Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain
Abstract
1. Introduction
2. Materials and Methods
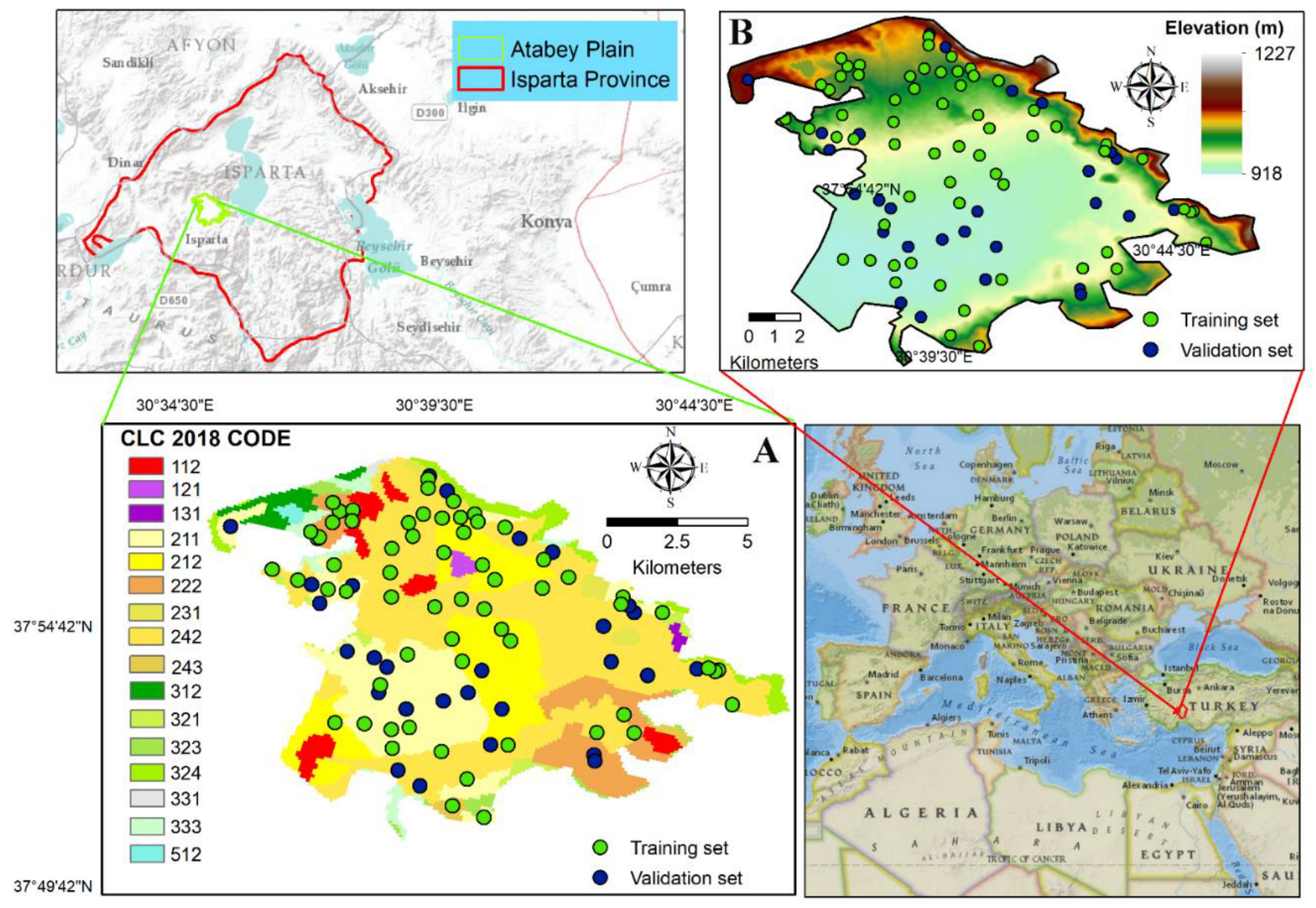
2.1. Study Area
2.2. Soil Sampling and Analysis
2.3. Environmental Covariates
2.3.1. Topographic Derivatives
2.3.2. Remote Sensing-Based Indices
| Soil-Forming Factors | Environmental Covariates | Definition | |
|---|---|---|---|
| Topography (R) | Elevation (m) | Often indicative of climatic processes. | |
| Slope (%) | The variable is the angle formed between any part of the earth and the horizontal. | ||
| Aspect (0° to 360°) | The variable was defined as the slope direction. | ||
| PRC | Profile curvature affects the deceleration and acceleration of the flow. | ||
| PLC | Planform curvature affects the decomposition and convergence of the flow. | ||
| TWI | (1) | ||
| As, specific catchment area; tanβ, specific catchment area slope; in radians. | |||
| Parent material (P) | TGSI [59] | (2) | |
| BI [60] | (3) | ||
| Parent material (P) | NDCI [61] | (4) | |
| Organism (O) | NDVI [61] | (5) | |
| Anthropogenic factor | CORINE LCC | Time series land cover class data. | |
2.3.3. Land Cover Class Data
2.4. Modelling Process
2.5. Spatial Prediction of Soil EC Using Machine Learning Algorithms
2.5.1. Random Forest Algorithm
2.5.2. Support Vector Regression
2.6. Spatial Prediction of EC Using Hybrid Methods (Regression-Kriging)
2.7. Model Accuracy of Regression-Based Algorithms
2.8. Salinization Risk Map
3. Results and Discussion
3.1. Statistical Description of the Sampled Data
3.2. Spatial Prediction of Model Residuals
3.3. Performance of Regression-Based Algorithms and Hybrid Methods
3.4. Spatial Patterns of Soil EC Dynamics
3.4.1. Random Forest
3.4.2. Support Vector Regression
3.5. Importance of Environmental Covariates for Different Machine Learning Algorithms
3.6. Salinity Risk Assessment
