; Ni, T.; Xun, W.; Huang, X.; Huang, Q.; Ran, W.; Shen, B.; Zhang, R.; Shen, Q. Influence of straw incorporation with and without straw decomposer on soil bacterial community structure and function in a rice-wheat cropping system. Appl. Microbiol. Biotechnol. 2017, 101, 4761–4773. [Google Scholar] [CrossRef] [PubMed]
- Kalivas, A.; Ganopoulos, I.; Psomopoulos, F.; Grigoriadis, I.; Xanthopoulou, A.; Hatzigiannakis, E.; Osathanunkul, M.; Tsaftaris, A.; Madesis, P. Comparative metagenomics reveals alterations in the soil bacterial community driven by N-fertilizer and Amino 16(R) application in lettuce. Genom. Data 2017, 14, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Quiza, L.; Tremblay, J. Microbial indicators are better predictors of wheat yield and quality than N fertilization. FEMS Microbiol. Ecol. 2020, 96, fiz205. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Wang, Y.; Hu, Y.; Li, X. Maize yield and soil fertility with combined use of compost and inorganic fertilizers on a calcareous soil on the North China Plain. Soil Tillage Res. 2016, 155, 85–94. [Google Scholar] [CrossRef]
- Zhang, B.Y.; Chen, T.L.; Wang, B. Effects of Long-term Uses of Chemical Fertilizers on Soil Quality. Chin. Agric. Ence Bull. 2010, 26, 182–187. [Google Scholar]
- Tayyab, M.; Islam, W.; Arafat, Y.; Pang, Z.; Zhang, C.; Lin, Y.; Waqas, M.; Lin, S.; Lin, W.; Zhang, H. Effect of Sugarcane Straw and Goat Manure on Soil Nutrient Transformation and Bacterial Communities. Sustainability 2018, 10, 2361. [Google Scholar] [CrossRef]
- Hester, E.R.; Harpenslager, S.F.; van Diggelen, J.M.H.; Lamers, L.L.; Jetten, M.S.M.; Lüke, C.; Lücker, S.; Welte, C.U. Linking Nitrogen Load to the Structure and Function of Wetland Soil and Rhizosphere Microbial Communities. Msystems 2018, 3, e00214–e00217. [Google Scholar] [CrossRef]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2017, 12, 212–224. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, Z.; Li, X.; Liu, J. Bacterial diversity as affected by application of manure in red soils of subtropical China. Biol. Fertil. Soils 2017, 53, 639–649. [Google Scholar] [CrossRef]
- Shi, Y.; Ziadi, N.; Hamel, C.; Bélanger, G.; Abdi, D.; Lajeunesse, J.; Lafond, J.; Lalande, R.; Shang, J. Soil microbial biomass, activity and community structure as affected by mineral phosphorus fertilization in grasslands. Appl. Soil Ecol. 2020, 146, 103391. [Google Scholar] [CrossRef]
- Gu, S.; Hu, Q.; Cheng, Y.; Bai, L.; Liu, Z.; Xiao, W.; Gong, Z.; Wu, Y.; Feng, K.; Deng, Y.; et al. Application of organic fertilizer improves microbial community diversity and alters microbial network structure in tea (Camellia sinensis) plantation soils. Soil Tillage Res. 2019, 195, 104356. [Google Scholar] [CrossRef]
- Su, J.Q.; Xia, Y.; Yao, H.Y.; Li, Y.Y.; An, X.L.; Singh, B.K.; Zhang, T.; Zhu, Y.G. Metagenomic assembly unravel microbial response to redox fluctuation in acid sulfate soil. Soil Biol. Biochem. 2017, 105, 244–252. [Google Scholar] [CrossRef]
- Chen, W.; Teng, Y.; Li, Z.; Liu, W.; Ren, W.; Luo, Y.; Christie, P. Mechanisms by which organic fertilizer and effective microbes mitigate peanut continuous cropping yield constraints in a red soil of south China. Appl. Soil Ecol. 2018, 128, 23–34. [Google Scholar] [CrossRef]
- Ahmed, V.; Verma, M.K.; Gupta, S.; Mandhan, V.; Chauhan, N.S. Metagenomic Profiling of Soil Microbes to Mine Salt Stress Tolerance Genes. Front. Microbiol. 2018, 9, 159. [Google Scholar] [CrossRef]
