Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate
Abstract
:1. Introduction
2. Impact of Climate Change on Soil Health
2.1. Effect of Elevated Temperatures
2.2. Effect of Elevated CO2 Levels
2.3. Effect of Drought
2.4. Effect of Increased Precipitation
3. Climate Change Adaptation and Soil Microbiome
3.1. Salinization
3.2. Drought
3.3. Soil Fertility
3.3.1. Bio-Fertilization with Nitrogen-Fixing Microorganisms
3.3.2. Bio-Fertilization with Nutrient Solubilizing and Mobilizing Microbial Inoculants
3.4. Bioremediation of Soil Pollutants
4. Bioengineered Microbes for Soil Health Restoration
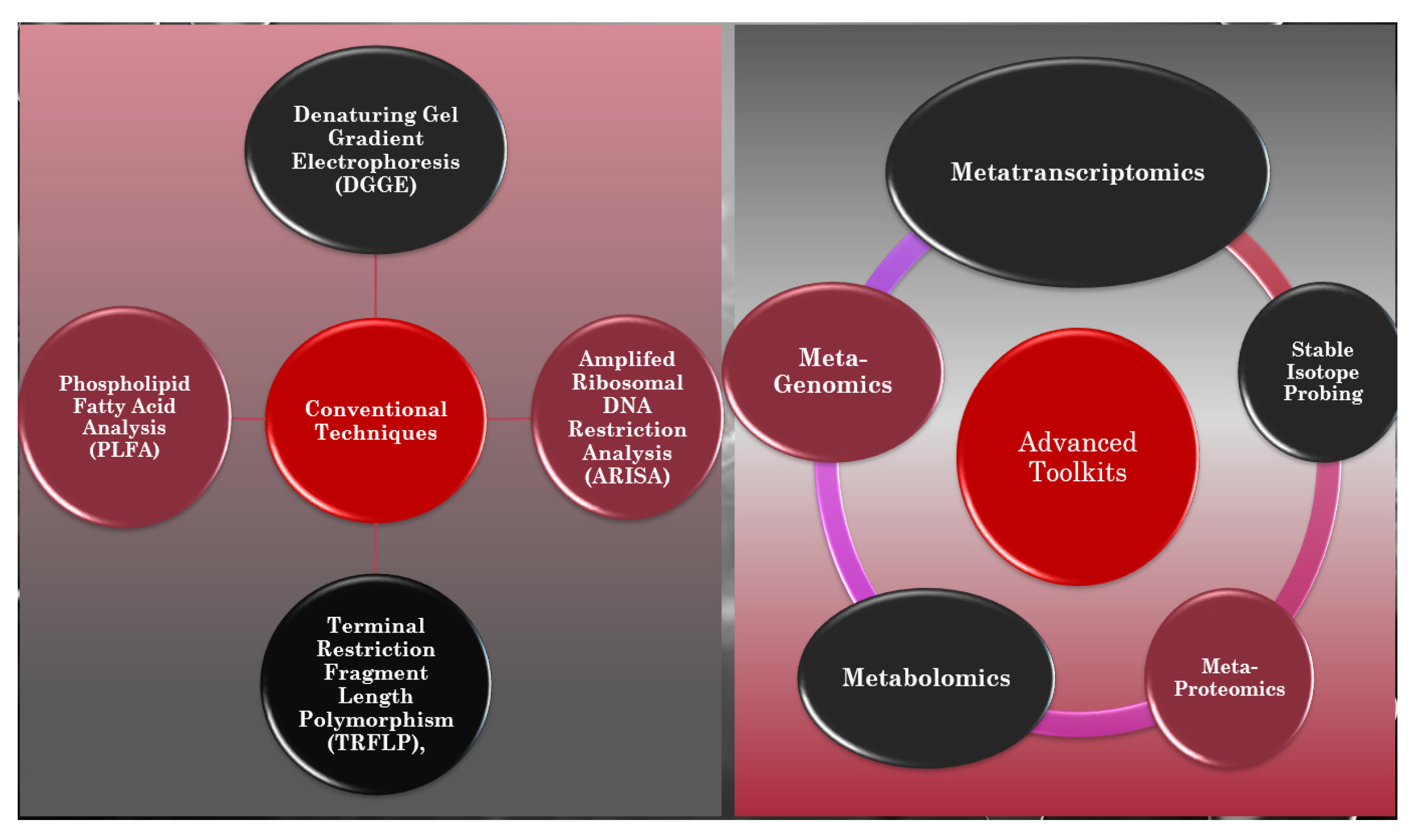
5. Advanced Tool Kits for Unveiling the Black Box of Soil
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lykogianni, M.; Bempelou, E.; Karamaouna, F.; Aliferis, K.A. Do pesticides promote or hinder sustainability in agriculture? The challenge of sustainable use of pesticides in modern agriculture. Sci. Total Environ. 2021, 795, 148625. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.G.; Dent, D.L.; Olsson, L.; Schaepman, M.E. Proxy global assessment of land degradation. Soil Use Manag. 2008, 24, 223–234. [Google Scholar] [CrossRef]
- Montanarella, L.; Pennock, D.J.; McKenzie, N.; Badraoui, M.; Chude, V.; Baptista, I.; Mamo, T.; Yemefack, M.; Singh Aulakh, M.; Yagi, K.; et al. World’s soils are under threat. Soil Discuss. 2015, 2, 1263–1272. [Google Scholar] [CrossRef] [Green Version]
- Van Meijl, J.C.M.; Havlik, P.; Lotze-Campen, H.; Stehfest, E.; Witzke, P.; Domínguez, I.P.; Bodirsky, B.; van Dijk, M.; Doelman, J.C.; Fellmann, T.; et al. Challenges of global agriculture in a climate change context by 2050 (AgCLIM50). In JRC Science for Policy Report; EUR 28649 EN; Publications Office of the European Union: Luxembourg. [CrossRef]
- Kopittke, P.M.; Menzies, N.W.; Wang, P.; McKenna, B.A.; Lombi, E. Soil and the intensification of agriculture for global food security. Environ. Int. 2019, 132, 105078. [Google Scholar] [CrossRef] [PubMed]
- Greiner, L.; Keller, A.; Gret-Regamey, A.; Papritz, A. Soil function assessment, review of methods for quantifying the contributions of soils to ecosystem services. Land Use Policy 2017, 69, 224–237. [Google Scholar] [CrossRef]
- MacLaren, C.; Storkey, J.; Menegat, A.; Metcalfe, H.; Dehnen-Schmutz, K. An ecological future for weed science to sustain crop production and the environment. A review. Agron. Sustain. Dev. 2020, 40, 1–29. [Google Scholar] [CrossRef]
- Rodriguez, R.; Duran, P. Natural holobiome engineering by using native extreme microbiome to counteract the climate change effects. Front. Bioeng. Biotechnol. 2020, 8, 568. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome, A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Poore, J.; Nemecek, T. Reducing food’s environmental impacts through producers and consumers. Science 2018, 360, 987. [Google Scholar] [CrossRef]
- Parajuli, R.; Thoma, G.; Matlock, M.D. Environmental sustainability of fruit and vegetable production supply chains in the face of climate change, A review. Sci. Total Environ. 2019, 650, 2863–2879. [Google Scholar] [CrossRef]
- Swaminathan, M.S.; Kesavan, P.C. Agricultural research in an era of climate change. Agric. Res. 2012, 1, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Blattner, C. Just transition for agriculture? A critical step in tackling climate change. J. Agric. Food Syst. Community Dev. 2020, 25, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Heidecke, C.; Montgomery, H.; Stalb, H.; Wollenberg, L. In Proceedings of the International Conference on Agricultural GHG Emissions and Food Security-Connecting Research to Policy and Practice, Berlin, Germany, 10–13 September 2018.
