Geodiversity as a Driver of Soil Microbial Community Diversity and Adaptation in a Mediterranean Landscape
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling Strategy and Geochemical Analyses
2.2. Geological Settings
2.3. DNA Extraction, qPCR Quantification, and Sequencing of 16S rRNA Gene
2.4. Bioinformatic Analyses
2.5. Geoinformatic Analyses
2.6. Machine Learning and Geostatistical Modeling
3. Results
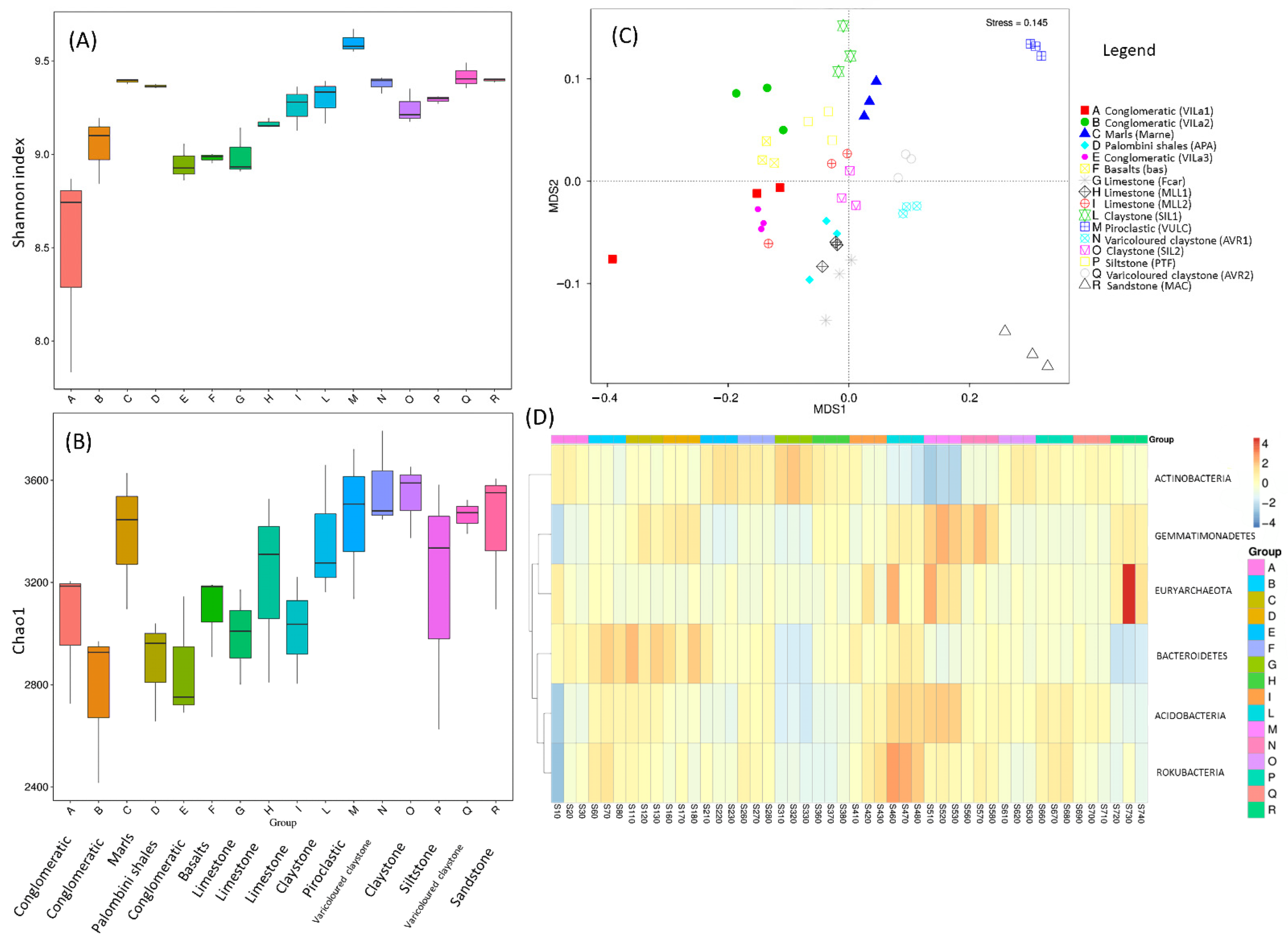
3.1. Geomicrobial Diversity Characterization and Community Composition for Ancient Olive Groves of the Tuscany Region
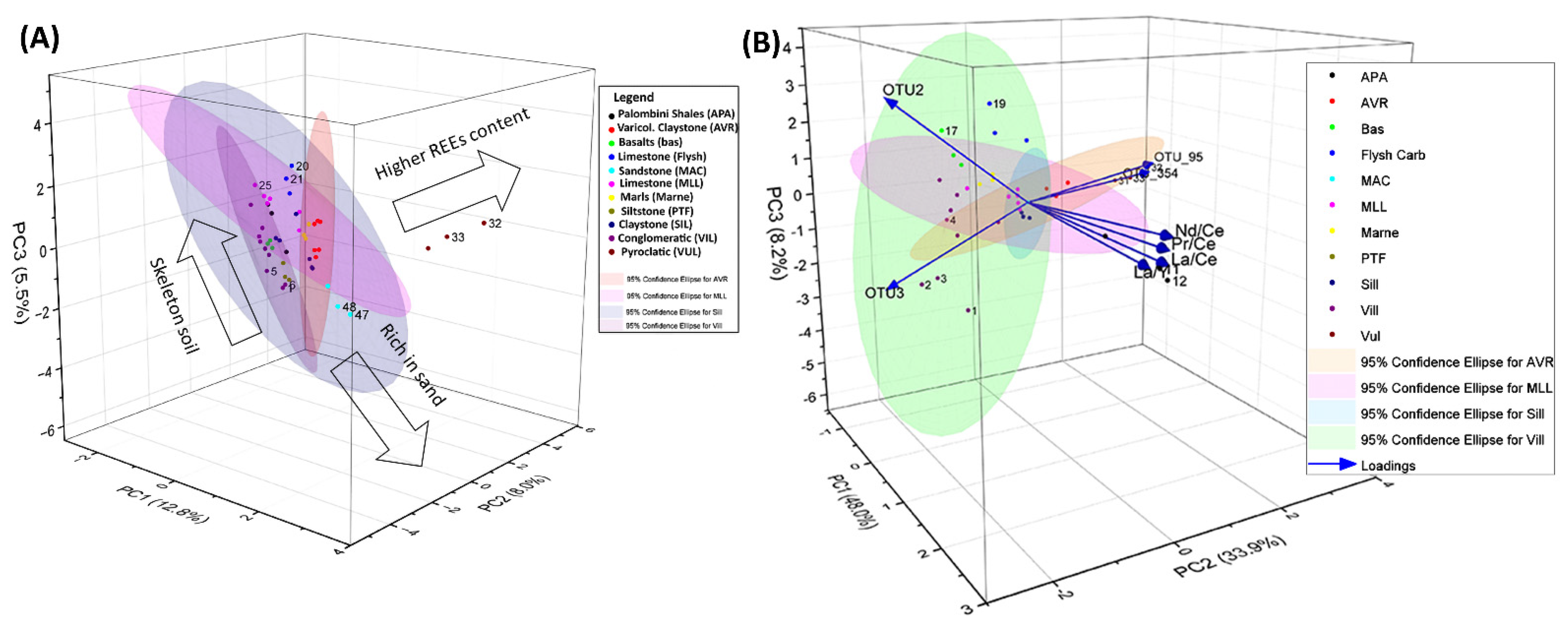
3.2. Relative Importance of Hydro-Geomorphometric and Geochemistry Factors on Soil Actinobacterial Diversity
3.3. Spatial Model Prediction of Biomarkers Based on the Most Dominant Actinobacteria Genera Across an Ancient Olive Grove Mediterranean Landscape
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OTU | Operational Taxonomic Unit |
| REE | Rare Earth Element |
| CNBL | Channel Network Base Level |
| VD | Valley Depth |
| CND | Vertical Distance to Channel Network |
| TRI | Terrain Ruggedness Index |
| TPI | Topographic Position Index |
| TWI | Topographic Wetness Index |
| DEM | Digital Elevation Model |
| LS Factor | Length Slope Factor |
| NDVI | Normalized Difference Vegetation Index |
| NDWI | Normalized Difference Water Index |
References
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive orchard microbiome: Characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Roccotelli, A.; Tommasini, S.; Ceccherini, M.T.; Calamai, L.; Ferrari, M.; Ghiotto, M.; Riccio, R.; Bonciani, L.; Pietramellara, G.; Moretti, S.; et al. Rare earth elements distribution and bacteriome to assess and characterize the soil landscapes of old olive orchards. Diversity 2024, 16, 427. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, D.; Vijayakumar, R.; Vadivalagan, C.; Karthika, P.; Khan MK, A. Isolation, structure elucidation and antibacterial activity of methyl-4,8-dimethylundecanate from the marine actinobacterium streptomyces albogriseolus ecr64. Microb. Pathog. 2018, 121, 166–172. [Google Scholar] [CrossRef]
- Riquelme, C.; Marshall Hathaway, J.J.; Enes Dapkevicius, M.d.L.N.; Miller, A.Z.; Kooser, A.; Northup, D.E.; Jurado, V.; Fernandez, O.; Saiz-Jimenez, C.; Cheeptham, N. Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Front. Microbiol. 2015, 6, 1342. [Google Scholar] [CrossRef]