- Das, S.; Gwon, H.S.; Khan, M.I.; Van Nostrand, J.D.; Alam, M.A.; Kim, P.J. Taxonomic and functional responses of soil microbial communities to slag-based fertilizer amendment in rice cropping systems. Environ. Int. 2019, 127, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Liang, B.; Yang, X.; He, X.; Murphy, D.V.; Zhou, J. Long-term combined application of manure and NPK fertilizers influenced nitrogen retention and stabilization of organic C in Loess soil. Plant Soil 2012, 353, 249–260. [Google Scholar] [CrossRef]
- Arp, D.J.; Stein, L.Y. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 471–495. [Google Scholar] [CrossRef]
- Tu, Q.; He, Z.; Wu, L.; Xue, K.; Xie, G.; Chain, P.; Reich, P.B.; Hobbie, S.E.; Zhou, J. Metagenomic reconstruction of nitrogen cycling pathways in a CO2-enriched grassland ecosystem. Soil Biol. Biochem. 2017, 106, 99–108. [Google Scholar] [CrossRef]
- Petersen, D.G.; Blazewicz, S.J.; Firestone, M.; Herman, D.J.; Turetsky, M.; Waldrop, M. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 2012, 14, 993–1008. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Romero, Y.; Navarro-Noya, Y.E.; Reynoso-Martínez, S.C.; Sarria-Guzmán, Y.; Govaerts, B.; Verhulst, N.; Dendooven, L.; Luna-Guido, M. 16S metagenomics reveals changes in the soil bacterial community driven by soil organic C, N-fertilizer and tillage-crop residue management. Soil Tillage Res. 2016, 159, 1–8. [Google Scholar] [CrossRef]
- Deng, Y.; Feng, K.; Wei, Z.; Liu, W.; Liang, Y.; Jin, D. Recent studies and applications of metagenomics in environmental engineering. Chin. J. Environ. Eng. 2016, 10, 3373–3382. [Google Scholar]
- Cui, J.T.; Li, Y.N.; Wang, C.Y.; Seok, K.K.; Wang, T.Y.; Liu, S. Characteristics of Rhizosphere Bacterial Community across Different Cultivation Years in Saline-alkaline Paddy Soils of Songnen Plain of China. Can. J. Microbiol. 2018, 64, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.F.; Zhang, S.; Li, N.; Chen, H.; Jia, H.; Song, X.; Liu, G.; Ni, C.; Wang, Z.; Shao, H. Metagenomic insights into effects of wheat straw compost fertliser application on microbial community composition and function in tobacco rihizoshere soil. Sci. Rep. 2019, 9, 6168. [Google Scholar]
- Tan, G.; Liu, K.; Kang, J.; Xu, K.; Zhang, Y.; Hu, L.; Zhang, J.; Li, C. Transcriptome analysis of the compatible interaction of tomato with Verticillium dahliae using RNA-sequencing. Front. Plant Sci. 2015, 6, 428. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, S.; Wang, J.; Zhang, X.; Mahamood, M.; Yu, J.; Zhang, X.; Liang, A.; Liang, W. Tillage and crop succession effects on soil microbial metabolic activity and carbon utilization in a clay loam soil. Eur. J. Soil Biol. 2018, 88, 97–104. [Google Scholar] [CrossRef]
- Hara-Yamamura, H.; Nakashima, K.; Hoque, A.; Miyoshi, T.; Kimura, K.; Watanabe, Y.; Okabe, S. Evaluation of Whole Wastewater Effluent Impacts on HepG2 using DNA Microarray-based Transcriptome Analysis. Environ. Sci. Technol. 2013, 47, 5425–5432. [Google Scholar] [CrossRef]
- Zhu, J.; Yu, Z.; He, L.; Cao, X.; Liu, S.; Song, X. Molecular Mechanism of Modified Clay Controlling the Brown Tide Organism Aureococcus anophagefferens Revealed by Transcriptome Analysis. Environ. Sci. Technol. 2018, 52, 7006–7014. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Liu, X.; Zhang, D.; Li, L.; Li, W.; Sheng, L. Effects of long-term application of organic fertilizer on improving organic matter content and retarding acidity in red soil from China. Soil Tillage Res. 2019, 195, 104382. [Google Scholar] [CrossRef]