- Del Buono, D. Can bio stimulants be used to mitigate the effect of anthropogenic climate change on agriculture? It is time to respond. Sci. Total Environ. 2021, 751, 141763. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 1, 35–46. [Google Scholar] [CrossRef]
- Guo, M. Soil Health Assessment and Management, Recent Development in Science and Practices. Soil Syst. 2021, 5, 61. [Google Scholar] [CrossRef]
- Bagnall, D.K.; Shanahan, J.F.; Flanders, A.; Morgan, C.L.; Honeycutt, C.W. Soil health considerations for global food security. Agronomy 2021, 113, 4581–4589. [Google Scholar] [CrossRef]
- Bisht, N.; Chauhan, P.S. Excessive and Disproportionate Use of Chemicals Cause Soil Contamination and Nutritional Stress. In Soil Contamination—Threats and Sustainable Solutions; Larramendy, M.L., Soloneski, S., Eds.; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Bargaz, A.; Lyamlouli, K.; Chtouki, M.; Zeroual, Y.; Dhiba, D. Soil microbial resources for improving fertilizers efficiency in an integrated plant nutrient management system. Front. Microbiol. 2018, 9, 1606. [Google Scholar] [CrossRef] [Green Version]
- Van Der Heijden, M.G.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority, soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil microbiome, a key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Norby, R.J.; De Kauwe, M.G.; Domingues, T.F.; Duursma, R.A.; Ellsworth, D.S.; Goll, D.S.; Lapola, D.M.; Luus, K.A.; MacKenzie, A.R.; Medlyn, B.E.; et al. Model–data synthesis for the next generation of forest free-air CO2 enrichment (FACE) experiments. New Phytol. 2016, 209, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Karmakar, R.; Das, I.; Dutta, D.; Rakshit, A. Potential effects of climate change on soil properties, a review. Sci. Int. 2016, 4, 51–73. [Google Scholar] [CrossRef] [Green Version]
- Pareek, N. Climate change impact on soils, adaptation and mitigation. MOJ Ecol. Environ. Sci. 2017, 2, 26. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.; Hussain, S.; Guo, R.; Sarwar, M.; Ren, X.; Krstic, D.; Aslam, Z.; Zulifqar, U.; Rauf, A.; Hano, C.; et al. Carbon Sequestration to Avoid Soil Degradation, A Review on the Role of Conservation Tillage. Plants 2021, 10, 2001. [Google Scholar] [CrossRef] [PubMed]
- Chander, S. Impact of Climate Change on Insects InClimate Change Impact, Adaptation and Mitigation in Agriculture, Methodology for Assessment and Application; Indian Agricultural Research Institute: New Delhi, India, 2012; Volume 302, pp. 111–130. [Google Scholar]
- DEFRA. Impact of Climate Change on Soil Functions; Final Project Report Research and Development: London, UK, 2005. [Google Scholar]
- Yi, F.; Liu, H.; Quan, Q. Impact of Climate Change on Chemical Inputs: Evidence of Pesticide Usage from China. In Proceedings of the 2021 Annual Meeting, Austin, Texas, 1–3 August 2021; Agricultural and Applied Economics Association: Milwaukee, WI, USA, 2021; p. 313374. [Google Scholar] [CrossRef]
- Medhi, K.; Bhardwaj, R.; Laxmi, R. Climate change with its impacts on soil and soil microbiome regulating biogeochemical nutrient transformations. In Climate Change and the Microbiome; Springer: Cham, Switzerland, 2021; pp. 95–138. [Google Scholar]
- Cai, A.; Feng, W.; Zhang, W.; Xu, M. Climate, soil texture, and soil types affect the contributions of fine-fraction-stabilized carbon to total soil organic carbon in different land uses across China. J. Environ. Manag. 2016, 172, 2–9. [Google Scholar] [CrossRef]
- He, A.L.; Niu, S.Q.; Zhao, Q.; Li, Y.S.; Gou, J.Y.; Gao, H.J.; Suo, S.Z.; Zhang, J.L. Induced salt tolerance of perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. Int. J. Mol. Sci. 2018, 19, 469. [Google Scholar] [CrossRef] [Green Version]
- Delgado, A.; Gomez, J.A. The soil. Physical, chemical and biological properties. In Principles of Agronomy for Sustainable Agriculture; Springer: Cham, Switzerland, 2016; pp. 15–26. [Google Scholar]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Alava, J.J.; Cheung, W.W.L.; Ross, P.S.; Sumaila, U.R. Climate change–contaminant interactions in marine food webs: Toward a conceptual framework. Glob. Chang. Biol. 2017, 23, 3984–4001. [Google Scholar] [CrossRef]
- Biswas, B.; Qi, F.; Biswas, J.K.; Wijayawardena, A.; Khan, M.A.I.; Naidu, R. The fate of chemical pollutants with soil properties and processes in the climate change paradigm—A review. Soil Syst. 2018, 2, 51. [Google Scholar] [CrossRef] [Green Version]
- Paltseva, A.A.; Neaman, A. An emerging frontier: Metal (loid) soil pollution threat under global climate change. Environ. Toxicol. Chem. 2020, 39, 1653–1654. [Google Scholar] [CrossRef]
- Haines, A.; Kovats, R.S.; Campbell-Lendrum, D.; Corvalan, C. Climate change and human health: Impacts, vulnerability and public health. Public Health 2006, 120, 585–596. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The role of soil microorganisms in plant mineral nutrition—current knowledge and future directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christy, C. Unearthing the Soil Microbiome, Climate Change, Carbon Storage Nexus. Am. Soc. Microbiol. 2021.