- Gousterova, A.; Paskaleva, D.; Vasileva-Tonkova, E. Characterization of culturable thermophilic actinobacteria from Livingston island, Antarctica. Int. Res. J. Biol. Sci. 2014, 3, 2278–3202. [Google Scholar]
- Crits-Christoph, A.; Robinson, C.K.; Barnum, T.; Fricke, W.F.; Davila, A.F.; Jedynak, B.; McKay, C.P.; DiRuggiero, J. Colonization patterns of soil microbial communities in the atacama desert. Microbiome 2013, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, S.N.; Thakur, D. Actinobacteria. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 443–476. [Google Scholar]
- Büntgen, U.; Reinig, F.; Verstege, A.; Piermattei, A.; Kunz, M.; Krusic, P.; Slavin, P.; Štěpánek, P.; Torbenson, M.; del Castillo, E.M. Recent summer warming over the western mediterranean region is unprecedented since medieval times. Glob. Planet. Change 2024, 232, 104336. [Google Scholar] [CrossRef]
- González-Ubierna, S.; de la Cruz, M.T.; Casermeiro, M.Á. Climate factors mediate soil respiration dynamics in mediterranean agricultural environments: An empirical approach. Soil Res. 2014, 52, 543–553. [Google Scholar] [CrossRef]
- Labouyrie, M.; Ballabio, C.; Romero, F.; Panagos, P.; Jones, A.; Schmid, M.W.; Mikryukov, V.; Dulya, O.; Tedersoo, L.; Bahram, M. Patterns in soil microbial diversity across europe. Nat. Commun. 2023, 14, 3311. [Google Scholar] [CrossRef]
- Dacal, M.; Garcia-Palacios, P.; Asensio, S.; Wang, J.; Singh, B.K.; Maestre, F.T. Climate change legacies contrastingly affect the resistance and resilience of soil microbial communities and multifunctionality to extreme drought. Funct. Ecol. 2022, 36, 908–920. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Bissett, A.; Eldridge, D.J.; Maestre, F.T.; He, J.-Z.; Wang, J.-T.; Hamonts, K.; Liu, Y.-R.; Singh, B.K.; Fierer, N. Palaeoclimate explains a unique proportion of the global variation in soil bacterial communities. Nat. Ecol. Evol. 2017, 1, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, E.M.; Mau, R.L.; Hayer, M.; Liu, X.-J.A.; Schwartz, E.; Dijkstra, P.; Koch, B.J.; Allen, K.; Blazewicz, S.J.; Hofmockel, K. Evolutionary history constrains microbial traits across environmental variation. Nat. Ecol. Evol. 2019, 3, 1064–1069. [Google Scholar] [CrossRef]
- De Vries, F.T.; Shade, A. Controls on soil microbial community stability under climate change. Front. Microbiol. 2013, 4, 265. [Google Scholar] [CrossRef]
