Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts
Abstract
1. Introduction
2. Materials and Methods
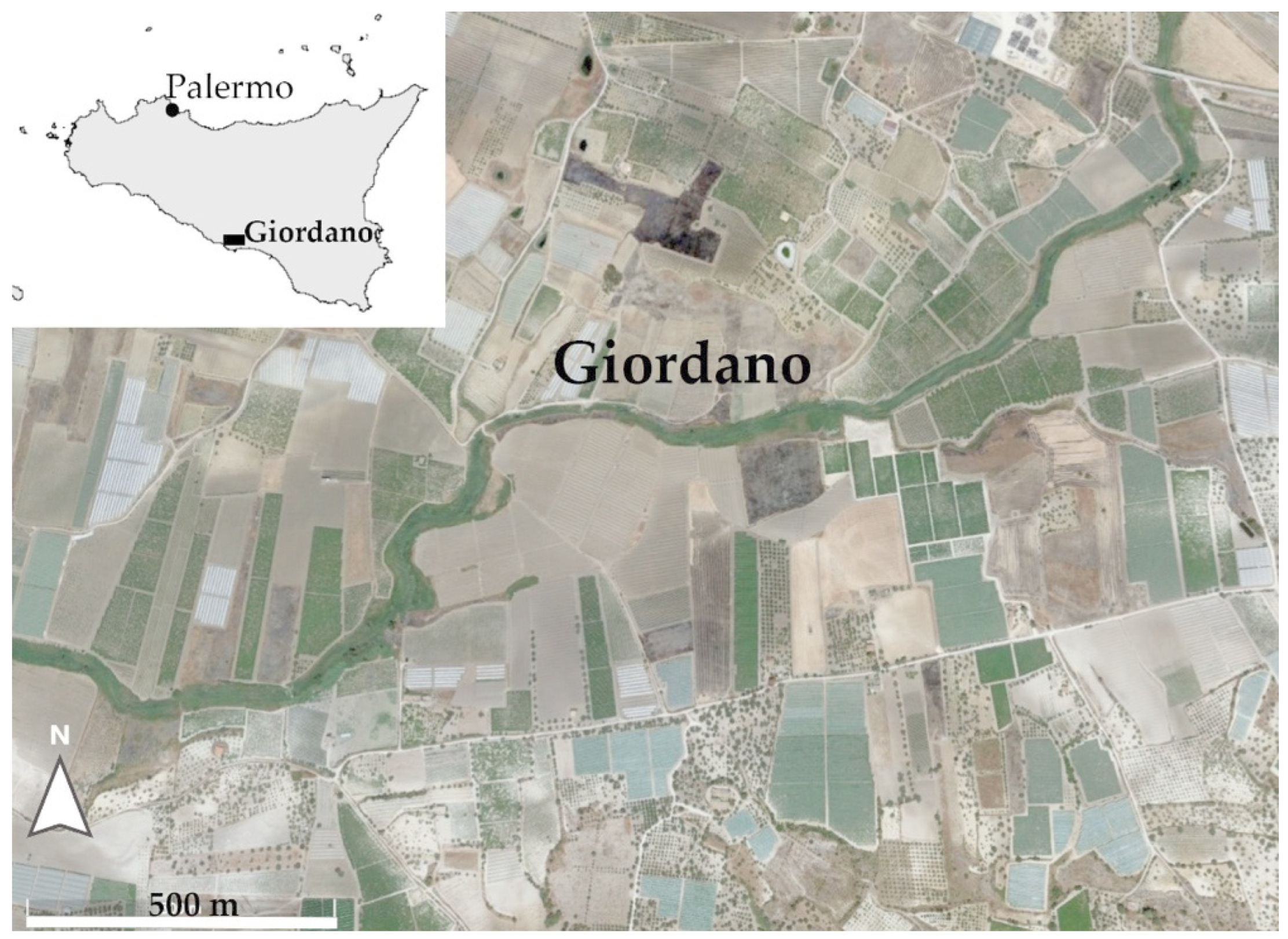
2.1. Study Area
2.2. Applied Pedotechnics
2.3. Sampling
2.4. Physicochemical Analyses
2.5. Culture-Dependent Microbiological Analysis
2.6. Culture-Independent Analysis
2.6.1. Amplicon Library Preparation
2.6.2. Illumina Data Analysis and Sequences Identification by QIIME2
2.7. Alpha Diversity Analysis
2.8. Statistical Analysis
3. Results
3.1. Physicochemical Characteristics of Soils
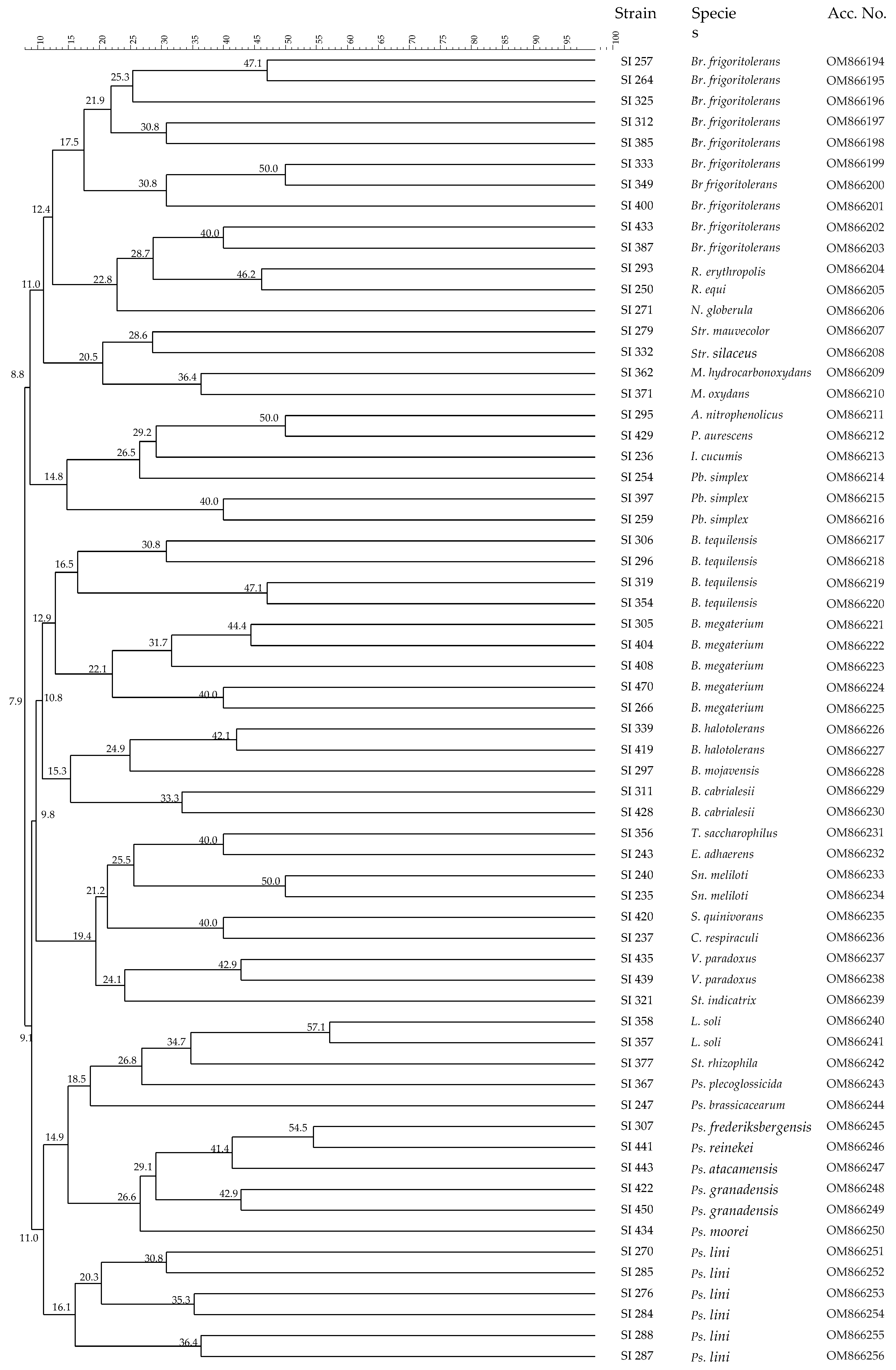
3.2. Analysis of Microbial Communities
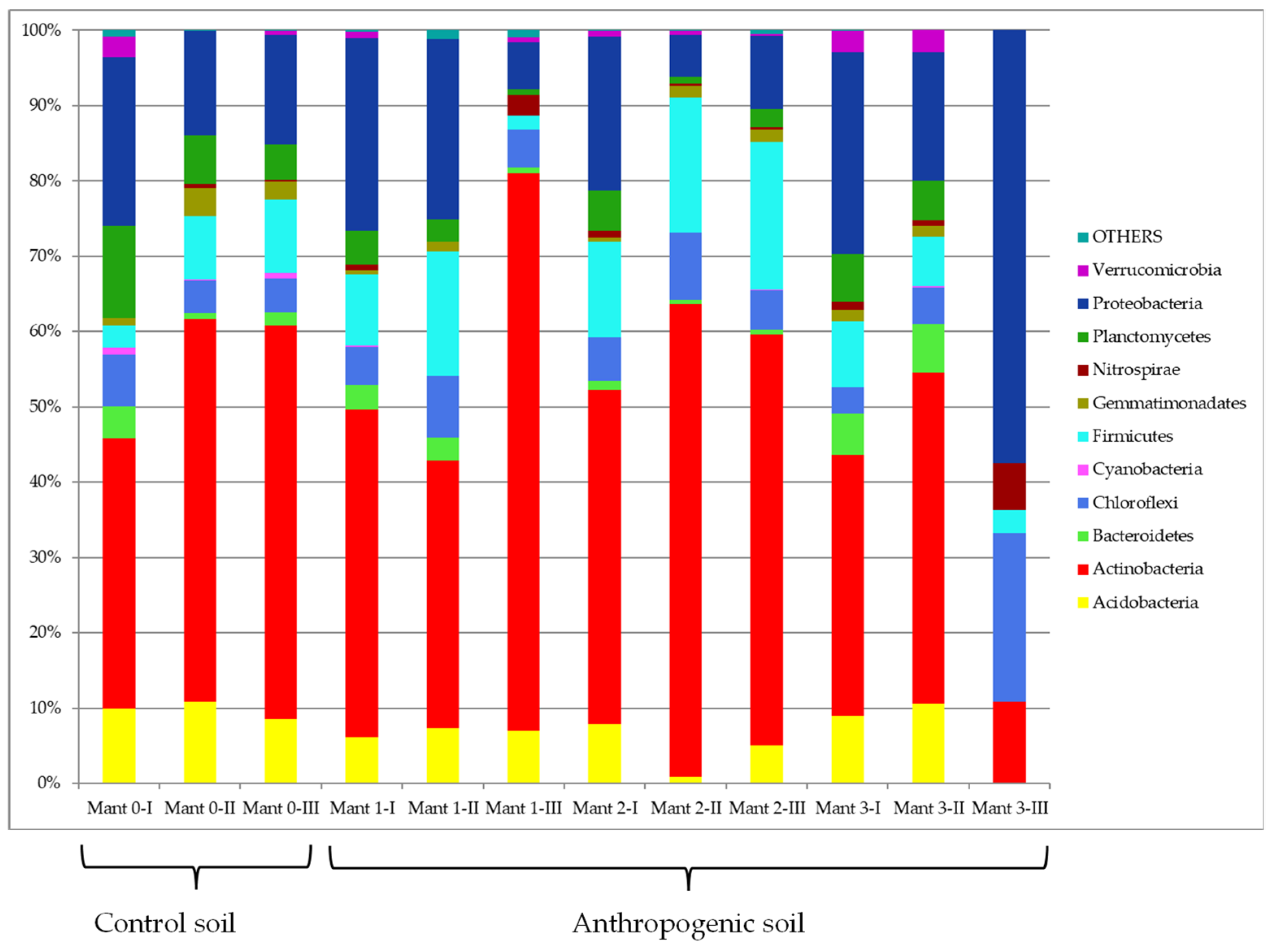
3.3. Culture-Independent Analysis
3.4. Analysis of Biodiversity Indexes (Alpha Diversity)
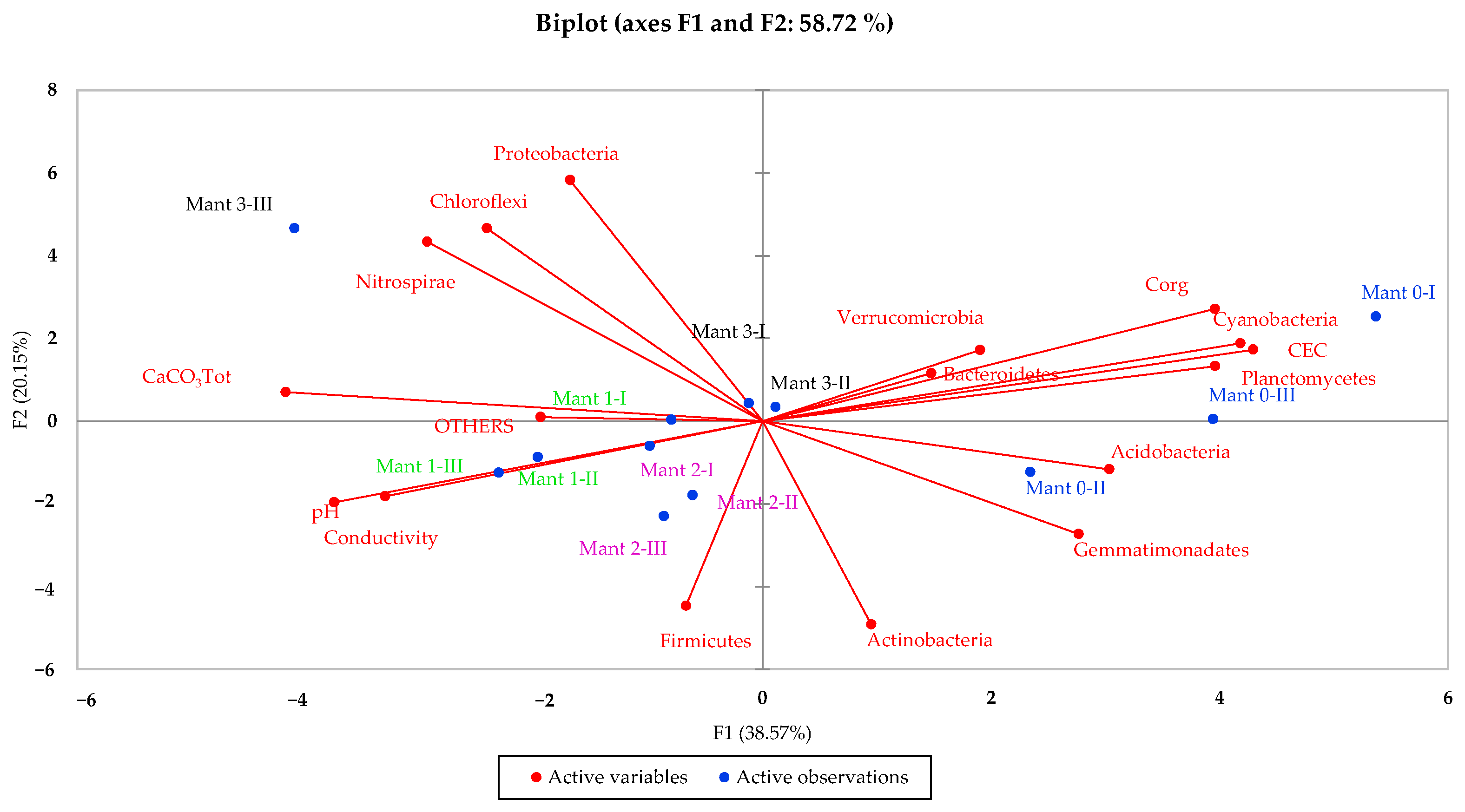
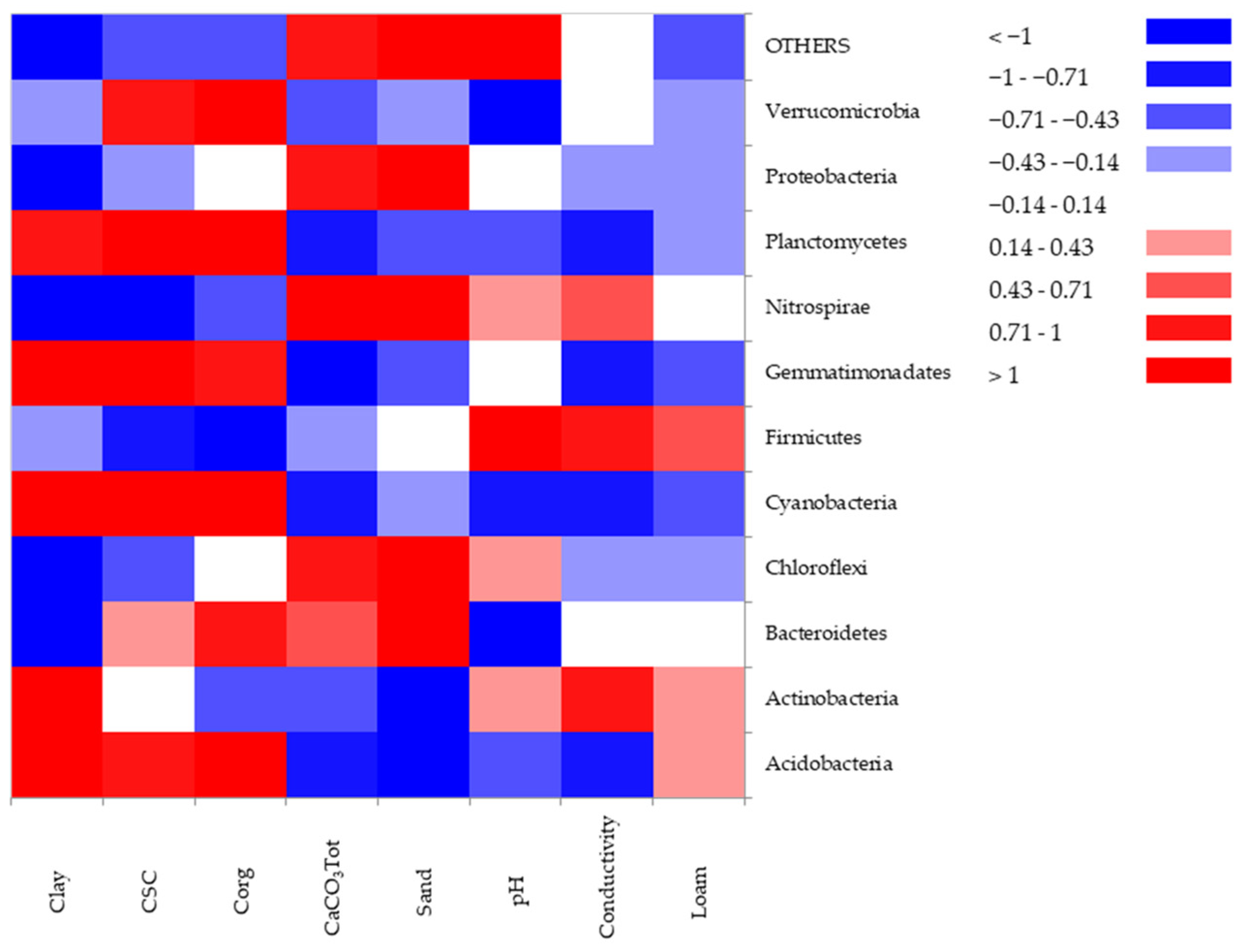
3.5. Multivariate Data Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Dudal, R.; Nachtergaele, F.O.; Purnell, M.F. Human factor of soil formation. In Proceedings of the 17th World Congress Soil Science, Bangkok, Thailand, 14–21 August 2002. [Google Scholar]
- Bockheim, J.G.; Gennadiyev, A.N. The role of soil-forming processes in the definition of taxa in Soil Taxonomy and the World Soil Reference Base. Geoderma 2000, 95, 53–72. [Google Scholar] [CrossRef]
- Bryant, R.B.; Galbraith, J.M.; Eswaran, H. Incorporating Anthropogenic Processes in Soil Classification; CRC Press: New York, NY, USA, 2003. [Google Scholar]
- Dazzi, C.; Lo Papa, G.; Palermo, V. Proposal for a new diagnostic horizon for WRB Anthrosols. Geoderma 2009, 151, 16–21. [Google Scholar] [CrossRef]
- Andrés-Abellán, M.; Del Álamo, J.B.; Landete-Castillejos, T.; López-Serrano, F.R.; García-Morote, F.A.; del Cerro-Barja, A. Impacts of visitors on soil and vegetation of the recreational area “Nacimiento del Rio Mundo” (Castilla-La Mancha, Spain). Environ. Mon. Assess. 2005, 101, 55–67. [Google Scholar]
- Dazzi, C.; Monteleone, S. Anthropogenic processes in the evolution of a soil chronosequence on marly-limestone substrata in an Italian Mediterranean environment. Geoderma 2007, 141, 201–209. [Google Scholar] [CrossRef]
- Hearing, K.C.; Daniels, W.L.; Galbraith, J.M. Mapping and classification of southwest Virginia mine soils. Soil Sci. Soc. Am. J. 2005, 69, 463–472. [Google Scholar] [CrossRef]
- Salvati, L.; Zitti, M.; Ceccarelli, T. Integrating economic and environmental indicators in the assessment of desertification risk: A case study. Appl. Ecol. Environ. Res. 2008, 6, 129–138. [Google Scholar] [CrossRef]
- Morgan, R.P.C. Soil Erosion and Conservation; John Wiley & Sons: Hoboken, New Zealand, 2009. [Google Scholar]
- Peña-Angulo, D.; Nadal-Romero, E.; González-Hidalgo, J.C.; Albaladejo, J.; Andreu, V.; Bagarello, V.; Barhi, H.; Batalla, R.J.; Bernal, S.; Bienes, R.; et al. Spatial variability of the relationships of runoff and sediment yield with weather types throughout the Mediterranean basin. J. Hydrol. 2019, 571, 390–405. [Google Scholar] [CrossRef]