3.7. Challenges of the Case-Based Studies and Outlook Perspectives for Salinity Status Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ivushkin, K.; Bartholomeus, H.; Bregt, A.K.; Pulatov, A.; Kempen, B.; de Sousa, L. Global Mapping of Soil Salinity Change. Remote Sens. Environ. 2019, 231, 111260. [Google Scholar] [CrossRef]
- Bouma, J.; Bonfante, A.; Basile, A.; van Tol, J.; Hack-ten Broeke, M.J.D.; Mulder, M.; Heinen, M.; Rossiter, D.G.; Poggio, L.; Hirmas, D.R. How Can Pedology and Soil Classification Contribute towards Sustainable Development as a Data Source and Information Carrier? Geoderma 2022, 424, 115988. [Google Scholar] [CrossRef]
- Omuto, C.T.; Vargas, R.R.; el Mobarak, A.M.; Mohamed, N.; Viatkin, K.; Yigini, Y. Mapping of Salt-Affected Soils: Technical Manual, 1st ed.; FAO: Rome, Italy, 2020. [Google Scholar]
- Omuto, C.T.; Vargas, R.R.; Elmobarak, A.A.; Mapeshoane, B.E.; Koetlisi, K.A.; Ahmadzai, H.; Abdalla Mohamed, N. Digital Soil Assessment in Support of a Soil Information System for Monitoring Salinization and Sodification in Agricultural Areas. Land Degrad. Dev. 2022, 33, 1204–1218. [Google Scholar] [CrossRef]
- Hopmans, J.W.; Qureshi, A.S.; Kisekka, I.; Munns, R.; Grattan, S.R.; Rengasamy, P.; Ben-Gal, A.; Assouline, S.; Javaux, M.; Minhas, P.S.; et al. Critical Knowledge Gaps and Research Priorities in Global Soil Salinity. Adv. Agron. 2021, 169, 1–191. [Google Scholar] [CrossRef]
- Ghiglieri, G.; Carletti, A.; Pittalis, D. Analysis of Salinization Processes in the Coastal Carbonate Aquifer of Porto Torres (NW Sardinia, Italy). J. Hydrol. 2012, 432–433, 43–51. [Google Scholar] [CrossRef]
- Konyushkova, M.; Alavipanah, S.; Heidari, A.; Kozlov, D.; Yu, M.; Semenkov, I. Spatial and Seasonal Salt Translocation in the Young Soils at the Coastal Plains of the Caspian Sea. Quat. Int. 2021, 590, 15–25. [Google Scholar] [CrossRef]
- FAO-ITPS Salt-Affected Soils Are a Global Issue. Available online: https://www.fao.org/3/cb4809en/cb4809en.pdf (accessed on 7 September 2022).