- Evans, S.E.; Wallenstein, M.D. Climate change alters ecological strategies of soil bacteria. Ecol. Lett. 2014, 17, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Pelacani, S.; Maerker, M.; Tommasini, S.; Moretti, S. Combining biodiversity and geodiversity on landscape scale: A novel approach using rare earth elements and spatial distribution models in an agricultural mediterranean landscape. Ecol. Indic. 2024, 158, 111583. [Google Scholar] [CrossRef]
- Melloni, R.; Cardoso, E.J.B.N. Microbiome associated with olive cultivation: A review. Plants 2023, 12, 897. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P.; Fausto, C.; Mininni, A.N.; Crecchio, C.; Scagliola, M.; Malerba, A.D.; Xiloyannis, C.; Dichio, B. The metabolic and genetic diversity of soil bacterial communities depends on the soil management system and c/n dynamics: The case of sustainable and conventional olive groves. Appl. Soil Ecol. 2019, 137, 21–28. [Google Scholar] [CrossRef]
- Karimi, B.; Terrat, S.; Dequiedt, S.; Saby NP, A.; Horrigue, W.; Lelièvre, M.; Nowak, V.; Jolivet, C.; Arrouays, D.; Wincker, P. Biogeography of soil bacteria and archaea across france. Sci. Adv. 2018, 4, eaat1808. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Ceccherini, M.T.; Ascher, J.; Pietramellara, G.; Mocali, S.; Viti, C.; Nannipieri, P. The effect of pharmaceutical waste-fungal biomass, treated to degrade dna, on the composition of eubacterial and ammonia oxidizing populations of soil. Biol. Fertil. Soils 2007, 44, 299–306. [Google Scholar] [CrossRef]
- Santoni, M.; Verdi, L.; Imran Pathan, S.; Napoli, M.; Dalla Marta, A.; Dani, F.R.; Pacini, G.C.; Ceccherini, M.T. Soil microbiome biomass, activity, composition and CO2 emissions in a long-term organic and conventional farming systems. Soil Use Manag. 2023, 39, 588–605. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16s rrna sequence formation and detection in sanger and 454-pyrosequenced pcr amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The silva ribosomal rna gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Chao, A.; Bunge, J. Estimating the number of species in a stochastic abundance model. Biometrics 2002, 58, 531–539. [Google Scholar] [CrossRef]
- Wang, L.; Liu, H. An efficient method for identifying and filling surface depressions in digital elevation models for hydrologic analysis and modelling. Int. J. Geogr. Inf. Sci. 2006, 20, 193–213. [Google Scholar] [CrossRef]