- Allison, S.D.; Martiny, J.B. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef] [Green Version]
- Wahl, T.; Ward, P.J.; Winsemius, H.C.; AghaKouchak, A.; Bender, J.; Haigh, I.D.; Jain, S.; Leonard, M.; Veldkamp, T.I.; Westra, S. When environmental forces collide. Eos 2018, 99. [Google Scholar] [CrossRef]
- Naylor, D.; Sadler, N.; Bhattacharjee, A.; Graham, E.B.; Anderton, C.R.; McClure, R.; Lipton, M.; Hofmockel, K.S.; Jansson, J.K. Soil microbiomes under climate change and implications for carbon cycling. Annu. Rev. Environ. Resour. 2020, 1, 29–59. [Google Scholar] [CrossRef]
- Classen, A.T.; Sundqvist, M.K.; Henning, J.A.; Newman, G.S.; Moore, J.A.; Cregger, M.A.; Moorhead, L.C.; Patterson, C.M. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions, What lies ahead? Ecosphere 2015, 6, 1–21. [Google Scholar] [CrossRef]
- Schindlbacher, A.; Rodler, A.; Kuffner, M.; Kitzler, B.; Sessitsch, A.; Zechmeister-Boltenstern, S. Experimental warming effects on the microbial community of a temperate mountain forest soil. Soil Biol. Biochem. 2011, 43, 1417–1425. [Google Scholar] [CrossRef]
- Hu, H.W.; Macdonald, C.A.; Trivedi, P.; Anderson, I.C.; Zheng, Y.; Holmes, B.; Bodrossy, L.; Wang, J.T.; He, J.Z.; Singh, B.K. Effects of climate warming and elevated CO2 on autotrophic nitrification and nitrifiers in dryland ecosystems. Soil Biol. Biochem. 2016, 92, 1–5. [Google Scholar] [CrossRef]
- Karhu, K.; Auffret, M.D.; Dungait, J.A.; Hopkins, D.W.; Prosser, J.I.; Singh, B.K.; Subke, J.A.; Wookey, P.A.; Agren, G.I.; Sebastia, M.T.; et al. Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 2014, 513, 81–84. [Google Scholar] [CrossRef]
- Bai, E.; Li, S.; Xu, W.; Li, W.; Dai, W.; Jiang, P. A meta-analysis of experimental warming effects on terrestrial nitrogen pools and dynamics. New Phytol. 2013, 199, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Melillo, J.M.; Frey, S.D.; DeAngelis, K.M.; Werner, W.J.; Bernard, M.J.; Bowles, F.P.; Pold, G.; Knorr, M.A.; Grandy, A.S. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science 2017, 358, 101–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeAngelis, K.M.; Pold, G.; Topcuoglu, B.D.; van Diepen, L.T.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 2015, 6, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, H.; Dutta, A. The microbial aspect of climate change. Energy Ecol. Environ. 2016, 1, 209–232. [Google Scholar] [CrossRef] [Green Version]
- Rosado-Porto, D.; Ratering, S.; Cardinale, M.; Maisinger, C.; Moser, G.; Deppe, M.; Müller, C.; Schnell, S. Elevated Atmospheric CO2 Modifies Mostly the Metabolic Active Rhizosphere Soil Microbiome in the Giessen FACE Experiment. Microb. Ecol. 2021, 19, 1–6. [Google Scholar] [CrossRef]
- Yu, H.; Deng, Y.; He, Z.; Van Nostrand, J.D.; Wang, S.; Jin, D.; Wang, A.; Wu, L.; Wang, D.; Tai, X.; et al. Elevated CO2 and warming altered grassland microbial communities in soil top-layers. Front. Microbiol. 2018, 9, 1790. [Google Scholar] [CrossRef]
- Qiao, N.A.; Schaefer, D.; Blagodatskaya, E.; Zou, X.; Xu, X.; Kuzyakov, Y. Labile carbon retention compensates for CO2 released by priming in forest soils. Glob. Change Biol. 2014, 20, 1943–1954. [Google Scholar] [CrossRef]
- Hayden, H.L.; Mele, P.M.; Bougoure, D.S.; Allan, C.Y.; Norng, S.; Piceno, Y.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Williams, A.L.; et al. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO2 and warming in an Australian native grassland soil. Environ. Microbiol. 2012, 14, 3081–3096. [Google Scholar] [CrossRef]
- Abdul Rahman, N.S.; Abdul Hamid, N.W.; Nadarajah, K. Effects of abiotic stress on soil microbiome. Int. J. Mol. Sci. 2021, 22, 9036. [Google Scholar] [CrossRef]
- Siebielec, S.; Siebielec, G.; Klimkowicz-Pawlas, A.; Galazka, A.; Grzadziel, J.; Stuczynski, T. Impact of water stress on microbial community and activity in sandy and loamy soils. Agronomy 2020, 10, 1429. [Google Scholar] [CrossRef]
- Sheik, C.S.; Beasley, W.H.; Elshahed, M.S.; Zhou, X.; Luo, Y.; Krumholz, L.R. Effect of warming and drought on grassland microbial communities. ISME J. 2011, 5, 1692–1700. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Osanai, Y.; Anderson, I.C.; Bange, M.P.; Tissue, D.T.; Singh, B.K. Flooding and prolonged drought have differential legacy impacts on soil nitrogen cycling, microbial communities and plant productivity. Plant Soil 2018, 1, 371–387. [Google Scholar] [CrossRef]
- Bogati, K.; Walczak, M. The impact of drought stress on soil microbial community, enzyme activities and plants. Agronomy 2022, 12, 189. [Google Scholar] [CrossRef]
- Breitkreuz, C.; Herzig, L.; Buscot, F.; Reitz, T.; Tarkka, M. Interactions between soil properties, agricultural management and cultivar type drive structural and functional adaptations of the wheat rhizosphere microbiome to drought. Environ. Microbiol. 2021, 23, 5866–5882. [Google Scholar] [CrossRef]
- Upton, R.N.; Bach, E.M.; Hofmockel, K.S. Below ground response of prairie restoration and resiliency to drought. Agric. Ecosyst. Environ. 2018, 266, 122–132. [Google Scholar] [CrossRef]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 1–2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treseder, K.K.; Berlemont, R.; Allison, S.D.; Martiny, A.C. Drought increases the frequencies of fungal functional genes related to carbon and nitrogen acquisition. PLoS ONE 2018, 13, e0206441. [Google Scholar] [CrossRef] [Green Version]
