UV Disinfection of Hand-Rinse Greywater and Performance Testing Using Indigenous Staphylococcus spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hand-Rinse Samples to Recover Skin-Bacteria and Evaluate Their UV-Resistance
2.2. Hand-Rinse Isolates
2.3. Oxybenzone (BP3)
2.4. Control Experiments
2.5. Statistical Analysis
3. Results and Discussion
3.1. Raw Hand-Rinse
- Pre-treatment prior to UV-C irradiation is likely necessary in order to achieve more than a 2-log10 reduction of endogenous bacteria due to particulate shielding and organic quenching effects; and
- Equation (1) describing the delivered UV-C intensity requires adjustment to account for shielding/quenching from high turbidity/organics in greywater samples.
3.2. Hand-Rinse Isolates
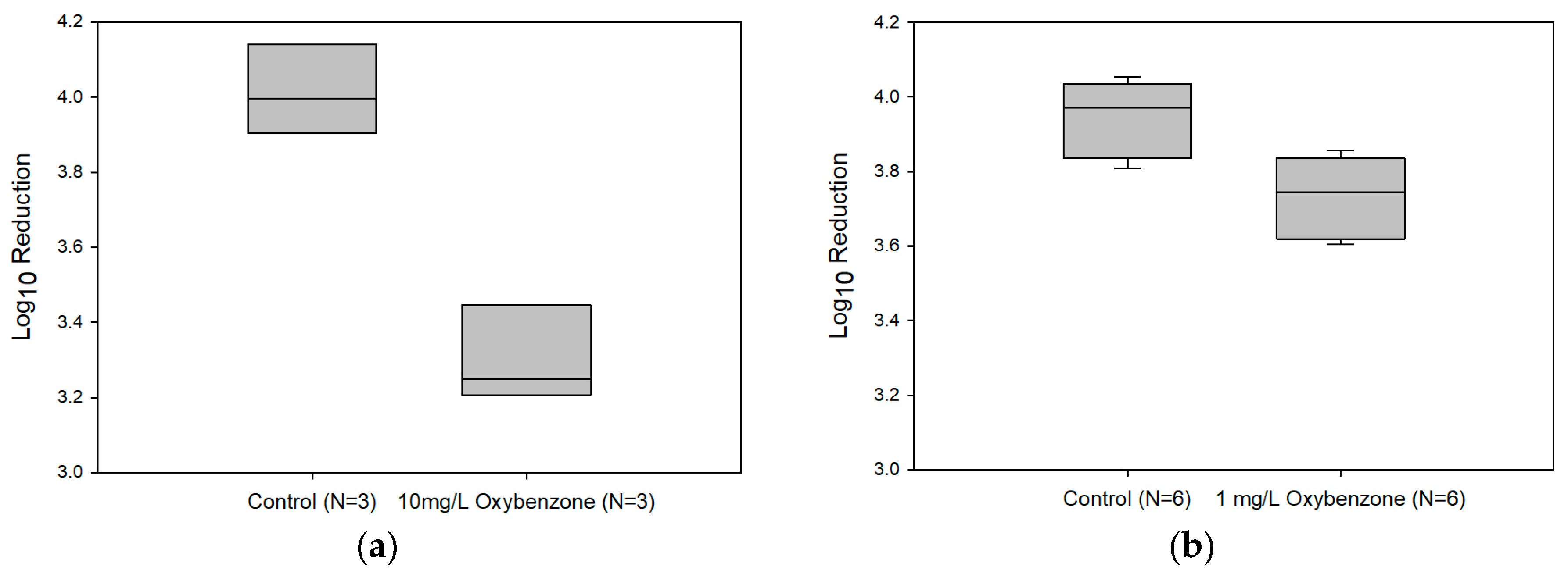
3.3. Affects of Oxybenzone on UV-C Efficacy
4. Summary and Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- UN Water. Coping with Water Scarcity: Challenge of the Twenty-First Century; Food and Agriculture Organization of the United Nations: Rome, Italy, 2007. [Google Scholar]
- Strengers, Y.; Maller, C. Materialising energy and water resources in everyday practices: Insights for securing supply systems. Glob. Environ. Chang. 2012, 22, 754–763. [Google Scholar] [CrossRef]
- Chang, N.B.; Qi, C.; Yang, Y.J. Optimal expansion of a drinking water infrastructure system with respect to carbon footprint, cost-effectiveness and water demand. J. Environ. Manag. 2012, 110, 194–206. [Google Scholar] [CrossRef] [PubMed]
- National Research Council of the National Academics. Using Graywater and Stormwater to Enhance Local Water Supplies: An Assessment of Risks, Costs, and Benefits; National Research Council of the National Academics: Washington, DC, USA, 2016; p. 420. [Google Scholar]
- Ashbolt, N.J. The short pipe path—Safe water, energy & nutrient recovery. Proc. Water Environ. Fed. 2011, 2011, 1233–1241. [Google Scholar]
- World Health Organization. WHO Guidelines for the Safe Use of Wastewater, Excreta and Greywater; World Health Organization: Geneva, Switzerland, 2006. [Google Scholar]
- Shoults, D.C.; Ashbolt, N.J. Total staphylococci as performance surrogate for greywater treatment. Environ. Sci. Pollut. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, B.D.; Ashbolt, N.J.; Garland, J.L.; Keely, S.; Wendell, D. Human mitochondrial DNA and endogenous bacterial surrogates for risk assessment of graywater reuse. Environ. Sci. Technol. 2014, 48, 7993–8002. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.P.; Brinkman, N.E.; Zimmerman, B.D.; Wendell, D.; Ekeren, K.M.; De Long, S.K.; Sharvelle, S.; Garland, J.L. Characterization of the relative importance of human- and infrastructure-associated bacteria in grey water: A case study. J. Appl. Microbiol. 