- Pankhurst, C.E.; Ophel-Keller, K.; Doube, B.M.; Gupta, V.V.S.R. Biodiversity of soil microbial communities in agricultural systems. Biodivers. Conserv. 1996, 5, 197–209. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs)-Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wingender, J.; Jaeger, K.E.; Flemming, H.C. Interaction between extracellular polysaccharides and enzymes. In Microbial Extracellular Polymeric Substances; Wingender, J., Neu, T.R., Flemming, H.C., Eds.; Springer: Berlin, Germany, 1999; pp. 231–251. [Google Scholar]
- Costa, O.Y.; Raaijmakers, J.M.; Kuramae, E.E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front. Microbiol. 2018, 9, 1636. [Google Scholar] [CrossRef] [PubMed]
- Vert, M.; Doi, Y.; Hellwich, K.H.; Hess, M.; Hodge, P.; Kubisa, P.; Rinaudo, M.; Schué, F. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 2012, 84, 377–410. [Google Scholar] [CrossRef]
- Saini, R.; Saini, S.; Sharma, S. Biofilm: A dental microbial infection. J. Nat. Sci. Biol. Med. 2011, 2, 71. [Google Scholar] [CrossRef]
- Vlastarakos, P.V.; Nikolopoulos, T.P.; Maragoudakis, P.; Tzagaroulakis, A.; Ferekidis, E. Biofilms in ear, nose, and throat infections: How important are they? Laryngoscope 2007, 117, 668–673. [Google Scholar] [CrossRef]
- Vardharajula, S.; Ali, S.Z. The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology 2015, 84, 512–519. [Google Scholar]
- Wang, X.; Sharp, C.E.; Jones, G.M.; Grasby, S.E.; Brady, A.L.; Dunfield, P.F. Stable-isotope probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl. Environ. Microbiol. 2015, 81, 4607–4615. [Google Scholar] [CrossRef]
- Bronick, C.J.; Lal, R. Soil structure and management: A review. Geoderma 2005, 124, 3–22. [Google Scholar] [CrossRef]
- Dinel, H.; Levesque, P.E.M.; Jambu, P.; Righi, D. Microbial activity and long-chain aliphatics in the formation of stable soil aggregates. Soil. Sci. Soc. Am. J. 1992, 56, 1455–1463. [Google Scholar] [CrossRef]
- Chenu, C. Extracellular polysaccharides: An interface between microorganisms and soil constituents. In Environmental Impact of Soil Component Interactions, 1st ed.; Huang, P.M., Berthelin, J., Bollag, J.M., McGill, W.B., Eds.; CRC Press: New York, NY, USA, 1995; pp. 217–233. [Google Scholar]
- Taubner, H.; Roth, B.; Tippkötter, R. Determination of soil texture: Comparison of the sedimentation method and the laser-diffraction analysis. J. Plant Nutr. Soil Sci. 2009, 172, 161–171. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Thomson, B.C.; Ostle, N.J.; McNamara, N.P.; Whiteley, A.S.; Griffiths, R.I. Effects of sieving, drying and rewetting upon soil bacterial community structure and respiration rates. J. Microbiol Meth. 2010, 83, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Miceli, A.; Francesca, N.; Moschetti, G. Investigation of the hygienic safety of aromatic plants cultivated in soil contaminated with Listeria monocytogenes. Food Control. 2012, 26, 213–219. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, L.; Gilliam, F.S.; Wang, Q.; Li, C. Effects of raised-bed planting for enhanced summer maize yield on rhizosphere soil microbial functional groups and enzyme activity in Henan Province, China. Field Crops Res. 2012, 130, 28–37. [Google Scholar] [CrossRef]