- Carson, J.K.; Gonzalez-Quinones, V.; Murphy, D.V.; Hinz, C.; Shaw, J.A.; Gleeson, D.B. Low pore connectivity increases bacterial diversity in soil. Appl. Environ. Microbiol. 2010, 76, 3936–3942. [Google Scholar] [CrossRef]
- Dechesne, A.; Wang, G.; Gulez, G.; Or, D.; Smets, B.F. Hydration-controlled bacterial motility and dispersal on surfaces. Proc. Natl. Acad. Sci. USA 2010, 107, 14369–14372. [Google Scholar] [CrossRef] [Green Version]
- Guhr, A.; Borken, W.; Spohn, M.; Matzner, E. Redistribution of soil water by a saprotrophic fungus enhances carbon mineralization. Proc. Natl. Acad. Sci. USA 2015, 112, 14647–14651. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, P.O.; Templer, P.H.; Finzi, A.C. Contrasting effects of winter snowpack and soil frost on growing season microbial biomass and enzyme activity in two mixed-hardwood forests. Biogeochemistry 2016, 128, 141–154. [Google Scholar] [CrossRef]
- Shim, J.H.; Pendall, E.; Morgan, J.A.; Ojima, D.S. Wetting and drying cycles drive variations in the stable carbon isotope ratio of respired carbon dioxide in semi-arid grassland. Oecologia 2009, 160, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Aanderud, Z.T.; Schoolmaster, D.R.; Lennon, J.T. Plants mediate the sensitivity of soil respiration to rainfall variability. Ecosystems 2011, 14, 156–167. [Google Scholar] [CrossRef]
- Grzyb, A.; Wolna-Maruwka, A.; Niewiadomska, A. Environmental factors affecting the mineralization of crop residues. Agronomy 2020, 10, 1951. [Google Scholar] [CrossRef]
- Sjogaard, K.S.; Valdemarsen, T.B.; Treusch, A.H. Responses of an agricultural soil microbiome to flooding with seawater after managed coastal realignment. Microorganisms 2018, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef]
- Babalola, O.O.; Glick, B.R. The use of microbial inoculants in African agriculture, current practice and future prospects. J. Food Agric. Environ. 2012, 10, 540–549. [Google Scholar]
- Kalayu, G. Phosphate solubilizing microorganisms, promising approach as biofertilizers. Int. J. Agron. 2019, 2019, 4917256. [Google Scholar] [CrossRef]
- Stamenkovic, S.; Beskoski, V.; Karabegovic, I.; Lazic, M.; Nikolic, N. Microbial fertilizers, A comprehensive review of current findings and future perspectives. Span. J. Agric. Res. 2018, 16, e09R01. [Google Scholar] [CrossRef]
- Kohl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases, relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, A.; Mor, V.S.; Tokas, J.; Punia, H.; Malik, S.; Malik, K.; Sangwan, S.; Tomar, S.; Singh, P.; Singh, N.; et al. Biostimulant-treated seedlings under sustainable agriculture, A global perspective facing climate change. Agronomy 2020, 11, 14. [Google Scholar] [CrossRef]
- Sun, X.G.; Tang, M. Effect of arbuscular mycorrhizal fungi inoculation on root traits and root volatile organic compound emissions of Sorghum bicolor. South Afr. J. Bot. 2013, 88, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Heil, M.; Bostock, R.M. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann. Bot. 2002, 89, 503–512. [Google Scholar] [CrossRef] [Green Version]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Gupta, V.V.; Zhang, B.; Penton, C.R.; Yu, J.; Tiedje, J.M. Diazotroph diversity and nitrogen fixation in summer active perennial grasses in a Mediterranean region agricultural soil. Front. Mol. Biosci. 2019, 6, 115. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants, a review. Rev. Environ. Sci. BioTechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.T.; Figueroa, C.R. Jasmonates and plant salt stress, Molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of plant growth promoting rhizobacteria in agricultural sustainability—a review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [Green Version]
- Neina, D. The role of soil pH in plant nutrition and soil remediation. Appl. Environ. Soil Sci. 2019, 2019, 5794869. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zhang, Q.; Ervin, E.; Yang, Z.; Zhang, X. Physiological mechanism of enhancing salt stress tolerance of perennial ryegrass by 24-epibrassinolide. Front. Plant Sci. 2017, 8, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, S.; Mishra, A.; Trenberth, K.E. Climate change and drought, a perspective on drought indices. Curr. Clim. Chang. Rep. 2018, 4, 145–163. [Google Scholar] [CrossRef]
- Mukhtar, S.; Malik, K.A.; Mehnaz, S. Microbiome of halophytes, Diversity and importance for plant health and productivity. Microbiol. Biotechnol. Lett. 2019, 47, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Malik, K.A. Isolation and characterization of bacteria associated with the rhizosphere of halophytes (Salsola stocksii and Atriplex amnicola) for production of hydrolytic enzymes. Braz. J. Microbiol. 2019, 50, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Bergi, J.; Trivedi, R. Bioremediation of Saline Soil by Cyanobacteria. In Microbial Bioremediation & Biodegradation; Springer: Singapore, 2020; pp. 447–465. [Google Scholar]
- Chakraborty, K.; Mondal, S.; Ray, S.; Samal, P.; Pradhan, B.; Chattopadhyay, K.; Kar, M.K.; Swain, P.; Sarkar, R.K. Tissue tolerance coupled with ionic discrimination can potentially minimize the energy cost of salinity tolerance in rice. Front. Plant Sci. 2020, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Csonka, L.N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 1989, 53, 121–147. [Google Scholar] [CrossRef]
- Dos Santos, R.M.; Diaz, P.A.; Lobo, L.L.; Rigobelo, E.C. Use of plant growth-promoting rhizobacteria in maize and sugarcane, Characteristics and applications. Front. Sustain. Food Syst. 2020, 4, 136. [Google Scholar] [CrossRef]