- Sörensen, R.; Zinko, U.; Seibert, J. On the calculation of the topographic wetness index: Evaluation of different methods based on field observations. Hydrol. Earth Syst. Sci. 2006, 10, 101–112. [Google Scholar] [CrossRef]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for automated geoscientific analyses (saga) v. 2.1. 4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Guisan, A.; Weiss, S.B.; Weiss, A.D. Glm versus cca spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Riley, S.J.; DeGloria, S.D.; Elliot, R. Index that quantifies topographic heterogeneity. Intermt. J. Sci. 1999, 5, 23–27. [Google Scholar]
- Bahrenberg, G.; Giese, E.; Nipper, J. Statistische Methoden in der Geographie. In Multivariate Statistik 2; Gebr. Borntraeger: Berlin, Germany, 1992; Volume 2. [Google Scholar]
- Brockett BF, T.; Prescott, C.E.; Grayston, S.J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Fairbanks, D.; Shepard, C.; Murphy, M.; Rasmussen, C.; Chorover, J.; Rich, V.; Gallery, R. Depth and topographic controls on microbial activity in a recently burned sub-alpine catchment. Soil Biol. Biochem. 2020, 148, 107844. [Google Scholar] [CrossRef]
- Siles, J.A.; Cajthaml, T.; Minerbi, S.; Margesin, R. Effect of altitude and season on microbial activity, abundance and community structure in alpine forest soils. FEMS Microbiol. Ecol. 2016, 92, fiw008. [Google Scholar] [CrossRef]
- Day, C.C.; von Strandmann, P.A.E.P.; Mason, A.J. Lithium isotopes and partition coefficients in inorganic carbonates: Proxy calibration for weathering reconstruction. Geochim. Et Cosmochim. Acta 2021, 305, 243–262. [Google Scholar] [CrossRef]
- von Strandmann, P.A.E.P.; Dellinger, M.; West, A.J. Lithium Isotopes: A Tracer of Past and Present Silicate Weathering; Cambridge University Press: Cambridge, UK, 2021. [Google Scholar]
- Vigier, N.; Decarreau, A.; Millot, R.; Carignan, J.; Petit, S.; France-Lanord, C. Quantifying li isotope fractionation during smectite formation and implications for the li cycle. Geochim. Et Cosmochim. Acta 2008, 72, 780–792. [Google Scholar] [CrossRef]
- Jiang, X.; Lin, X.; Yao, D.; Zhai, S.; Guo, W. Geochemistry of lithium in marine ferromanganese oxide deposits. Deep Sea Res. Part I Oceanogr. Res. Pap. 2007, 54, 85–98. [Google Scholar] [CrossRef]
- Yeager, C.M. Life on the Edge: Microbes in Rock Varnish. October 2019. Available online: https://doi.org/10.2172/1570605 (accessed on 15 January 2025).