- Sun, L.N.; Yang, E.D.; Wei, J.C.; Tang, X.Y.; Cao, Y.Y.; Han, G.M. Caulobacter flavus sp. nov., a stalked bacterium isolated from rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 4374–4380. [Google Scholar] [CrossRef]
- Huang, H.D.; Wang, W.; Ma, T.; Li, G.Q.; Liang, F.L.; Liu, R.L. Sphingomonas sanxanigenens sp. Nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 719–723. [Google Scholar] [CrossRef]
- Rippka, R.; Waterbury, J.B.; Stanier, R.Y. Isolation and purification of cyanobacteria: Some general principles. In The Prokaryotes; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin, Germany, 1981; pp. 212–220. [Google Scholar]
- Gaglio, R.; Francesca, N.; Di Gerlando, R.; Mahony, J.; De Martino, S.; Stucchi, C.; Moschetti, G.; Settanni, L. Enteric bacteria of food ice and their survival in alcoholic beverages and soft drinks. Food Microbiol. 2017, 67, 17–22. [Google Scholar] [CrossRef]
- Weisburg, W.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bact. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Wang, Q.; O′sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Spellerberg, I.F.; Fedor, P.J. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’Index. Global Ecol. Biogeog. 2003, 12, 177–179. [Google Scholar] [CrossRef]
- Chao, A.; Jost, L.; Chiang, S.C.; Jiang, Y.H.; Chazdon, R.L. A two-stage probabilistic approach to multiple-community similarity indices. Biometrics 2008, 64, 1178–1186. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis for Special Types of Data; Springer: New York, NY, USA, 2002. [Google Scholar]
- Mazzei, P.; Francesca, N.; Moschetti, G.; Piccolo, A. NMR spectroscopy evaluation of direct relationship between soils and molecular composition of red wines from Aglianico grapes. Anal. Chim. Acta 2010, 673, 167–172. [Google Scholar] [CrossRef]
- Todeschini, R. Introduzione Alla Chemiometria: Strategie, Metodi e Algoritmi Per L’analisi e il Modellamento di Dati Chimici, Farmacologici e Ambientali; Edises: Naples, Italy, 1998. [Google Scholar]
- Wen, C.; Wu, L.; Qin, Y.; Van Nostrand, J.D.; Ning, D.; Sun, B.; Xue, K.; Liu, F.; Deng, Y.; Liang, Y.; et al. Evaluation of the reproducibility of amplicon sequencing with Illumina MiSeq platform. PLoS ONE 2017, 12, e0176716. [Google Scholar] [CrossRef]
- Moran, E.F. Socio-economic aspects of acid soil management. In Plant-Soil Interactions at Low pH: Principles and Management; Date, R.A., Grundon, N.J., Rayment, G.E., Probert, M.E., Eds.; Springer: Berlin, Germany, 1995; pp. 663–669. [Google Scholar]