- Manuel, R.; Machado, A.; Serralheiro, R.P.; Alvino, A.; Freire, M.I.; Ferreira, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Boudibi, S.; Sakaa, B.; Benguega, Z.; Fadlaoui, H.; Othman, T.; Bouzidi, N. Spatial Prediction and Modeling of Soil Salinity Using Simple Cokriging, Artificial Neural Networks, and Support Vector Machines in El Outaya Plain, Biskra, Southeastern Algeria. Acta Geochim. 2021, 40, 390–408. [Google Scholar] [CrossRef]
- Iglesias, A.; Quiroga, S.; Moneo, M.; Garrote, L. From Climate Change Impacts to the Development of Adaptation Strategies: Challenges for Agriculture in Europe. Clim. Chang. 2012, 112, 143–168. [Google Scholar] [CrossRef]
- Haddeland, I.; Heinke, J.; Biemans, H.; Eisner, S.; Flörke, M.; Hanasaki, N.; Konzmann, M.; Ludwig, F.; Masaki, Y.; Schewe, J.; et al. Global Water Resources Affected by Human Interventions and Climate Change. Proc. Natl. Acad. Sci. USA 2014, 111, 3251–3256. [Google Scholar] [CrossRef]
- Cramer, W.; Guiot, J.; Fader, M.; Garrabou, J.; Gattuso, J.P.; Iglesias, A.; Lange, M.A.; Lionello, P.; Llasat, M.C.; Paz, S.; et al. Climate Change and Interconnected Risks to Sustainable Development in the Mediterranean. Nat. Clim. Chang. 2018, 8, 972–980. [Google Scholar] [CrossRef]
- Hassani, A.; Azapagic, A.; Shokri, N. Predicting Long-Term Dynamics of Soil Salinity and Sodicity on a Global Scale. Proc. Natl. Acad. Sci. USA 2020, 117, 33017–33027. [Google Scholar] [CrossRef] [PubMed]
- Okur, B.; Örçen, N. Soil Salinization and Climate Change. In Climate Change and Soil Interactions; Elsevier: New York, NY, USA, 2020; pp. 331–350. [Google Scholar] [CrossRef]
- Kendirli, B.; Cakmak, B.; Ucar, Y. Salinity in the Southeastern Anatolia Project (GAP), Turkey: Issues and Options. Irrig. Drain. 2005, 54, 115–122. [Google Scholar] [CrossRef]
- Çullu, M.A.; Aydemir, S.; Qadir, M.; Almaca, A.; Öztürkmen, A.R.; Bilgiç, A.; Aǧca, N. Implication of Groundwater Fluctuation on the Seasonal Salt Dynamic in the Harran Plain, South-Eastern Turkey. Irrig. Drain. 2010, 59, 465–476. [Google Scholar] [CrossRef]
- Bilgili, A.V. Spatial Assessment of Soil Salinity in the Harran Plain Using Multiple Kriging Techniques. Environ. Monit Assess 2013, 185, 777–795. [Google Scholar] [CrossRef] [PubMed]
- FAO GSASmap v1.0, Global Map of Salt-Affected Soils. Available online: https://www.fao.org/3/cb7247en/cb7247en.pdf (accessed on 7 September 2022).
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On Digital Soil Mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Mponela, P.; Snapp, S.; Villamor, G.B.; Tamene, L.; Le, Q.B.; Borgemeister, C. Digital Soil Mapping of Nitrogen, Phosphorus, Potassium, Organic Carbon and Their Crop Response Thresholds in Smallholder Managed Escarpments of Malawi. Appl. Geogr. 2020, 124, 102299. [Google Scholar] [CrossRef]
- Bakacsi, Z.; Tóth, T.; Makó, A.; Barna, G.; Laborczi, A.; Szabó, J.; Szatmári, G.; Pásztor, L. National Level Assessment of Soil Salinization and Structural Degradation Risks under Irrigation. Hung. Geogr. Bull. 2019, 68, 141–156. [Google Scholar] [CrossRef]
- Aksoy, S.; Yildirim, A.; Gorji, T.; Hamzehpour, N.; Tanik, A.; Sertel, E. Assessing the Performance of Machine Learning Algorithms for Soil Salinity Mapping in Google Earth Engine Platform Using Sentinel-2A and Landsat-8 OLI Data. Adv. Space Res. 2022, 69, 1072–1086. [Google Scholar] [CrossRef]
- Bannari, A.; Abuelgasim, A. Potential and Limits of Vegetation Indices Compared to Evaporite Mineral Indices for Soil Salinity Discrimination and Mapping. SOIL Discuss. 2021, 1–48. [Google Scholar] [CrossRef]
- Karra, K.; Kontgis, C.; Statman-Weil, Z.; Mazzariello, J.C.; Mathis, M.; Brumby, S.P. Global Land Use/Land Cover with Sentinel 2 and Deep Learning. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 4704–4707. [Google Scholar] [CrossRef]
- Zhang, T.T.; Zeng, S.L.; Gao, Y.; Ouyang, Z.T.; Li, B.; Fang, C.M.; Zhao, B. Assessing Impact of Land Uses on Land Salinization in the Yellow River Delta, China Using an Integrated and Spatial Statistical Model. Land Use Policy 2011, 28, 857–866. [Google Scholar] [CrossRef]
- Poggio, L.; de Sousa, L.; Genova, G.; D’Angelo, P.; Schwind, P.; Heiden, U. Soil organic carbon modelling with digital soil mapping and remote sensing for permanently vegetated areas. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium, 11–16 July 2021; pp. 492–494. [Google Scholar] [CrossRef]
- FAO Soil Organic Carbon Mapping Cookbook. Available online: https://www.fao.org/documents/card/en/c/I8895EN/ (accessed on 7 September 2022).