2015, 119, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Benami, M.; Busgang, A.; Gillor, O.; Gross, A. Quantification and risks associated with bacterial aerosols near domestic greywater-treatment systems. Sci. Total Environ. 2016, 562, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Song, I.; Oh, H.; Jong, J.; Park, J.; Choung, Y. A laboratory-scale graywater treatment system based on a membrane filtration and oxidation process—Characteristics of graywater from a residential complex. Desalination 2009, 238, 347–357. [Google Scholar] [CrossRef]
- Casanova, L.M.; Gerba, C.P.; Karpiscak, M. Chemical and microbial characterization of household graywater. Environ. Sci. Health 2001, A36, 395–401. [Google Scholar] [CrossRef]
- Gilboa, Y.; Friedler, E. UV disinfection of rbc-treated light greywater effluent: Kinetics, survival and regrowth of selected microorganisms. Water Res. 2007, 42, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Burrows, W.D.; Schmidt, M.O.; Carnevale, R.M.; Schaub, S.A. Nonpotable reuse: Development of health criteria and technologies for shower water recycle. Water Sci. Technol. 1991, 24, 81–88. [Google Scholar]
- Gross, A.; Kaplan, D.; Baker, K. Removal of chemical and microbiological contaminants from domestic greywater using a recycled vertical flow bioreactor (rvfb). Ecol. Eng. 2007, 31, 107–114. [Google Scholar] [CrossRef]
- Maimon, A.; Friedler, E.; Gross, A. Parameters affecting greywater quality and its safety for reuse. Sci. Total Environ. 2014, 487, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.M.; Freire, M.O.; Gabrilska, R.A.; Rumbaugh, K.P.; Lemon, K.P. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 2016, 7, 1230. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, J.; Ishibashi, M.; Kawashima, M.; Takagi, Y.; Ichikawa, Y.; Imokawa, G. Decreased levels of sphingosine, a natural antimicrobial agent, may be associated with vulnerability of the stratum corneum from patients with atopic dermatitis to colonization by Staphylococcus aureus. J. Investig. Dermatol. 2002, 119, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Coates, R.; Moran, J.; Horsburgh, M.J. Staphylococci: Colonizers and pathogens of human skin. Future Microbiol. 2014, 9, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Winward, G.P.; Avery, L.M.; Stephenson, T.; Jefferson, B. Ultraviolet (UV) disinfection of grey water: Particle size effects. Environ. Technol. 2008, 29, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Homem, V.; Alves, A.; Santos, L. A review of organic UV-filters in wastewater treatment plants. Environ. Int. 2016, 86, 24–44. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Ultraviolet Disinfection Guidance Manual for the Final Long Term 2 Enhanced Surface Water Treatment Rule; U.S. Environmental Protection Agency: Washington, DC, USA, 2006.