- Lo Verde, F.M. Agricoltura e Mutamento Sociale; Éditions L’Harmattan: Paris, France, 1995. [Google Scholar]
- Lo Papa, G.; Antisari, L.V.; Vianello, G.; Dazzi, C. Soil interpretation in the context of anthropedogenic transformations and pedotechniques application. Catena 2018, 166, 240–248. [Google Scholar] [CrossRef]
- Asio, V.B.; Cabunos, C.C., Jr.; Chen, Z.S. Morphology, physiochemical characteristics, and fertility of soils from Quaternary limestone in Leyte, Philippines. Soil Sci. 2006, 171, 648–661. [Google Scholar] [CrossRef]
- Loeppert, R.H.; Suarez, D.L. Carbonate and gypsum. In Methods of Soil Analysis Part 3. Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 1996; pp. 437–474. [Google Scholar]
- Ulrich, B. Natural and anthropogenic components of soil acidification. J. Plant Nutr. Soil Sci. 1986, 149, 702–717. [Google Scholar] [CrossRef]
- Ghorbani, H.; Kashi, H.; Hafezi Moghadas, N.; Emamgholizadeh, S. Estimation of soil cation exchange capacity using multiple regression, artificial neural networks, and adaptive neuro-fuzzy inference system models in Golestan Province, Iran. Commun. Soil Sci. Plant Anal. 2015, 46, 763–780. [Google Scholar] [CrossRef]
- Khaledian, Y.; Brevik, E.C.; Pereira, P.; Cerdà, A.; Fattah, M.A.; Tazikeh, H. Modeling soil cation exchange capacity in multiple countries. Catena 2017, 158, 194–200. [Google Scholar] [CrossRef]
- McBratney, A.B.; Minasny, B.; Cattle, S.R.; Vervoort, R.W. From pedotransfer functions to soil inference systems. Geoderma 2002, 109, 41–73. [Google Scholar] [CrossRef]
- Ulusoy, Y.; Tekin, Y.; Tümsavas, Z.; Mouazen, A.M. Prediction of soil cation exchange capacity using visible and near infrared spectroscopy. Biosyst. Eng. 2016, 152, 79–93. [Google Scholar] [CrossRef]
- Dazzi, C.; Galati, A.; Crescimanno, M.; Lo Papa, G. Pedotechnique applications in large-scale farming: Economic value, soil ecosystems services and soil security. Catena 2019, 181, 104072. [Google Scholar] [CrossRef]
- Rech, M.; Pansera, M.R.; Sartori, V.C.; Ribeiro, R.T.D.S. Microbiota do solo em vinhedos agroecológico e convencional e sob vegetação nativa em Caxias do Sul. RS. Rev. Bras. Agroecol. 2013, 8, 141–151. [Google Scholar]
- Silva, J.G.D.; da Luz, J.M.R.; Henrique, J.; de Carvalho, J.J.; da Silva, J.E.C. Domestic wastewater for forage cultivation in Cerrado soil. J. Agric. Sci. 2018, 10, 248. [Google Scholar] [CrossRef]
- Carvalho, J.J.; da Luz, J.M.R.; Henrique, J.; Silva, J.G.D.; Silva, J.E.C.; Santos, E.A. Biofertigation of forage with effluents of green line of a cattle slaughterhouse: Microbial diversity and leaf dry mass productivity. J. Agric. Sci. 2018, 10, 353. [Google Scholar] [CrossRef][Green Version]
- Neeraj, K.S. Organic amendments to soil inoculated arbuscular mycorrhizal fungi and Pseudomonas fluorescens treatments reduce the development of root-rot disease and enhance the yield of Phaseolus vulgaris L. Eur. J. Soil Biol. 2011, 47, 288–295. [Google Scholar] [CrossRef]