- Davis, E.; Wang, C.; Dow, K. Comparing Sentinel-2 MSI and Landsat 8 OLI in Soil Salinity Detection: A Case Study of Agricultural Lands in Coastal North Carolina. Int. J. Remote Sens. 2019, 40, 6134–6153. [Google Scholar] [CrossRef]
- Maleki, S.; Fathizad, H.; Karimi, A.; Taghizadeh-Mehrjardi, R.; Pourghasemi, H.R. Monitoring of Spatiotemporal Changes of Soil Salinity and Alkalinity in Eastern and Central Parts of Iran. In Computers in Earth and Environmental Sciences; Elsevier: New York, NY, USA, 2022; pp. 547–561. [Google Scholar]
- Naimi, S.; Ayoubi, S.; Zeraatpisheh, M.; Dematte, J.A.M. Ground Observations and Environmental Covariates Integration for Mapping of Soil Salinity: A Machine Learning-Based Approach. Remote Sens. 2021, 13, 4825. [Google Scholar] [CrossRef]
- Zhao, D.; Arshad, M.; Wang, J.; Triantafilis, J. Soil Exchangeable Cations Estimation Using Vis-NIR Spectroscopy in Different Depths: Effects of Multiple Calibration Models and Spiking. Comput. Electron. Agric. 2021, 182, 105990. [Google Scholar] [CrossRef]
- Zhao, D.; Li, N.; Zare, E.; Wang, J.; Triantafilis, J. Mapping Cation Exchange Capacity Using a Quasi-3d Joint Inversion of EM38 and EM31 Data. Soil Tillage Res. 2020, 200, 104618. [Google Scholar] [CrossRef]
- Zhao, D.; Zhao, X.; Khongnawang, T.; Arshad, M.; Triantafilis, J. A Vis-NIR Spectral Library to Predict Clay in Australian Cotton Growing Soil. Soil Sci. Soc. Am. J. 2018, 82, 1347–1357. [Google Scholar] [CrossRef]
- Hengl, T.; Nussbaum, M.; Wright, M.N.; Heuvelink, G.B.M.; Gräler, B. Random Forest as a Generic Framework for Predictive Modeling of Spatial and Spatio-Temporal Variables. PeerJ 2018, 2018, e5518. [Google Scholar] [CrossRef]
- Yang, S.H.; Liu, F.; Song, X.D.; Lu, Y.Y.; Li, D.C.; Zhao, Y.G.; Zhang, G.L. Mapping Topsoil Electrical Conductivity by a Mixed Geographically Weighted Regression Kriging: A Case Study in the Heihe River Basin, Northwest China. Ecol. Indic. 2019, 102, 252–264. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Román-Dobarco, M.; McBratney, A.B. Perspectives on Data-Driven Soil Research. Eur. J. Soil Sci. 2021, 72, 1675–1689. [Google Scholar] [CrossRef]
- Kaya, F.; Başayiğit, L. Using Machine Learning Algorithms to Mapping of the Soil Macronutrient Elements Variability with Digital Environmental Data in an Alluvial Plain. In Artificial Intelligence and Smart Agriculture Applications; Kose, U., Prasath, V.B.S., Mondal, M.R.H., Podder, P., Subrato, B., Eds.; Auerbach Publications: Boca Raton, FL, USA, 2022; pp. 107–136. [Google Scholar]
- Kaya, F.; Başayiğit, L.; Keshavarzi, A.; Francaviglia, R. Digital Mapping for Soil Texture Class Prediction in Northwestern Türkiye by Different Machine Learning Algorithms. Geoderma Reg. 2022, 31, e00584. [Google Scholar] [CrossRef]
- van Wambeke, A.R. JNSM—Java Newhall Simulation Model JNSM—The Newhall Simulation Model for Estimating Soil Moisture & Temperature Regimes; USDA: Indianapolis, IN, USA, 2000; pp. 1–9.
- GDSHW. Isparta Atabey Plain Irrigation Rehabilitation Project Resettlement Action Plan. 2020. Available online: https://jagomart.net/item/25154/isparta-atabey-plain-irrigation-rehabilitation-project-first-phase-resettlement-action-plan-may-2021 (accessed on 7 September 2022).
- TSMS. Climate of Turkey According to Köppen Climate Classification. Available online: https://www.mgm.gov.tr/FILES/iklim/iklim_siniflandirmalari/koppen.pdf (accessed on 7 September 2022).
- Soil Survey Staff Keys to Soil Taxonomy, 12th ed.; USDA-Natural Resources Conservation Service: Washington, DC, USA, 2014.
- European Union. C.L.M.S. Corine Land Cover Class 2018 V 2.0 Data. Available online: https://land.copernicus.eu/pan-european/corine-land-cover (accessed on 7 September 2022).