- Qaswar, M.; Jing, H.; Ahmed, W.; Li, D.C.; Liu, S.J.; Lu, Z.; Cai, A.D.; Liu, L.S.; Xu, Y.M.; Gao, J.S.; et al. Yield sustainability, soil organic carbon sequestration and nutrients balance under long-term combined application of manure and inorganic fertilizers in acidic paddy soil. Soil Tillage Res. 2020, 198, 104569. [Google Scholar] [CrossRef]
- Wang, S.; Adhikari, K.; Wang, Q.; Jin, X.; Li, H. Role of environmental variables in the spatial distribution of soil carbon (C), nitrogen (N), and C:N ratio from the northeastern coastal agroecosystems in China. Ecol. Indic. 2018, 84, 263–272. [Google Scholar] [CrossRef]
- Zhou, G.; Qiu, X.; Zhang, J.; Tao, C. Effects of seaweed fertilizer on enzyme activities, metabolic characteristics, and bacterial communities during maize straw composting. Bioresour. Technol. 2019, 286, 121375. [Google Scholar] [CrossRef] [PubMed]
- Agehara, S.; Warncke, D.D. Soil Moisture and Temperature Effects on Nitrogen Release from Organic Nitrogen Sources. Soil Sci. Soc. Am. J. 2005, 69, 1844–1855. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.X.; Guo, X.; Wang, D.; Chu, H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Sci. Found. China 2015, 88, 9–18. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Roy, K.S.; Nayak, A.K.; Shahid, M.; Lal, B.; Gautam, P.; Mohapatra, T. Metagenomic assessment of methane production-oxidation and nitrogen metabolism of long term manured systems in lowland rice paddy. Sci. Total Environ. 2017, 586, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.C.; Cantão, M.E.; Vasconcelos, A.T.R.; Nogueira, M.A.; Hungria, M. Soil metagenomics reveals differences under conventional and no-tillage with crop rotation or succession. Appl. Soil Ecol. 2013, 72, 49–61. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, J.; Min, L.; Qin, C.; Shen, Q. Microbial communities of an arable soil treated for 8 years with organic and inorganic fertilizers. Biol. Fertil. Soils 2016, 52, 455–467. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Baugher, C.M.; Kahl, K.; Huggins, D.R.; Johnson-Maynard, J.L.; Paulitz, T.C. Bacterial communities of soil and earthworm casts of native Palouse Prairie remnants and no-till wheat cropping systems. Soil Biol. Biochem. 2019, 139, 107625. [Google Scholar] [CrossRef]
- Starke, R.; Kermer, R.; Ullmann-Zeunert, L.; Baldwin, I.T.; Seifert, J.; Bastida, F.; von Bergen, M.; Jehmlich, N. Bacteria dominate the short-term assimilation of plant-derived N in soil. Soil Biol. Biochem. 2016, 96, 30–38. [Google Scholar] [CrossRef]
- Zhang, N.; Li, Y.; Wubet, T.; Bruelheide, H.; Liang, Y.; Purahong, W.; Buscot, F.; Ma, K. Tree species richness and fungi in freshly fallen leaf litter: Unique patterns of fungal species composition and their implications for enzymatic decomposition. Soil Biol. Biochem. 2018, 127, 120–126. [Google Scholar] [CrossRef]
- De Carvalho, T.S.; da Conceicao Jesus, E.; Barlow, J.; Gardner, T.A.; Soares, I.C.; Tiedje, J.M.; de Souza Moreira, F.M. Land use intensification in the humid tropics increased both alpha and beta diversity of soil bacteria. Ecology 2016, 97, 2760–2771. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.F.A.; Pan, Y.; Bloem, J.; Berge, H.t.; Kuramae, E.E. Organic nitrogen rearranges both structure and activity of the soil-borne microbial seedbank. Sci. Rep. 2017, 7, 42634. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Wilkinson, M.J.; Wang, Y.; Li, J.; Paull, J.G. Gene Function Expression Profile of Faba bean (Vicia faba) Seeds. J. Appl. Microbiol. Biochem. 2018, 1, 3–11. [Google Scholar] [CrossRef]