- NSF International. NSF/ANSI 55: Ultraviolet Microbiological Water Treatment Systems; NSF International: Ann Arbor, MI, USA, 2014. [Google Scholar]
- Ewald, S. Evaluation of a rapid tube lysostaphin test to differentiate between staphylococci and micrococci. Int. J. Food Microbiol. 1986, 3, 31–41. [Google Scholar] [CrossRef]
- Hardy Diagnostics. Lysostaphin Differentiation Disks: Instructions for Use. Available online: https://catalog.hardydiagnostics.com/cp_prod/Content/hugo/LysostaphinDiffDisks.htm (accessed on 10 March 2017).
- Palmquist, H.; Hanæus, J. Hazardous substances in separately collected grey- and blackwater from ordinary swedish households. Sci. Total Environ. 2005, 348, 151–163. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. USEPA Manual of Methods for Virology; Environmental Protection Agency: Washington, DC, USA, 2001; Chapter 16.
- Liu, W.J.; Zhang, Y.J. Effects of UV intensity and water turbidity on microbial indicator inactivation. J. Environ. Sci. (China) 2006, 18, 650–653. [Google Scholar] [PubMed]
- Abshire, R.L.; Dunton, H. Resistance of selected strains of Pseudomonas aeruginosa to low-intensity ultravioloet radiation. Appl. Environ. Microbiol. 1981, 41, 1419–1423. [Google Scholar] [PubMed]
- Rodríguez, M.; Núñez, F.; Córdoba, J.J.; Sanabria, C.; Bermúdez, E.; Asensio, M.A. Characterization of Staphylococcus spp. and Micrococcus spp. isolated from Iberian ham throughout the ripening process. Int. J. Food Microbiol. 1994, 24, 329–335. [Google Scholar] [CrossRef]
- Quiloan, M.L.G.; Vu, J.; Carvalho, J. Enterococcus faecalis can be distinguished from Enterococcus faecium via differential susceptibility to antibiotics and growth and fermentation characteristics on mannitol salt agar. Front. Biol. 2012, 7, 167–177. [Google Scholar] [CrossRef]
- Kloos, W.E.; Musselwhite, M.S. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 1975, 30, 381–395. [Google Scholar] [PubMed]