- Kaya, F.; Başayiğit, L. Spatial Prediction and Digital Mapping of Soil Texture Classes in a Floodplain Using Multinomial Logistic Regression. In Intelligent and Fuzzy Techniques for Emerging Conditions and Digital Transformation; Kahraman, C., Cebi, S., Onar Cevik, S., Oztaysi, B., Tolga, A.C., Sari, I.U., Eds.; Springer: Berlin/Heidelberg, Germany, 2022; pp. 463–473. [Google Scholar]
- Soil Science Division Staff. Soil Survey Manual; Ditzler, C., Scheffe, K., Monger, H.C., Eds.; USDA Handbook 18; Government Printing Office: Washington, DC, USA, 2017.
- Rhoades, J.D.; Chanduvi, F.; Lesch, S. Soil Salinity Assessment: Methods and Interpretation of Electrical Conductivity Measurements. Available online: https://www.fao.org/3/x2002e/x2002e.pdf (accessed on 7 September 2022).
- Bouyoucos, G.J. Hydrometer Method Improved for Making Particle Size Analyses of Soils1. Agron. J. 1962, 54, 464–465. [Google Scholar] [CrossRef]
- EPA. Available online: https://www.epa.gov/sites/default/files/2015-12/documents/9081.pdf (accessed on 21 November 2022).
- Kacar, B. Fiziksel ve Kimyasal Toprak Analizler, 1st ed.; Nobel: Ankara, Turkey, 2016; ISBN 9786053204305. [Google Scholar]
- Schillaci, C.; Braun, A.; Kropáček, J. Terrain Analysis and Landform Recognition. In Geomorphological Techniques (Online Edition); Clarke, N.E., Nield, J.M., Eds.; British Society for Geomorphology: London, UK, 2015. [Google Scholar]
- Mulder, V.L.; de Bruin, S.; Schaepman, M.E.; Mayr, T.R. The Use of Remote Sensing in Soil and Terrain Mapping—A Review. Geoderma 2011, 162, 1–19. [Google Scholar] [CrossRef]
- Silva, B.P.C.; Silva, M.L.N.; Avalos, F.A.P.; de Menezes, M.D.; Curi, N. Digital Soil Mapping Including Additional Point Sampling in Posses Ecosystem Services Pilot Watershed, Southeastern Brazil. Sci. Rep. 2019, 9, 13763. [Google Scholar] [CrossRef] [PubMed]
- Yahiaoui, I.; Douaoui, A.; Zhang, Q.; Ziane, A. Soil Salinity Prediction in the Lower Cheliff Plain (Algeria) Based on Remote Sensing and Topographic Feature Analysis. J. Arid Land 2015, 7, 794–805. [Google Scholar] [CrossRef]
- Ma, G.; Ding, J.; Han, L.; Zhang, Z.; Ran, S. Digital Mapping of Soil Salinization Based on Sentinel-1 and Sentinel-2 Data Combined with Machine Learning Algorithms. Reg. Sustain. 2021, 2, 177–188. [Google Scholar] [CrossRef]
- ASF DAAC. ALOS PALSAR_Radiometric_Terrain_Corrected_high_res; Includes Material © JAXA/METI 2007. Available online: https://asf.alaska.edu/data-sets/derived-data-sets/alos-palsar-rtc/alos-palsar-radiometric-terrain-correction/ (accessed on 5 September 2021). [CrossRef]
- Gruber, S.; Peckham, S. Chapter 7 Land-Surface Parameters and Objects in Hydrology. In Developments in Soil Science; Elsevier: New York, NY, USA, 2009; Volume 33, pp. 171–194. [Google Scholar]
- Sentinel-2 User Handbook; 1.2; European Space Agency (ESA): Paris, France, 2015.
- Xiao, J.; Shen, Y.; Tateishi, R.; Bayaer, W. Development of Topsoil Grain Size Index for Monitoring Desertification in Arid Land Using Remote Sensing. Int. J. Remote Sens. 2007, 27, 2411–2422. [Google Scholar] [CrossRef]
- Hounkpatin, K.O.L.; Schmidt, K.; Stumpf, F.; Forkuor, G.; Behrens, T.; Scholten, T.; Amelung, W.; Welp, G. Predicting Reference Soil Groups Using Legacy Data: A Data Pruning and Random Forest Approach for Tropical Environment (Dano Catchment, Burkina Faso). Sci. Rep. 2018, 8, 9959. [Google Scholar] [CrossRef]
- Brown, K.S.; Libohova, Z.; Boettinger, J. Digital Soil Mapping. In Soil Survey Manual; Ditzler, C., Scheffe, K., Monger, H.C., Eds.; USDA Handbook 18; Government Printing Office: Washington, DC, USA, 2017. [Google Scholar]
- Ließ, M.; Gebauer, A.; Don, A. Machine Learning With GA Optimization to Model the Agricultural Soil-Landscape of Germany: An Approach Involving Soil Functional Types With Their Multivariate Parameter Distributions Along the Depth Profile. Front. Environ. Sci. 2021, 9, 212. [Google Scholar] [CrossRef]
- ESRI. ArcGIS User’s Guide. 2022. Available online: http://www.esri.com (accessed on 15 September 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.r-project.org/ (accessed on 6 July 2022).