- Aslam, F.; Ali, B. Halotolerant bacterial diversity associated with Suaeda fruticosa (L.) forssk improved growth of maize under salinity stress. Agronomy 2018, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signalling and abiotic stress tolerance in plants, a review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Otieno, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ye, J.; Kong, Q. Potassium-solubilizing activity of bacillus aryabhattai SK1-7 and its growth-promoting effect on Populus alba L. Forests 2020, 11, 1348. [Google Scholar] [CrossRef]
- Fiodor, A.; Singh, S.; Pranaw, K. The contrivance of plant growth promoting microbes to mitigate climate change impact in agriculture. Microorganisms 2021, 9, 1841. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current progress in nitrogen fixing plants and microbiome research. Plants 2020, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Mus, F.; Alleman, A.B.; Pence, N.; Seefeldt, L.C.; Peters, J.W. Exploring the alternatives of biological nitrogen fixation. Metallomics 2018, 10, 523–538. [Google Scholar] [CrossRef]
- Raimi, A.R.; Ezeokoli, O.T.; Adeleke, R.A. High-throughput sequence analysis of bacterial communities in commercial biofertilizers products marketed in South Africa, an independent snapshot quality assessment. 3 Biotech 2019, 9, 1–2. [Google Scholar]
- Mabrouk, Y.; Hemissi, I.; Salem, I.B.; Mejri, S.; Saidi, M.; Belhadj, O. Potential of rhizobia in improving nitrogen fixation and yields of legumes. Symbiosis 2018, 107, 73495. [Google Scholar]
- Menge, E.M.; Njeru, E.M.; Koskey, G.; Maingi, J. Rhizobial inoculation methods affect the nodulation and plant growth traits of host plant genotypes, a case study of common bean Phaseolus vulgaris L. germplasms cultivated by smallholder farmers in Eastern Kenya. Adv. Agric. Sci. 2018, 6, 77–94. [Google Scholar]
- Lengwati, D.M.; Mathews, C.; Dakora, F.D. Rotation benefits from N2-fixing grain legumes to cereals, from increases in seed yield and quality to greater household cash-income by a following maize crop. Front. Sustain. Food Syst. 2020, 4, 94. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability, an overview. Chem. Biol. Technol. Agric. 2017, 4, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Thilakarathna, M.S.; McElroy, M.S.; Chapagain, T.; Papadopoulos, Y.A.; Raizada, M.N. Belowground nitrogen transfer from legumes to non-legumes under managed herbaceous cropping systems. A review. Agron. Sustain. Dev. 2016, 36, 1–6. [Google Scholar]
- Rashid, M.I.; Mujawar, L.H.; Shahzad, T.; Almeelbi, T.; Ismail, I.M.; Oves, M. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 2016, 183, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Nasto, M.K.; Alvarez-Clare, S.; Lekberg, Y.; Sullivan, B.W.; Townsend, A.R.; Cleveland, C.C. Interactions among nitrogen fixation and soil phosphorus acquisition strategies in lowland tropical rain forests. Ecol. Lett. 2014, 17, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Lingua, G.; Manassero, P.; Cantamessa, S.; Marsano, F.; Todeschini, V.; Copetta, A.; D’Agostino, G.; Massa, N.; Avidano, L.; et al. AM fungi and PGP pseudomonads increase flowering, fruit production, and vitamin content in strawberry grown at low nitrogen and phosphorus levels. Mycorrhiza 2015, 25, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, L.; Palla, M.; Agnolucci, M.; Avio, L.; Sbrana, C.; Turrini, A.; Giovannetti, M. Arbuscular mycorrhizal fungi and associated microbiota as plant biostimulants, research strategies for the selection of the best performing inocula. Agronomy 2020, 10, 106. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Kumar, A.; Patel, H. Role of microbes in phosphorus availability and acquisition by plants. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1344–1347. [Google Scholar] [CrossRef]
- Yousefi, A.A.; Khavazi, K.; Moezi, A.A.; Rejali, F.; Nadian, H.A. Phosphate solubilizing bacteria and arbuscular mycorrhizal fungi impacts on inorganic phosphorus fractions and wheat growth. World Appl. Sci. J. 2011, 15, 1310–1318. [Google Scholar]
- Santana, E.B.; Marques, E.L.; Dias, J.C. Effects of phosphate-solubilizing bacteria, native microorganisms, and rock dust on Jatropha curcas L. growth. Genet. Mol. Res. 2016, 15, 15048729. [Google Scholar] [CrossRef]
- Park, J.; Bolan, N.; Megharaj, M.; Naidu, R. Isolation of Phosphate-Solubilizing Bacteria and Characterization of Their Effects on Lead Immobilization (Special Issue: International Symposium, Challenges to Soil Degradation Towards Sustaining Life and Environment, Tokyo Metropolitan University Symposium Series No. 2, 2009). Pedologist 2010, 53, 67–75. [Google Scholar]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: London, UK, 2008; pp. 1–637. [Google Scholar]
- Tawaraya, K.; Naito, M.; Wagatsuma, T. Solubilization of insoluble inorganic phosphate by hyphal exudates of arbuscular mycorrhizal fungi. J. Plant Nutr. 2006, 29, 657–665. [Google Scholar] [CrossRef]
- Richardson, A.E.; Barea, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Musyoka, D.M.; Njeru, E.M.; Nyamwange, M.M.; Maingi, J.M. Arbuscular mycorrhizal fungi and Bradyrhizobium co-inoculation enhances nitrogen fixation and growth of green grams (Vigna radiata L.) under water stress. J. Plant Nutr. 2020, 43, 1036–1047. [Google Scholar] [CrossRef]
- Khurshid, M.Y. Potassium as A Key Fertilizer in Combating Climate Change And Malnutrition. 2020. Available online: www.technologytimes.pk. (accessed on 31 December 2020).