- Smith, S.E.; Facelli, E.; Pope, S.; Smith, F.A. Plant performance in stressful environments: Interpreting new and established knowledge of the roles of arbuscular mycorrhizas. Plant Soil. 2010, 326, 3–20. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Lennon, J.T.; Aanderud, Z.T.; Lehmkuhl, B.K.; Schoolmaster, D.R., Jr. Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 2012, 93, 1867–1879. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.R.; Manna, S.; Saha, P.; Basak, R.K.; Sen, R.; Roy, D.; Adhikari, B. Composition analysis and material characterization of an emulsifying extracellular polysaccharide (EPS) produced by Bacillus megaterium RB-05: A hydrodynamic sediment-attached isolate of freshwater origin. J. Appl. Microb. 2011, 111, 1381–1393. [Google Scholar] [CrossRef]
- Caesar-TonThat, T.C.; Caesar, A.J.; Gaskin, J.F.; Sainju, U.M.; Busscher, W.J. Taxonomic diversity of predominant culturable bacteria associated with microaggregates from two different agroecosystems and their ability to aggregate soil in vitro. Appl. Soil Ecol. 2007, 36, 10–21. [Google Scholar] [CrossRef]
- Fiodor, A.; Singh, S.; Pranaw, K. The contrivance of plant growth promoting microbes to mitigate climate change impact in agriculture. Microorganisms 2021, 9, 1841. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a sensitive indicator of agricultural soil usage revealed by culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue–Bacterial families and genera: A metagenomic approach. Ecol. Ind. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Legrand, F.; Picot, A.; Cobo-Díaz, J.F.; Carof, M.; Chen, W.; Le Floch, G. Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 2018, 132, 135–145. [Google Scholar] [CrossRef]
- Chang, Y.J.; Land, M.; Hauser, L.; Chertkov, O.; Glavina Del Rio, T.; Nolan, M.; Copeland, A.; Tice, H.; Cheng, J.-F.; Lucas, S.; et al. Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21T). Stand. Genomic Sci. 2011, 5, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Koyama, A.; Wallenstein, M.D.; Simpson, R.T.; Moore, J.C. Soil bacterial community composition altered by increased nutrient availability in Arctic tundra soils. Front. Microbiol. 2014, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Aislabie, J.; Deslippe, J.R. Soil microbes and their contribution in soil services. In Ecosystem Services in New Zealand-Conditions and Trends; Dyamond, J.R., Ed.; Manaaki Whenua Press: Lincoln, New Zeland, 2013; pp. 143–161. [Google Scholar]
- Koch, A.L. Oligotrophs versus copiotrophs. Bioessays 2001, 23, 657–661. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, H.F.; Cury, J.C.; do Carmo, F.L.; dos Santos, A.L.; Tiedje, J.; van Elsas, J.D.; Rosado, A.S.; Peixoto, R.S. Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: Bacterial proxies for oil pollution. PLoS ONE 2011, 6, e16943. [Google Scholar] [CrossRef] [PubMed]