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. 2022. Available online: https://cran.r-project.org/web/packages/raster/raster.pdf (accessed on 6 July 2022).
- Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research. 2022. Available online: https://www.scholars.northwestern.edu/en/publications/psych-procedures-for-personality-and-psychological-research (accessed on 6 July 2022).
- Ismayilov, A.I.; Mamedov, A.I.; Fujimaki, H.; Tsunekawa, A.; Levy, G.J. Soil Salinity Type Effects on the Relationship between the Electrical Conductivity and Salt Content for 1:5 Soil-to-Water Extract. Sustainability 2021, 13, 3395. [Google Scholar] [CrossRef]
- Wang, F.; Shi, Z.; Biswas, A.; Yang, S.; Ding, J. Multi-Algorithm Comparison for Predicting Soil Salinity. Geoderma 2020, 365, 114211. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Biau, G.; Scornet, E. A Random Forest Guided Tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V.; Saitta, L. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support Vector Regression Machines. Adv. Neural Inf. Process. Syst. 1996, 9, 155–161. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM. ACM Trans. Intell. Syst. Technol. (TIST) 2011, 2, 27. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. E1071: Misc Functions of the Department of Statistics; Probability Group (Formerly: E1071), TU Wien: Vienna, Austria, 2022. [Google Scholar]
- Hengl, T.; Macmillan, R.A. Predictive Soil Mapping with R; OpenGeoHub Foundation: Wageningen, The Netherlands, 2019; pp. 227–273. [Google Scholar]
- Tziachris, P.; Aschonitis, V.; Chatzistathis, T.; Papadopoulou, M. Assessment of Spatial Hybrid Methods for Predicting Soil Organic Matter Using DEM Derivatives and Soil Parameters. Catena 2019, 174, 206–216. [Google Scholar] [CrossRef]
- Gräler, B.; Pebesma, E.; Heuvelink, G. Spatio-Temporal Interpolation Using Gstat. R J. 2016, 8, 204. [Google Scholar] [CrossRef]
- Keshavarzi, A.; Tuffour, H.O.; Oppong, J.C.; Zeraatpisheh, M.; Kumar, V. Dealing with Soil Organic Carbon Modeling: Some Insights from an Agro-Ecosystem in Northeast Iran. Earth Sci. Inf. 2021, 14, 1833–1845. [Google Scholar] [CrossRef]
- Zambrano-Bigiarini, M. HydroGOF: Goodness-of-Fit Functions for Comparison of Simulated and Observed Hydrological Time Series. 2020. Available online: https://www.rforge.net/hydroGOF/git.html (accessed on 6 July 2022).
- Lin, L.I.-K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255. [Google Scholar] [CrossRef] [PubMed]
- Wilding, L.P. Spatial Variability: Its Documentation, Accommodation, and Implication to Soil Surveys. In Soil Spatial Variability; Nielsen, D.R., Bouma, J., Eds.; Pudoc: Wageningen, The Netherlands, 1985; pp. 166–193. [Google Scholar]
- Fathizad, H.; Ali Hakimzadeh Ardakani, M.; Sodaiezadeh, H.; Kerry, R.; Taghizadeh-Mehrjardi, R. Investigation of the Spatial and Temporal Variation of Soil Salinity Using Random Forests in the Central Desert of Iran. Geoderma 2020, 365, 114233. [Google Scholar] [CrossRef]
- Nosetto, M.D.; Acosta, A.M.; Jayawickreme, D.H.; Ballesteros, S.I.; Jackson, R.B.; Jobbágy, E.G. Land-Use and Topography Shape Soil and Groundwater Salinity in Central Argentina. Agric. Water Manag. 2013, 129, 120–129. [Google Scholar] [CrossRef]