- Dijkstra, P.; Dalder, J.J.; Selmants, P.C.; Hart, S.C.; Koch, G.W.; Schwartz, E.; Hungate, B.A. Modeling soil metabolic processes using isotopologue pairs of position-specific 13C-labeled glucose and pyruvate. Soil Biol. Biochem. 2011, 43, 1848–1857. [Google Scholar] [CrossRef]
- Graham, M.H.; Haynes, R.J. Organic matter accumulation and fertilizer-induced acidification interact to affect soil microbial and enzyme activity on a long-term sugarcane management experiment. Biol. Fertil. Soils 2005, 41, 249–256. [Google Scholar] [CrossRef]
- Li, W.L.; Wang, J.F.; Lv, Y.; Dong, H.J.; Wang, L.L.; He, T.; Li, Q.S. Improving cadmium mobilization by phosphate-solubilizing bacteria via regulating organic acids metabolism with potassium. Chemosphere 2019, 244, 125475. [Google Scholar] [CrossRef] [PubMed]
- Meidute, S.; Demoling, F.; Bååth, E. Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol. Biochem. 2008, 40, 2334–2343. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The Evolution and Future of Earth’s Nitrogen Cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef]
- Chang, N.; Zhai, Z.; Li, H.; Wang, L.; Deng, J. Impacts of nitrogen management and organic matter application on nitrous oxide emissions and soil organic carbon from spring maize fields in the North China Plain. Soil Tillage Res. 2020, 196, 104441. [Google Scholar] [CrossRef]
- Guo, J.; Wang, Y.; Lai, J.; Pan, C.; Wang, S.; Fu, H.; Zhang, B.; Cui, Y.; Zhang, L. Spatiotemporal distribution of nitrogen biogeochemical processes in the coastal regions of northern Beibu Gulf, south China sea. Chemosphere 2020, 239, 124803. [Google Scholar] [CrossRef] [PubMed]
- Ziadi, N.; Angers, D.A.; Gagnon, B.; Lalande, R.; Morel, C.; Rochette, P.; Chantigny, M.H. Long-term tillage and synthetic fertilization affect soil functioning and crop yields in a corn–soybean rotation in eastern Canada. Can. J. Soil Sci. 2014, 94, 365–376. [Google Scholar] [CrossRef]
- Blankenau, K.; Olfs, H.-W.; Kuhlmann, H. Effect of microbial nitrogen immobilization during the growth period on the availability of nitrogen fertilizer for winter cereals. Biol. Fertil. Soils 2000, 32, 157–165. [Google Scholar] [CrossRef]
- Soni, R.; Suyal, D.C.; Sai, S.; Goel, R. Exploration of nifH gene through soil metagenomes of the western Indian Himalayas. 3 Biotech 2016, 6, 25. [Google Scholar] [CrossRef]
- Shao, T.; Zhao, J.; Zhu, T.; Chen, M.; Wu, Y.; Long, X.; Gao, X. Relationship between rhizosphere soil properties and blossom-end rot of tomatoes in coastal saline-alkali land. Appl. Soil Ecol. 2018, 127, 96–101. [Google Scholar] [CrossRef]
- Yang, Y.D.; Ren, Y.-F.; Wang, X.-Q.; Hu, Y.-G.; Wang, Z.-M.; Zeng, Z.-H. Ammonia-oxidizing archaea and bacteria responding differently to fertilizer type and irrigation frequency as revealed by Illumina Miseq sequencing. J. Soils Sediments 2017, 18, 1029–1040. [Google Scholar] [CrossRef]
- Chen, C.L.; Wu, M.N.; Wei, W.X. Effect of Long-Term Application of Nitrogen Fertilizer on the Diversity of Nitrifying Genes (amoA and hao) in Paddy Soil. Environ. Sci. 2011, 32, 1489–1496. [Google Scholar]