- Fairén, A.G.; Losa-Adams, E.; Gil-Lozano, C.; Gago-Duport, L.; Uceda, E.R.; Squyres, S.W.; Rodríguez, J.A.P.; Davila, A.F.; McKay, C.P. Tracking the weathering of basalts on mars using lithium isotope fractionation models. Geochem. Geophys. Geosystems 2015, 16, 1172–1197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, X.; Tai, X.; Sun, L.; Wu, M.; Zhang, W.; Chen, X.; Zhang, G.; Chen, T.; Liu, G.; et al. Variation in actinobacterial community composition and potential function in different soil ecosystems belonging to the arid heihe river basin of northwest china. Front. Microbiol. 2019, 10, 2209. [Google Scholar] [CrossRef]
- Severino, R.; Froufe, H.J.C.; Barroso, C.; Albuquerque, L.; Lobo-da-Cunha, A.; da Costa, M.S.; Egas, C. High-quality draft genome sequence of gaiella occulta isolated from a 150 meter deep mineral water borehole and comparison with the genome sequences of other deep-branching lineages of the phylum actinobacteria. MicrobiologyOpen 2019, 8, e00840. [Google Scholar] [CrossRef]
- Yang, Q.; Peng, J.; Ni, S.; Zhang, C.; Wang, J.; Cai, C. Erosion and deposition significantly affect the microbial diversity, co-occurrence network, and multifunctionality in agricultural soils of northeast china. J. Soils Sediments 2024, 24, 888–900. [Google Scholar] [CrossRef]
- Mickan, B.S.; Abbott, L.K.; Solaiman, Z.M.; Mathes, F.; Siddique, K.H.M.; Jenkins, S.N. Soil disturbance and water stress interact to influence arbuscular mycorrhizal fungi, rhizosphere bacteria and potential for n and c cycling in an agricultural soil. Biol. Fertil. Soils 2019, 55, 53–66. [Google Scholar] [CrossRef]
- Jaeger, A.C.H.; Hartmann, M.; Conz, R.F.; Six, J.; Solly, E.F. Prolonged water limitation shifts the soil microbiome from copiotrophic to oligotrophic lifestyles in scots pine mesocosms. Environ. Microbiol. Rep. 2024, 16, e13211. [Google Scholar] [CrossRef]
- Djemiel, C.; Dequiedt, S.; Bailly, A.; Tripied, J.; Lelièvre, M.; Horrigue, W.; Jolivet, C.; Bispo, A.; Saby, N.; Valé, M.; et al. Biogeographical patterns of the soil fungal:Bacterial ratio across france. mSphere 2023, 8, e00365-23. [Google Scholar] [CrossRef]
- Laveuf, C.; Cornu, S. A review on the potentiality of Rare Earth Elements to trace pedogenetic processes. Geoderma 2009, 154, 1–9. [Google Scholar] [CrossRef]
- Cederlund, H.; Wessén, E.; Enwall, K.; Jones, C.M.; Juhanson, J.; Pell, M.; Philippot, L.; Hallin, S. Soil carbon quality and nitrogen fertilization structure bacterial communities with predictable responses of major bacterial phyla. Appl. Soil Ecol. 2014, 84, 62–68. [Google Scholar] [CrossRef]