- Militon, C.; Boucher, D.; Vachelard, C.; Perchet, G.; Barra, V.; Troquet, J.; Peyretaillade, E.; Peyret, P. Bacterial community changes during bioremediation of aliphatic hydrocarbon-contaminated soil. FEMS Microbiol. Ecol. 2010, 74, 669–681. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef]
- Roesch, L.F.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.M.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.O.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef]
- Anderson, C.R.; Condron, L.M.; Clough, T.J.; Fiers, M.; Stewart, A.; Hill, R.A.; Shrelock, R.R. Biochar induced soil microbial community change: Implications for biogeochemical cycling of carbon, nitrogen and phosphorus. Pedobiologia 2011, 54, 309–320. [Google Scholar] [CrossRef]
- Reis, V.M.; Teixeira, K.R. Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. J. Basic Microbiol. 2015, 55, 931–949. [Google Scholar] [CrossRef]
- Ofek, M.; Hadar, Y.; Minz, D. Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 2012, 7, e40117. [Google Scholar] [CrossRef]
- Hu, S.J.; Van Bruggen, A.H.C.; Grünwald, N.J. Dynamics of bacterial populations in relation to carbon availability in a residue-amended soil. Appl. Soil Ecol. 1999, 13, 21–30. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.P.; Atekwana, E.A.; Duris, J.W.; Werkema, D.D.; Rossbach, S. The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl. Environ. Microbiol. 2007, 73, 2860–2870. [Google Scholar] [CrossRef]
- Saul, D.J.; Aislabie, J.M.; Brown, C.E.; Harris, L.; Foght, J.M. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 2005, 53, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, E.; Déziel, E.; Villemur, R.; Juteau, P.; Lépine, F.; Beaudet, R. Initial characterization of new bacteria degrading high-molecular weight polycyclic aromatic hydrocarbons isolated from a 2-year enrichment in a two-liquid-phase culture system. J. Appl. Microbiol. 2003, 94, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Mehrshad, M.; Salcher, M.M.; Okazaki, Y.; Nakano, S.I.; Šimek, K.; Andrei, A.S.; Ghai, R. Hidden in plain sight—highly abundant and diverse planktonic freshwater Chloroflexi. Microbiome 2018, 6, 176. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Whitman, W.B. Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley Online Library: New York, NY, USA, 2015. [Google Scholar]
- Guida, G.; Palmeri, V.; Settanni, L.; Gaglio, R.; Tolone, M.; Ferro, V. Ability of soil bacterial composition as an indicator of levels of soil erosion in a badland. Int. J. Sediment Res. 2022, 37, 493–504. [Google Scholar] [CrossRef]
- Golobočanin, D.D.; Škrbić, B.D.; Miljević, N.R. Principal component analysis for soil contamination with PAHs. Chemometr. Intell. Lab. 2004, 72, 219–223. [Google Scholar] [CrossRef]