- Jiang, Q.; Peng, J.; Biswas, A.; Hu, J.; Zhao, R.; He, K.; Shi, Z. Characterising Dryland Salinity in Three Dimensions. Sci. Total Environ. 2019, 682, 190–199. [Google Scholar] [CrossRef]
- Kargas, G.; Londra, P.; Sotirakoglou, K. The Effect of Soil Texture on the Conversion Factor of 1:5 Soil/Water Extract Electrical Conductivity (EC1:5) to Soil Saturated Paste Extract Electrical Conductivity (ECe). Water 2022, 14, 642. [Google Scholar] [CrossRef]
- Cambardella, C.A.; Moorman, T.B.; Novak, J.M.; Parkin, T.B.; Karlen, D.L.; Turco, R.F.; Konopka, A.E. Field-Scale Variability of Soil Properties in Central Iowa Soils. Soil Sci. Soc. Am. J. 1994, 58, 1501–1511. [Google Scholar] [CrossRef]
- Malone, B.P.; Minasny, B.; McBratney, A.B. Continuous Soil Attribute Modeling and Mapping. In Using R for Digital Soil Mapping; Springer: Cham, Switzerland, 2017; pp. 117–149. [Google Scholar]
- Mishra, G.; Sulieman, M.M.; Kaya, F.; Francaviglia, R.; Keshavarzi, A.; Bakhshandeh, E.; Loum, M.; Jangir, A.; Ahmed, I.; Elmobarak, A.; et al. Machine Learning for Cation Exchange Capacity Prediction in Different Land Uses. Catena 2022, 216, 106404. [Google Scholar] [CrossRef]
- Taghadosi, M.M.; Hasanlou, M.; Eftekhari, K. Retrieval of Soil Salinity from Sentinel-2 Multispectral Imagery. Eur. J. Remote Sens. 2019, 52, 138–154. [Google Scholar] [CrossRef]
- Wadoux, A.M.J.C.; Heuvelink, G.B.M.; de Bruin, S.; Brus, D.J. Spatial Cross-Validation Is Not the Right Way to Evaluate Map Accuracy. Ecol. Model. 2021, 457, 109692. [Google Scholar] [CrossRef]
- Wang, J.; Peng, J.; Li, H.; Yin, C.; Liu, W.; Wang, T.; Zhang, H. Soil Salinity Mapping Using Machine Learning Algorithms with the Sentinel-2 MSI in Arid Areas, China. Remote Sens. 2021, 13, 305. [Google Scholar] [CrossRef]
- Habibi, V.; Ahmadi, H.; Jafari, M.; Moeini, A. Machine Learning and Multispectral Data-Based Detection of Soil Salinity in an Arid Region, Central Iran. Environ. Monit. Assess. 2020, 192, 759. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Toomanian, N.; Heung, B.; Behrens, T.; Mosavi, A.; Band, S.; Amirian-Chakan, A.; Fathabadi, A.; Scholten, T. Improving the Spatial Prediction of Soil Salinity in Arid Regions Using Wavelet Transformation and Support Vector Regression Models. Geoderma 2021, 383, 114793. [Google Scholar] [CrossRef]
- Burke, M.; Driscoll, A.; Lobell, D.B.; Ermon, S. Using Satellite Imagery to Understand and Promote Sustainable Development. Science 2021, 371, 6535. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.Y.M.; Biswas, R.K.; Chowdhury, A.I.; Billah, S.M. Modeling Soil Salinity Using Direct and Indirect Measurement Techniques: A Comparative Analysis. Environ. Dev. 2019, 29, 67–80. [Google Scholar] [CrossRef]
- Szatmári, G.; Bakacsi, Z.; Laborczi, A.; Petrik, O.; Pataki, R.; Tóth, T.; Pásztor, L. Elaborating Hungarian Segment of the Global Map of Salt-Affected Soils (GSSmap): National Contribution to an International Initiative. Remote Sens. 2020, 12, 4073. [Google Scholar] [CrossRef]
- Keesstra, S.; Mol, G.; de Leeuw, J.; Okx, J.; Molenaar, C.; de Cleen, M.; Visser, S. Soil-Related Sustainable Development Goals: Four Concepts to Make Land Degradation Neutrality and Restoration Work. Land 2018, 7, 133. [Google Scholar] [CrossRef]
- Planet Team Planet Application Program Interface. Available online: https://www.planet.com/explorer/ (accessed on 8 September 2022).