- Parmar, P.; Sindhu, S.S. Potassium solubilization by rhizosphere bacteria, influence of nutritional and environmental conditions. J. Microbiol Res. 2013, 3, 25–31. [Google Scholar]
- Kavadia, A.; Omirou, M.; Fasoula, D.; Ioannides, I.M. The importance of microbial inoculants in a climate-changing agriculture in eastern Mediterranean region. Atmosphere 2020, 11, 1136. [Google Scholar] [CrossRef]
- Liu, D.; Lian, B.; Dong, H. Isolation of Paenibacillus sp. and assessment of its potential for enhancing mineral weathering. Geomicrobiol. J. 2012, 29, 413–421. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Goswami, M.P.; Bhattacharyya, L.H. Perspective of beneficial microbes in agriculture under changing climatic scenario, A review. J. Phytol. 2016, 8, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Novo, L.A.; Castro, P.M.; Alvarenga, P.; da Silva, E. Plant growth–promoting rhizobacteria-assisted phytoremediation of mine soils. In Bio-Geotechnologies for Mine Site Rehabilitation; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 281–295. [Google Scholar]
- Kannahi, M.; Senbagam, N. Studies on siderophore production by microbial isolates obtained from rhizosphere soil and its antibacterial activity. J. Chem. Pharm. Res. 2014, 6, 1142–1145. [Google Scholar]
- Mahmud, A.A.; Upadhyay, S.K.; Srivastava, A.K.; Bhojiya, A.A. Biofertilizers, A Nexus between soil fertility and crop productivity under abiotic stress. Curr. Res. Environ. Sustain. 2021, 3, 100063. [Google Scholar] [CrossRef]
- Saha, M.; Maurya, B.R.; Bahadur, I.; Kumar, A.; Meena, V.S. Can potassium-solubilising bacteria mitigate the potassium problems in India? In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 127–136. [Google Scholar]
- Landrigan, P.J.; Fuller, R.; Acosta, N.J.R.; Adeyi, O.; Arnold, R.; Basu, N.; Baldé, A.B.; Bertollini, R.; Bose-O’Reilly, S.; Boufford, J.I. The Lancet Commission on pollution and health. Lancet 2018, 391, 462–512. [Google Scholar] [CrossRef] [Green Version]
- Khatoon, Z.; Huang, S.; Rafique, M.; Fakhar, A.; Kamran, M.A.; Santoyo, G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 2020, 273, 111–118. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Chanotiya, C.S.; Kalra, A. Plant Growth-Promoting Rhizobacteria Enhance Wheat Salt and Drought Stress Tolerance by Altering Endogenous Phytohormone Levels and TaCTR1/TaDREB2 Expression. Physiol. Plant 2017, 161, 502–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.M.; Haque, E.; Paul, N.C.; Khaleque, M.A.; Al-Garni, S.M.; Rahman, M.; Islam, M.T. Enhancement of growth and grain yield of rice in nutrient deficient soils by rice probiotic bacteria. Rice Sci. 2017, 24, 264–273. [Google Scholar] [CrossRef]
- Rima, F.S.; Biswas, S.; Sarker, P.K.; Islam, M.D.; Seraj, Z.I. Bacteria endemic to saline coastal belt and their ability to mitigate the effects of salt stress on rice growth and yields. Ann. Microbiol. 2018, 68, 525–535. [Google Scholar] [CrossRef]
- Vives-Peris, V.; Gomez-Cadenas, A.; Perez-Clemente, R.M. Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria Pseudomonas putida and Novosphingobium sp. Plant Cell Rep. 2018, 37, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Vimal, S.R.; Patel, V.K.; Singh, J.S. Plant growth promoting Curtobacterium albidum strain SRV4, an agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol. Indic. 2019, 105, 553–562. [Google Scholar] [CrossRef]
- El-Akhdar, I.; Elsakhawy, T.; Abo-Koura, H.A. Alleviation of salt stress on wheat (Triticum aestivum L.) by plant growth promoting bacteria strains Bacillus halotolerans MSR-H4 and Lelliottia amnigena MSR-MJ. Adv. Microbiol. 2020, 20, 44–58. [Google Scholar] [CrossRef] [Green Version]
- El-Nahrawy, S.; Yassin, M. Response of different cultivars of wheat plants (Triticum aestivum L.) to inoculation by Azotobacter sp. under salinity stress conditions. J. Adv. Microbiol. 2020, 20, 59–79. [Google Scholar] [CrossRef] [Green Version]
- Sapre, S.; Gontia-Mishra, I.; Tiwari, S. Plant growth-promoting rhizobacteria ameliorates salinity stress in pea (Pisum sativum). J. Plant Growth Regul. 2021, 41, 647–656. [Google Scholar] [CrossRef]
- Saravanakumar, D.; Kavino, M.; Raguchander, T.; Subbian, P.; Samiyappan, R. Plant growth promoting bacteria enhance water stress resistance in green gram plants. Acta Physiol. Plant. 2011, 33, 203–209. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, S.D. Induction of drought stress resistance by multi-functional PGPR Bacillus licheniformis K11 in pepper. Plant Pathol. J. 2013, 29, 201. [Google Scholar] [CrossRef]
- Castillo, P.; Escalante, M.; Gallardo, M.; Alemano, S.; Abdala, G. Effects of bacterial single inoculation and co-inoculation on growth and phytohormone production of sunflower seedlings under water stress. Acta Physiol. Plant. 2013, 35, 2299–2309. [Google Scholar] [CrossRef]
- Grover, M.; Madhubala, R.; Ali, S.Z.; Yadav, S.K.; Venkateswarlu, B. Influence of Bacillus spp. strains on seedling growth and physiological parameters of sorghum under moisture stress conditions. J. Basic Microbiol. 2014, 54, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Abd El-Daim, I.A.; Copolovici, L.; Tanilas, T.; Kannaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenstrom, E.; Niinemets, U. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments, enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 2014, 9, e96086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarma, R.K.; Saikia, R. Alleviation of drought stress in mung bean by strain Pseudomonas aeruginosa GGRJ. Plant Soil 2014, 377, 111–126. [Google Scholar] [CrossRef]