- Wu, T.; Chellemi, D.O.; Graham, J.H.; Martin, K.J.; Rosskopf, E.N. Comparison of soil bacterial communities under diverse agricultural land management and crop production practices. Microb. Ecol. 2008, 55, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Tripathi, B.; Kim, M.; Singh, D.; Lee-Cruz, L.; Lai-Hoe, A.; Ainuddin, A.N.; Go, R.; Abdul Rahim, R.; Husni, M.H.A.; Chun, J.; et al. Tropical soil bacterial communities in Malaysia: pH dominates in the equatorial tropics too. Microb. Ecol. 2012, 64, 474–484. [Google Scholar] [CrossRef]
- Dhakal, D.; Pokhrel, A.R.; Shrestha, B.; Sohng, J.K. Marine rare actinobacteria: Isolation, characterization, and strategies for harnessing bioactive compounds. Front. Microbiol. 2017, 8, 1106. [Google Scholar] [CrossRef]
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare taxa and dark microbial matter: Novel bioactive actinobacteria abound in Atacama Desert soils. Antonie Van Leeuw. 2018, 111, 1315–1332. [Google Scholar] [CrossRef]
- Qin, S.; Xing, K.; Jiang, J.H.; Xu, L.H.; Li, W.J. Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 2011, 89, 457–473. [Google Scholar] [CrossRef]


| Profile | Depth | CaCO3 Tot (g/kg) | Clay (g/kg) | Silt (g/kg) | Sand (g/kg) | pH (H2O) | EC (µS/cm) | CEC (Cmol(+)/kg) | Corg (g/kg) |
|---|---|---|---|---|---|---|---|---|---|
| Mant 0 | I | 200 | 446 | 269 | 285 | 7.83 | 82.0 | 33.75 | 15.11 |
| Mant 0 | II | 190 | 452 | 304 | 244 | 7.89 | 93.2 | 25.00 | 8.72 |
| Mant 0 | II | 200 | 498 | 259 | 243 | 7.76 | 95.5 | 31.25 | 8.52 |
| Mant 1 | I | 550 | 371 | 308 | 321 | 7.87 | 147.6 | 22.50 | 3.10 |
| Mant 1 | II | 640 | 295 | 328 | 377 | 7.90 | 145.5 | 20.00 | 2.13 |
| Mant 1 | III | 640 | 369 | 342 | 289 | 7.89 | 192.5 | 18.75 | 2.90 |
| Mant 2 | I | 550 | 363 | 418 | 219 | 7.87 | 172.5 | 21.30 | 5.03 |
| Mant 2 | II | 360 | 402 | 317 | 281 | 7.88 | 150.0 | 25.00 | 6.97 |
| Mant 2 | III | 420 | 390 | 307 | 303 | 7.90 | 160.5 | 21.30 | 3.87 |
| Mant 3 | I | 460 | 363 | 299 | 338 | 7.86 | 178.9 | 22.50 | 5.62 |
| Mant 3 | II | 640 | 333 | 373 | 294 | 7.88 | 154.9 | 20.00 | 5.23 |
| Mant 3 | III | 640 | 314 | 313 | 373 | 7.88 | 141.1 | 21.30 | 6.00 |
| Soil | TMA | FF | AB | TMAn | NF | Sphingomonas | Caulobacter |
|---|---|---|---|---|---|---|---|
| Control soil | |||||||
| Mant 0-I | 6.78 ± 0.10 b | 5.74 ± 0.04 a | 4.65 ± 0.05 a | 6.61 ± 0.11 a | 6.90 ± 0.18 ab | 6.42 ± 0.06 c | 6.87 ± 0.07 ab |
| Mant 0-II | 6.34 ± 0.08 b | 5.30 ± 0.00 a | 4.39 ± 0.09 a | 6.19 ± 0.05 a | 6.30 ± 0.08 a | 6.20 ± 0.10 a | 6.52 ± 0.20 ab |
| Mant 0-III | 6.14 ± 0.02 c | <2 b | 4.00 ± 0.00 b | 6.12 ± 0.03 a | 5.78 ± 0.17 b | 6.00 ± 0.08 b | 6.31 ± 0.01 b |
| Statistical significance | *** | *** | *** | *** | *** | ** | ** |
| Anthropogenic soil | |||||||
| Mant 1-I | 7.00 ± 0.07 b | 4.74 ± 0.04 b | 4.60 ± 0.20 a | 6.45 ± 0.09 a | 6.59 ± 0.30 b | 6.23 ± 0.03 d | 6.64 ± 0.28 b |
| Mant 1-II | 6.22 ± 0.01 bc | 4.30 ± 0.30 b | 4.30 ± 0.20 a | 5.71 ± 0.13 b | 6.03 ± 0.30 a | 5.97 ± 0.15 b | 6.20 ± 0.09 ab |
| Mant 1-III | 6.15 ± 0.05 c | <2 b | 4.45 ± 0.15 a | 5.75 ± 0.04 b | 5.97 ± 0.11 b | 5.76 ± 0.14 c | 6.29 ± 0.05 b |