- Davis, K.E.R.; Sangwan, P.; Janssen, P.H. Acidobacteria, rubrobacteridae and chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jing, Q.; Liu, X.; Zhou, X.; Fang, C.; Li, B.; Zhou, S.; Nie, M. Microbial respiratory thermal adaptation is regulated by r-/k-strategy dominance. Ecol. Lett. 2022, 25, 2489–2499. [Google Scholar] [CrossRef] [PubMed]




| Landform | Site Location | Zone | Soil Samples | Soil Classification WRB | Lithology | Aspect | Sand (2 mm) | Silt (50 μm) |
Clay (2 μm) | pH | SOM (%) | Mg [mg/kg] | Li [µg/kg] | Fe [µg/kg] | Mn [mg/kg] | Sb [µg/kg] | P [mg/kg] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low gradient slope | Tosteto—Pitigliano | Maremma | 31-32-33 | Eutric Andosols | Pyroclastic deposits (Vulc—PIT) | SE | 80.6 | 1.3 | 18.0 | 6.28 | 1.2 | 3.1 | <0.1 | 101.0 | 2.3 | <0.1 | 0.2 |
| Graded | Faggeto—Anghiari (Arezzo) | Alta Valtiberina | 1-2-3 | Calcari Epileptic Cambisols | Fluvial-lacustrine deposits (VILa1) | NE | 46.1 | 42.6 | 11.3 | 7.41 | 1.5 | 34.8 | 14.1 | 22.3 | 1.7 | 0.37 | 12.9 |
| Faggeto—Anghiari | Alta Valtiberina | 4-5-6 | Calcari Epileptic Cambisols | Fluvial-lacustrine deposits (VILa2) | NW | 46.1 | 42.6 | 11.3 | 7.41 | 1.5 | 48.1 | 12.8 | 14.3 | 2.6 | 0.41 | 23.3 | |
| Lamole—Greve in Chianti | Chianti | 46-47-48 | Eutric Cambisols | Macigno Formation (MAC) | S-SW | 76.8 | 15.1 | 8.1 | 7.46 | 0.5 | 65.6 | 16.3 | 533.2 | 13.3 | 0.14 | 5.1 | |
| Midslope ridges | Torriano—Montefiridolfi, San Casciano | Chianti | 13-14-15 | Endoskeleti Calcaric Cambisols | Fluvial deposits (VILa) | NW | 52.6 | 27.1 | 20.3 | 7.97 | 2.2 | 129.0 | 121.6 | 19.7 | 50.5 | 3.8 | 2.8 |
| Pruneti—Chiocchio, | Chianti | 10-11-12 | Calcari Endoleptic Cambisols | Palombini Shales (APA) | NE | 26.4 | 42.4 | 31.2 | 8.15 | 1.6 | 208.0 | 97.4 | 17.0 | 3.0 | 0.87 | 0.3 | |
| Pruneti—S. Polo in Chianti | Chianti | 7-8-9 | Calcari Endoleptic Cambisols | Marls (Marne) | SE | 64.0 | 18.5 | 17.5 | 7.59 | 2.1 | 94.0 | 97.8 | 22.9 | 14.5 | 0.95 | 2.1 | |
| Upper slopes | Rignana—Greve in Chianti | Chianti | 40-41-42 | Eutri Epileptic Regosol | Pietraforte Formation (PTF) | S-SW | 39.8 | 51.8 | 8.4 | 7.78 | 1.2 | 183.0 | 95.7 | 80.2 | 14.8 | 0.51 | 11.2 |
| Rignana—Greve in Chianti | Chianti | 43-44-45 | Calcaric Regosols | Varicolori Shales (AVR) | S-SW | 29.5 | 66.0 | 4.5 | 7.62 | 1.3 | 33.0 | 45.8 | 27.7 | 9.1 | 0.6 | 2.2 | |