- Avdan, U.; Kaplan, G.; Küçük Matcı, D.; Yiğit Avdan, Z.; Erdem, F.; Tuğba Mızık, E.; Demirtaş, İ. Soil Salinity Prediction Models Constructed by Different Remote Sensors. Phys. Chem. Earth Parts A/B/C 2022, 128, 103230. [Google Scholar] [CrossRef]



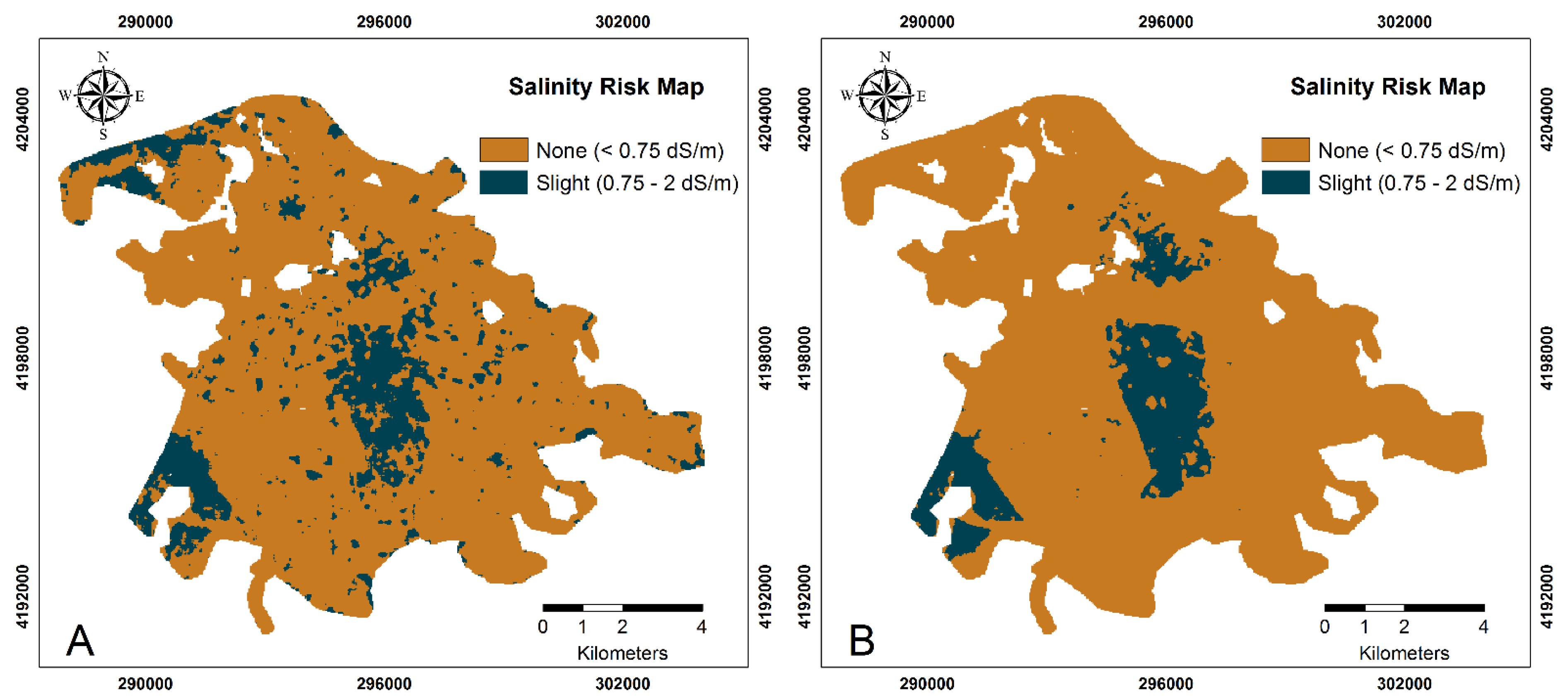
| Variable | Dataset | Mean | SD | CV | Minimum | Maximum | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|
| EC | Training | 616.30 | 249.60 | 40.49 | 110.0 | 1692.0 | 1.29 | 4.47 |
| Validation | 602.80 | 366.60 | 60.82 | 152.0 | 2068.0 | 2.64 | 9.14 | |
| Clay (%) | All | 36.91 | 13.68 | 37.06 | 13.28 | 73.76 | 0.82 | −0.22 |
| Sand (%) | 37.29 | 15.01 | 40.25 | 6.35 | 69.30 | −0.34 | −0.87 | |
| Silt (%) | 25.81 | 7.16 | 27.74 | 12.47 | 51.94 | 1.13 | 1.64 | |
| CEC (cmolc kg−1) | 22.75 | 6.75 | 29.67 | 9.34 | 40.24 | 0.71 | 0.05 | |
| SSP (%) | 36.76 | 5.93 | 16.13 | 20.50 | 50.00 | −0.18 | −0.22 |
| Soil Property | Model | Training | Validation | Hybrid Model Validation | ||||
|---|---|---|---|---|---|---|---|---|
| LCCC | RMSE | NRMSE | LCCC | NRMSE | LCCC | NRMSE | ||
| EC | SVR | 0.50 | 198.05 | 79.4 | 0.25 | 89.6 | 0.26 | 88.7 |
| RF | 0.87 | 102.83 | 41.2 | 0.28 | 85.6 | 0.29 | 85.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaya, F.; Schillaci, C.; Keshavarzi, A.; Başayiğit, L. Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain. Land 2022, 11, 2148. https://doi.org/10.3390/land11122148
Kaya F, Schillaci C, Keshavarzi A, Başayiğit L. Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain. Land. 2022; 11(12):2148. https://doi.org/10.3390/land11122148
Chicago/Turabian StyleKaya, Fuat, Calogero Schillaci, Ali Keshavarzi, and Levent Başayiğit. 2022. "Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain" Land 11, no. 12: 2148. https://doi.org/10.3390/land11122148
APA StyleKaya, F., Schillaci, C., Keshavarzi, A., & Başayiğit, L. (2022). Predictive Mapping of Electrical Conductivity and Assessment of Soil Salinity in a Western Türkiye Alluvial Plain. Land, 11(12), 2148. https://doi.org/10.3390/land11122148