| Statistical significance | *** | *** | *** | * | ** | * | |
| Anthropogenic soil | |||||||
| Mant 2-I | 7.09 ± 0.09 ab | 4.69 ± 0.09 b | 5.00 ± 0.25 a | 6.52 ± 0.06 a | 7.08 ± 0.04 a | 6.63 ± 0.1 b | 6.78 ± 0.00 ab |
| Mant 2-II | 6.06 ± 0.01 c | 4.24 ± 0.24 b | <2 b | 5.43 ± 0.05 c | 5.44 ± 0.05 b | 5.64 ± 0.00 c | 6.5 ± 0.30 b |
| Mant 2-III | 6.37 ± 0.06 b | 2.00 ± 0.30 a | <2 c | 5.77 ± 0.01 b | 6.00 ± 0.02 b | 6.10 ± 0.01 b | 6.31 ± 0.08 b |
| Statistical significance | *** | *** | *** | *** | *** | *** | * |
| Anthropogenic soil | |||||||
| Mant 3-I | 7.56 ± 0.35 a | 5.70 ± 0.00 a | 4.59 ± 0.11 a | 6.66 ± 0.03 a | 7.29 ± 0.04 a | 6.81 ± 0.07 a | 7.06 ± 0.01 a |
| Mant 3-II | 6.77 ± 0.17 a | 5.39 ± 0.09 a | 4.30 ± 0.25 a | 6.06 ± 0.09 a | 6.39 ± 0.16 a | 6.33 ± 0.03 a | 6.59 ± 0.07 a |
| Mant 3-III | 6.74 ± 0.00 a | <2 b | 4.00 ± 0.00 b | 6.14 ± 0.02 a | 6.43 ± 0.17 a | 6.52 ± 0.01 a | 6.82 ± 0.13 a |
| Statistical significance | ** | *** | * | *** | *** | *** | ** |
| Statistical significance I | ** | *** | ** | *** | * | ||
| Statistical significance II | *** | *** | *** | *** | *** | *** | * |
| Statistical significance III | *** | *** | *** | *** | ** | *** | *** |
| Samples | No. of Phyla | HSh | ESh | HSi | ESi |
|---|---|---|---|---|---|
| Mant 0-I | 11 | 1.84 | 0.74 | 0.79 | 0.86 |
| Mant 0-II | 11 | 1.58 | 0.64 | 0.70 | 0.76 |
| Mant 0-III | 12 | 1.58 | 0.64 | 0.68 | 0.75 |
| Mant 1-I | 12 | 1.64 | 0.66 | 0.73 | 0.79 |
| Mant 1-II | 9 | 1.73 | 0.69 | 0.78 | 0.85 |
| Mant 1-III | 10 | 1.06 | 0.42 | 0.44 | 0.48 |
| Mant 2-I | 12 | 1.64 | 0.66 | 0.73 | 0.80 |
| Mant 2-II | 11 | 1.20 | 0.48 | 0.56 | 0.61 |
| Mant 2-III | 12 | 1.44 | 0.58 | 0.65 | 0.71 |
| Mant 3-I | 11 | 1.82 | 0.73 | 0.78 | 0.85 |
| Mant 3-II | 11 | 1.77 | 0.71 | 0.75 | 0.82 |
| Mant 3-III | 5 | 1.17 | 0.47 | 0.60 | 0.66 |
| Reference value for a perfect even community | 2.48 | 1 | 0.92 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbaccia, P.; Dazzi, C.; Franciosi, E.; Di Gerlando, R.; Settanni, L.; Lo Papa, G. Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts. Land 2022, 11, 748. https://doi.org/10.3390/land11050748
Barbaccia P, Dazzi C, Franciosi E, Di Gerlando R, Settanni L, Lo Papa G. Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts. Land. 2022; 11(5):748. https://doi.org/10.3390/land11050748
Chicago/Turabian StyleBarbaccia, Pietro, Carmelo Dazzi, Elena Franciosi, Rosalia Di Gerlando, Luca Settanni, and Giuseppe Lo Papa. 2022. "Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts" Land 11, no. 5: 748. https://doi.org/10.3390/land11050748
APA StyleBarbaccia, P., Dazzi, C., Franciosi, E., Di Gerlando, R., Settanni, L., & Lo Papa, G. (2022). Microbiological Analysis and Metagenomic Profiling of the Bacterial Community of an Anthropogenic Soil Modified from Typic Haploxererts. Land, 11(5), 748. https://doi.org/10.3390/land11050748