- Kasprzyk-Hordern, B.; Dinsdale, R.M.; Guwy, A.J. The removal of pharmaceuticals, personal care products, endocrine disruptors and illicit drugs during wastewater treatment and its impact on the quality of receiving waters. Water Res. 2009, 43, 363–380. [Google Scholar] [CrossRef] [PubMed]
- Christian, D. Pharmaceuticals and personal care products: An overview. Pipeline 2007, 18, 1–8. [Google Scholar]
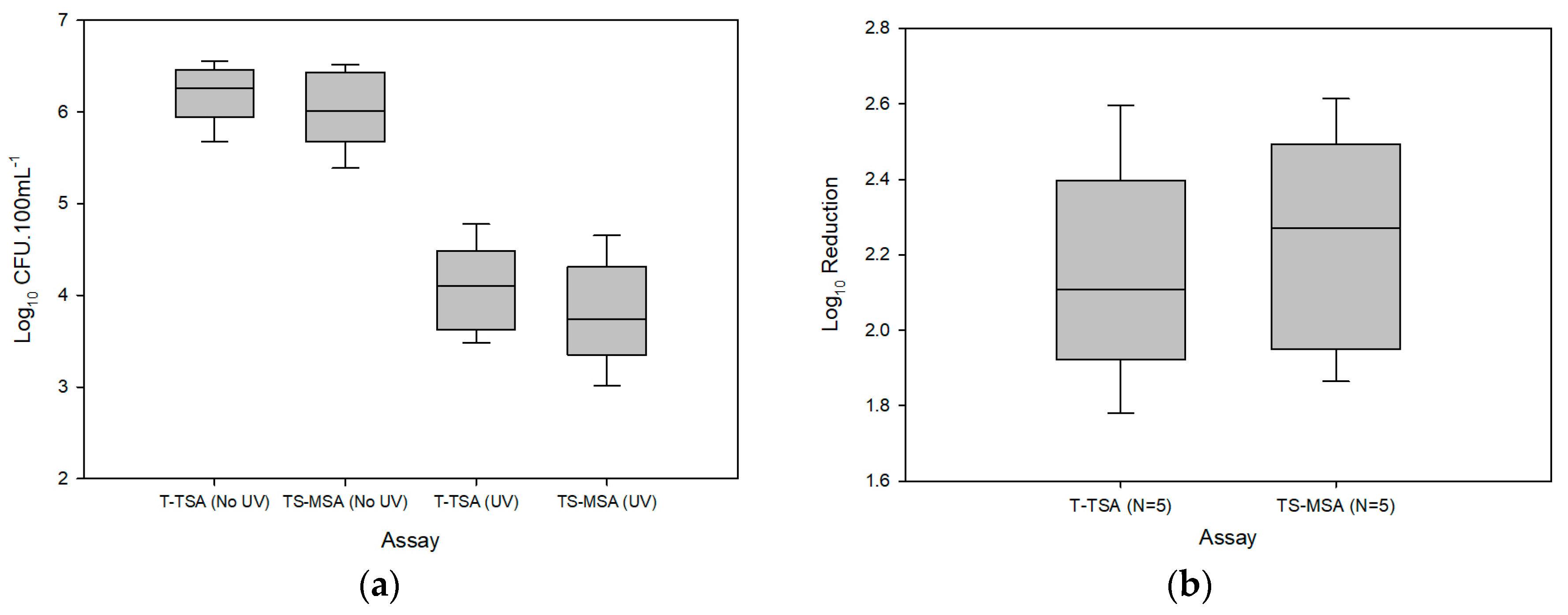
| Assay | Log10 CFU·100 mL−1 ± (SD) | ||
|---|---|---|---|
| 0 mJ·cm−2 | 220 mJ·cm−2 | Reduction | |
| T-TSA | 6.2 ± (0.3) | 4.1 ± (0.5) | 2.1 ± (0.3) |
| TS-MSA | 6.0 ± (0.4) | 3.8 ± (0.6) | 2.2 ± (0.3) |
| Paired t-test | 0.04 a | 0.04 a | 0.112 a |
| Bacteria | Log10 Reduction at 11.0 mJ·cm−2 ± (SD) |
|---|---|
| S. aureus (ATCC 25923) | 4.9 ± (0.0) |
| S. aureus (i) | >5.2 |
| S. capitis (ii) | >5.7 |
| S. capitis (iii) | >6.3 |
| S. lentus (unknown) | 5.0 ± (0.0) |
| S. epidermidis (ii) | >5.7 |
| S. epidermidis (iiia) | >6.1 |
| S. epidermidis (iiib) | >5.6 |
| S. haemolyticus (iv) | 3.4 ± (0.1) |
| S. haemolyticus (v) | 4.4 ± (0.1) |
| S. hominis (v) | 1.6 ± (0.1) |
| S. pasteuri (iii) | >5.8 |
| S. pasteuri (iv) | >5.5 |
| S. pasteuri (v) | 5.1 ± (0.1) |
| S. warneri (i) | >5.7 |
| S. warneri (ii) | >5.5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shoults, D.C.; Ashbolt, N.J. UV Disinfection of Hand-Rinse Greywater and Performance Testing Using Indigenous Staphylococcus spp. Water 2017, 9, 963. https://doi.org/10.3390/w9120963
Shoults DC, Ashbolt NJ. UV Disinfection of Hand-Rinse Greywater and Performance Testing Using Indigenous Staphylococcus spp. Water. 2017; 9(12):963. https://doi.org/10.3390/w9120963
Chicago/Turabian StyleShoults, David C., and Nicholas J. Ashbolt. 2017. "UV Disinfection of Hand-Rinse Greywater and Performance Testing Using Indigenous Staphylococcus spp." Water 9, no. 12: 963. https://doi.org/10.3390/w9120963
APA StyleShoults, D. C., & Ashbolt, N. J. (2017). UV Disinfection of Hand-Rinse Greywater and Performance Testing Using Indigenous Staphylococcus spp. Water, 9(12), 963. https://doi.org/10.3390/w9120963