- Fan, X.; Hu, H.; Huang, G.; Huang, F.; Li, Y.; Palta, J. Soil inoculation with Burkholderia sp. LD-11 has positive effect on water-use efficiency in inbred lines of maize. Plant Soil 2015, 390, 337–349. [Google Scholar] [CrossRef]
- Cohen, A.C.; Bottini, R.; Pontin, M.; Berli, F.J.; Moreno, D.; Boccanlandro, H.; Travaglia, C.N.; Piccoli, P.N. Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 2015, 153, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Rawat, S. Rhizoremediation of heavy metal-and xenobiotic-contaminated soil, an eco-friendly approach. In Removal of Emerging Contaminants through Microbial Processes; Springer: Singapore, 2021; pp. 95–113. [Google Scholar]
- Chitara, M.K.; Chauhan, S.; Singh, R.P. Bioremediation of Polluted Soil by Using Plant Growth–Promoting Rhizobacteria. In Microbial Rejuvenation of Polluted Environment; Springer: Singapore, 2021; pp. 203–226. [Google Scholar]
- Pires, C.; Franco, A.R.; Pereira, S.I.; Henriques, I.; Correia, A.; Magan, N.; Castro, P.M. Metal (loid)-contaminated soils as a source of culturable heterotrophic aerobic bacteria for remediation applications. Geomicrobiol. J. 2017, 34, 760–768. [Google Scholar] [CrossRef]
- Mishra, J.; Singh, R.; Arora, N.K. Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front. Microbiol. 2017, 8, 1706. [Google Scholar] [CrossRef] [Green Version]
- Seneviratne, M.; Seneviratne, G.; Madawala, H.M.; Vithanage, M. Role of rhizospheric microbes in heavy metal uptake by plants. In Agro-Environmental Sustainability; Springer: Cham, Switzerland, 2017; pp. 147–163. [Google Scholar]
- Fomina, M.; Hillier, S.; Charnock, J.M.; Melville, K.; Alexander, I.J.; Gadd, G.M. Role of oxalic acid overexcretion in transformations of toxic metal minerals by Beauveria caledonica. Appl. Environ. Microbiol. 2005, 71, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Manko-Jurkowska, D.; Ostrowska-Ligeza, E.; Gorska, A.; Głowacka, R. The role of bio surfactants in soil remediation. Zesz. Probl. Postępów Nauk. Rol. 2019, 596, 33–43. [Google Scholar]
- Yuanfan, H.; Jin, Z.; Qing, H.; Qian, W.; Jiandong, J.; Shunpeng, L. Characterization of a fenpropathrin-degrading strain and construction of a genetically engineered microorganism for simultaneous degradation of methyl parathion and fenpropathrin. J. Environ. Manag. 2010, 91, 2295–2300. [Google Scholar] [CrossRef]
- Rebello, S.; Nathan, V.K.; Sindhu, R.; Binod, P.; Awasthi, M.K.; Pandey, A. Bioengineered microbes for soil health restoration: Present status and future. Bioengineered 2021, 12, 12839–12853. [Google Scholar] [CrossRef] [PubMed]
- Phour, M.; Sindhu, S.S. Mitigating abiotic stress: Microbiome engineering for improving agricultural production and environmental sustainability. Planta 2022, 256, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.E.; Teo, W.F.A.; Teoh, E.Y.; Tan, B.C. Microbiome engineering and plant biostimulants for sustainable crop improvement and mitigation of biotic and abiotic stresses. Discov. Food 2022, 2, 1–23. [Google Scholar] [CrossRef]
- Kumar, N.M.; Muthukumaran, C.; Sharmila, G.; Gurunathan, B. Genetically modified organisms and its impact on the enhancement of bioremediation. In Bioremediation, Applications for Environmental Protection and Management; Springer: Singapore, 2018; pp. 53–76. [Google Scholar]
- Bell-Dereske, L.; Takacs-Vesbach, C.; Kivlin, S.N.; Emery, S.M.; Rudgers, J.A. Leaf endophytic fungus interacts with precipitation to alter belowground microbial communities in primary successional dunes. FEMS Microbiol. Ecol. 2017, 93, fix036. [Google Scholar] [CrossRef] [Green Version]
- Biswas, R.; Sarkar, A. ‘Omics’ tools in soil microbiology, the state of the art. Advances in soil microbiology, Recent trends and future prospects. Microorg. Sustain. 2018, 3, 35–64. [Google Scholar] [CrossRef]
- Feng, Y.Y. Omics breakthroughs for environmental microbiology. Omics Env. Microbiol. 2013, 40, 18–33. [Google Scholar]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—from metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Zaheer, R.; Noyes, N.; Ortega Polo, R.; Cook, S.R.; Marinier, E.; Van Domselaar, G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 2018, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Deng, Y.; Zhang, X.; Wang, X.; Kang, G.; Bai, L.; Huang, H. Biases in prokaryotic community amplicon sequencing affected by DNA extraction methods in both saline and non-saline soil. Front. Microbiol. 2018, 9, 1796. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, C.; Li, X.; Yang, X.; Zhao, L.; Liu, L.; Zhu, C.; Li, R. Linking plant ecological stoichiometry with soil nutrient and bacterial communities in apple orchards. Appl. Soil Ecol. 2018, 126, 1–10. [Google Scholar] [CrossRef]