| Castel Ruggero Monta Taurina | Chianti | 37-38-39 | Calcaric Regosols | Claystone (SIL) | S-SW | 33.1 | 42.6 | 24.3 | 7.95 | 1.1 | 153.5 | 67.5 | 42.9 | 7.7 | 0.30 | 0.2 | |
| Monteoriolo, Impruneta | Chianti | 28-28-30 | Calcaric Regosols | Claystone (SIL) | E | 61.3 | 22.5 | 16.2 | 7.05 | 2.6 | 51.6 | <0.1 | 385.5 | 4.3 | 1.5 | 7.5 | |
| Pruneti—I Tinti, Strada in Chianti | Chianti | 16-17-18 | Calcari Endoleptic Cambisols | Basalts (bas) | N-NW | 61.9 | 31.5 | 6.6 | 7.90 | 2.3 | 408.8 | 24.12 | 43.7 | 3.9 | 1.7 | 2.0 | |
| Open slopes | Pruneti—Lizzano | Chianti | 22-23-24 | Calcaric Regosols | Monte Morello Formation (MLL) | N-NW | 36.7 | 38.9 | 24.4 | 8.07 | 1.2 | 98.7 | 41.1 | 94.6 | 5.8 | 0.6 | 1.4 |
| La Querce—Impruneta | Chianti | 25-26-27 | Calcaric Regosols | Monte Morello Formation (MLL) | W | 22.7 | 68.4 | 8.9 | 8.12 | 1.1 | 132.8 | 9.3 | 91.5 | 4.9 | 0.7 | 1.6 | |
| Erta di Quintole—Impruneta | Chianti | 19-20-21 | Endoskeleti Calcaric Cambisols | Carbonatic flysch (Flysh) | S | 57.6 | 22.4 | 20.0 | 7.54 | 2.8 | 235.8 | 76.5 | 128.7 | 7.6 | 1.1 | 3.3 | |
| Castel Ruggero Poggio Fontaccia | Chianti | 34-35-36 | Calcaric Regosols | Varicolori Shales (AVR1) | S-SW | 34.5 | 51.3 | 14.2 | 8.09 | 1.1 | 119.2 | 79.1 | 104.7 | 3.1 | 0.36 | 1.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pelacani, S.; Ceccherini, M.T.; Barbadori, F.; Moretti, S.; Tommasini, S. Geodiversity as a Driver of Soil Microbial Community Diversity and Adaptation in a Mediterranean Landscape. Land 2025, 14, 583. https://doi.org/10.3390/land14030583
Pelacani S, Ceccherini MT, Barbadori F, Moretti S, Tommasini S. Geodiversity as a Driver of Soil Microbial Community Diversity and Adaptation in a Mediterranean Landscape. Land. 2025; 14(3):583. https://doi.org/10.3390/land14030583
Chicago/Turabian StylePelacani, Samuel, Maria Teresa Ceccherini, Francesco Barbadori, Sandro Moretti, and Simone Tommasini. 2025. "Geodiversity as a Driver of Soil Microbial Community Diversity and Adaptation in a Mediterranean Landscape" Land 14, no. 3: 583. https://doi.org/10.3390/land14030583
APA StylePelacani, S., Ceccherini, M. T., Barbadori, F., Moretti, S., & Tommasini, S. (2025). Geodiversity as a Driver of Soil Microbial Community Diversity and Adaptation in a Mediterranean Landscape. Land, 14(3), 583. https://doi.org/10.3390/land14030583