- Meng, L.; Ding, W.; Cai, Z. Long-term application of organic manure and nitrogen fertilizer on N2O emissions, soil quality and crop production in a sandy loam soil. Soil Biol. Biochem. 2005, 37, 2037–2045. [Google Scholar] [CrossRef]


| Samples | Clean Reads (107) | Base Sum (1010) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| CK | 3.65 ± 0.02 a | 1.09 ± 0.05 a | 97.38 ± 0.04 a | 93.15 ± 0.08 a | 63.67 ± 0.08 a |
| NPK | 3.35 ± 0.02 a | 0.98 ± 0.04 a | 97.42 ± 0.01 a | 93.25 ± 0.02 a | 63.69 ± 0.18 a |
| NPK+S0.5 | 3.81 ± 0.02 a | 1.14 ± 0.06 a | 97.39 ± 0.05 a | 93.16 ± 0.12 a | 64.14 ± 0.49 a |
| S0.5 | 3.40 ± 0.02 a | 1.02 ± 0.05 a | 97.43 ± 0.06 a | 93.17 ± 0.05 a | 65.13 ± 1.52 a |
| CK | NPK | NPK+S0.5 | S0.5 | ||
|---|---|---|---|---|---|
| (%) | (%) | (%) | (%) | ||
| Bacteria | Proteobacteria | 51.89 ± 0.47 b | 57.32 ± 1.26 a | 51.85 ± 3.65 b | 51.71 ± 1.99 c |
| Acidobacteria | 42.53 ± 0.27 b | 37.12 ± 1.36 c | 43.43 ± 3.66 a | 43.26 ± 1.78 a | |
| Firmicutes | 1.16 ± 0.03 b | 1.24 ± 0.08 a | 1.22 ± 0.30 a | 1.13 ± 0.11 b | |
| Fungi | Ascomycota | 87.40 ± 0.22 d | 87.81 ± 1.04 c | 89.77 ± 0.98 a | 88.62 ± 0.55 b |
| Basidiomycota | 10.39 ± 0.43 b | 10.57 ± 0.83 a | 8.43 ± 0.93 d | 9.71 ± 0.41 c | |
| Mucoromycota | 0.94 ± 0.26 a | 1.41 ± 0.22 a | 1.20 ± 0.33 a | 0.99 ± 0.21 a | |
| Archaea | Crenarchaeota | 95.87 ± 0.03 c | 97.78 ± 0.05 a | 97.39 ± 0.09 b | 96.16 ± 0.22 bc |
| Euryarchaeota | 2.56 ± 0.20 a | 1.08 ± 0.12 c | 1.45 ± 0.28 c | 2.17 ± 0.37 b | |
| Thaumarchaeota | 1.46 ± 0.16 b | 1.01 ± 0.12 c | 1.05 ± 0.19 c | 1.51 ± 0.16 a | |
| Bacteria | Bradyrhizobium | 7.66 ± 0.08 b | 10.33 ± 0.10 a | 10.75 ± 0.11 a | 6.75 ± 0.07 b |
| Streptomyces | 7.34 ± 0.07 a | 6.8 ± 0.07 a | 7.86 ± 0.08 a | 7.64 ± 0.08 a | |
| Mycobacterium | 3.78 ± 0.04 b | 4.13 ± 0.04 b | 4.96 ± 0.05 a | 3.84 ± 0.04 b | |
| Fungi | Sordaria | 5.83 ± 0.06 b | 11.02 ± 0.11 a | 7.03 ± 0.07 b | 5.63 ± 0.06 b |
| Fusarium | 2.41 ± 0.02 a | 2.64 ± 0.03 a | 2.58 ± 0.03 a | 3.15 ± 0.03 a | |
| Paracoccidioides | 2.8 ± 0.03 a | 1.95 ± 0.02 b | 1.85 ± 0.02 b | 2.2 ± 0.02 b | |
| Archaea | Halorubrum | 13.84 ± 0.14 a | 14.36 ± 0.14 a | 14.47 ± 0.12 a | 13.92 ± 0.09 a |
| Halococcus | 9.48 ± 0.09 ab | 9.01 ± 0.09 b | 9.62 ± 0.10 a | 9.27 ± 0.09 ab | |
| Haloferax | 6.85 ± 0.07 a | 7.1 ± 0.07 a | 6.88 ± 0.05 a | 7.04 ± 0.07 a | |
| Viruses | Alphabaculovirus | 10.67 ± 0.11 a | 13.33 ± 0.13 a | 14.67 ± 0.15 a | 13 ± 0.13 a |
| Jd18virus | 11.00 ± 0.11 a | 14.33 ± 0.14 a | 10.33 ± 0.10 a | 14.33 ± 0.14 a | |
| Betabaculovirus | 1.00 ± 0.01 a | 2.00 ± 0.02 a | 1.33 ± 0.01 a | 1.33 ± 0.01 a | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, C.; Wang, T.; Liu, Y.; Jia, S.; Gao, Y.; Liu, S. Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions. Land 2020, 9, 329. https://doi.org/10.3390/land9090329
Li Y, Wang C, Wang T, Liu Y, Jia S, Gao Y, Liu S. Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions. Land. 2020; 9(9):329. https://doi.org/10.3390/land9090329
Chicago/Turabian StyleLi, Yanan, Chengyu Wang, Tianye Wang, Yutao Liu, Shuxia Jia, Yunhang Gao, and Shuxia Liu. 2020. "Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions" Land 9, no. 9: 329. https://doi.org/10.3390/land9090329
APA StyleLi, Y., Wang, C., Wang, T., Liu, Y., Jia, S., Gao, Y., & Liu, S. (2020). Effects of Different Fertilizer Treatments on Rhizosphere Soil Microbiome Composition and Functions. Land, 9(9), 329. https://doi.org/10.3390/land9090329