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordonez, A. Biotechnological applications of functional metagenomics in the food and pharmaceutical industries. Front. Microbiol. 2015, 6, 672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-throughput metagenomic technologies for complex microbial community analysis, open and closed formats. MBio 2015, 6, e02288-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wooley, J.C.; Godzik, A.; Friedberg, I. A primer on metagenomics. PLOS Comput. Biol. 2010, 6, e1000667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakya, M.; Lo, C.C.; Chain, P.S. Advances and challenges in metatranscriptomic analysis. Front Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [Green Version]
- Wolf, J.B. Principles of transcriptome analysis and gene expression quantification, an RNA-seq tutorial. Mol. Ecol. Resour. 2013, 13, 559–572. [Google Scholar] [CrossRef]
- Callister, S.J.; Fillmore, T.L.; Nicora, C.D.; Shaw, J.B.; Purvine, S.O.; Orton, D.J.; White, R.A., III; Moore, R.J.; Burnet, M.C.; Nakayasu, E.S.; et al. Addressing the challenge of soil metaproteome complexity by improving metaproteome depth of coverage through two-dimensional liquid chromatography. Soil Biol. Biochem. 2018, 125, 290–299. [Google Scholar] [CrossRef]
- Heaven, M.W.; Benheim, D. Soil microbial metabolomics. In Microbial Metabolomics, Applications in Clinical, Environmental, and Industrial Microbiology; Beale, D.J., Kouremenos, K.A., Palombo, E.A., Eds.; Springer: Cham, Switzerland, 2016; pp. 147–198. [Google Scholar]
- Nesme, J.; Achouak, W.; Agathos, S.N.; Bailey, M.; Baldrian, P.; Brunel, D.; Frostegard, A.; Heulin, T.; Jansson, J.K.; Jurkevitch, E.; et al. Back to the future of soil metagenomics. Front. Microbiol. 2016, 7, 73. [Google Scholar] [CrossRef]


| Stress | Microbes | Mechanism of Mitigation of Abiotic Stress | Beneficial Host | Reference |
|---|---|---|---|---|
| Drought and Salinity | B. subtilis; A. protophormiae; D. Natronolimnaea | Production of IAA, abscisic acid/ACC deaminase level regulation, and modulation of gene expression encoding for CTR1/DREB2 proteins | T. aestivum | Barnawal et al., 2017 [133] |
| Salinity | Sphingomonas sp. | Endogenous phytohormone regulation (salicylic acid, abscisic acid, and jasmonic acid) | Solanum pimpinellifolium | Khan et al., 2017 [134] |
| Salinity | Halobacillus dabanensis; Halobacillus sp. | Physiological modulation and Osmo-regulation | Oryza sativa | Rima et al., 2018 [135] |
| Salinity | P. putida Novosphingobium sp. | Reduction of ABA and SA levels, inhibition of proline and chloride accretion | Citrus | Vives-Peris et al., 2018 [136] |
| Salinity | Curtobacterium albidum | Inducing systemic tolerance | O. sativa | Vimal et al., 2019 [137] |
| Salinity | B. halotolerans; Lelliottia amnigena | Judicious employment of K+ and Na+ in root and shoot uptake | T. aestivum | El-Akhdar et al., 2020 [138] |
| Salinity | Azotobacter sp. | Improved physiological attributes and perked-up growth aspects | T. aestivum | El-Nahrawy et al., 2020 [139] |
| Salinity | Acinetobacter bereziniae; Enterobacter ludwigii; Alcaligenes faecalis | Inflection of proline content, chlorophyll, total soluble sugars | Pisum sativum | Sapre et al., 2021 [140] |
| Drought | P. fluorescens; B. Subtilis | Proline accretion, Enzyme activation | Vigna radiata | Saravanakumar et al., 2010 [141] |
| Drought | B. licheniformis | upregulation of stress-related genes and Stress protein activation | Capsicum annuum | Lim et al., 2013 [142] |
| Drought | Achromobacter xylosoxidans; B. Pumilis | Phytohormone Secretion and regulation | Helianthus annuus | Castillo et al., 2013 [143] |
| Drought | Bacillus spp. | Enriched relative water content, higher retention of soil moisture, better plant physiology and proline contents | Sorghum bicolor | Grover et al., 2014 [144] |
| Drought | B. thuringiensis; P. polymyxa | Enhanced volatile products such as β-pinene, benzaldehyde, and geranyl acetone | Triticum aestivum | Timmusk et al., 2014 [145] |
| Drought | P. aeruginosa | Elevated antioxidant levels, enriched cell osmolytes and upregulating the stress-responsive genes | V. radiata | Sarma et al., 2013 [146] |
| Drought | Burkholderia sp. | Improved plant physiology and heightened plant growth regulators | Zea mays | Fan et al., 2015 [147] |
| Drought | A. brasilense | Physiological and biochemical alterations encircling the accent of photosynthetic pigments, abscisic acid levels, lipid peroxidation and proline content | A. Thaliana | Cohen et al., 2015 [148] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, A.M.; Khan, I.M.; Shah, T.I.; Bangroo, S.A.; Kirmani, N.A.; Nazir, S.; Malik, A.R.; Aezum, A.M.; Mir, Y.H.; Hilal, A.; et al. Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate. Land 2022, 11, 1887. https://doi.org/10.3390/land11111887
Shah AM, Khan IM, Shah TI, Bangroo SA, Kirmani NA, Nazir S, Malik AR, Aezum AM, Mir YH, Hilal A, et al. Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate. Land. 2022; 11(11):1887. https://doi.org/10.3390/land11111887
Chicago/Turabian StyleShah, Aanisa Manzoor, Inayat Mustafa Khan, Tajamul Islam Shah, Shabir Ahmed Bangroo, Nayar Afaq Kirmani, Shaista Nazir, Abdul Raouf Malik, Aziz Mujtaba Aezum, Yasir Hanif Mir, Aatira Hilal, and et al. 2022. "Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate" Land 11, no. 11: 1887. https://doi.org/10.3390/land11111887
APA StyleShah, A. M., Khan, I. M., Shah, T. I., Bangroo, S. A., Kirmani, N. A., Nazir, S., Malik, A. R., Aezum, A. M., Mir, Y. H., Hilal, A., & Biswas, A. (2022). Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate. Land, 11(11), 1887. https://doi.org/10.3390/land11111887







