Microbial Source Tracking in a Watershed Dominated by Swine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
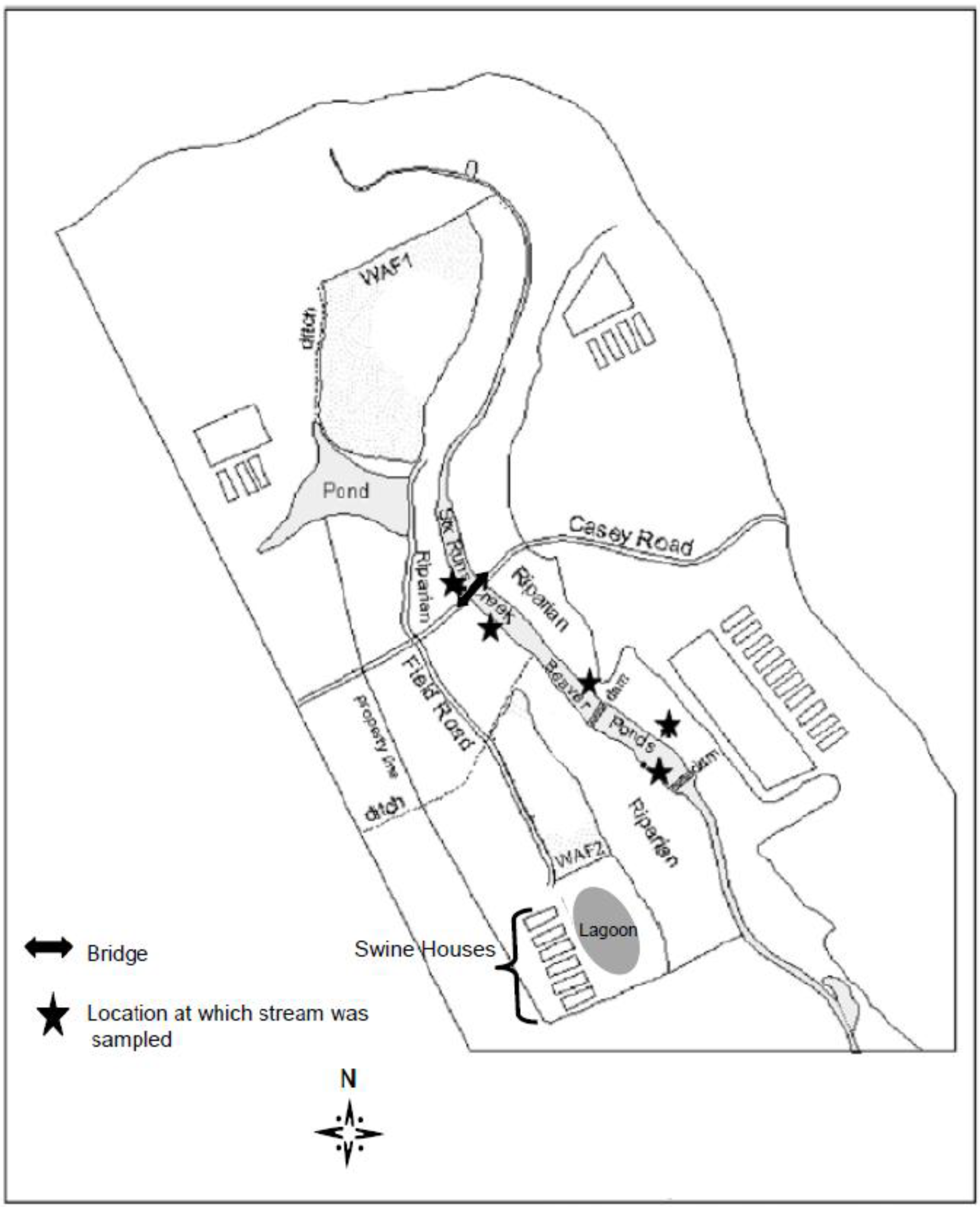
2.2. Host-origin Isolates
2.3. Water Sample Isolates
2.4. Antibiotic Resistance Analysis (ARA)
| Antibiotics | Concentrations (µg/mL) | No. of Variables |
|---|---|---|
| Erythromycin | 60, 70, 90 and 100 | 4 |
| Neomycin | 2.5, 5.0 and 10 | 3 |
| Oxytetracycline | 2.5, 5.0, 7.5, 10 and 15 | 5 |
| Streptomycin | 2.5, 5.0, 7.5, 10 and 15 | 5 |
| Tetracycline | 2.5, 5.0, 7.5, 10 and 15 | 5 |
| Cephalothin | 15, 25 and 35 | 4 |
| Apramycin | 8, 16, 32, 64 and 128 | 3 |
| Trimethoprim | 8, 16, 32, 64 and 128 | 5 |
| Rifamicin | 60, 75 and 90 | 3 |
| Total antibiotic variables | 38 | |
| Control | No antibiotic | 2 |
2.5. Host-Origin Library
2.6. Calculation of ARCC, AFM and MDP
3. Results
3.1. Fecal Coliform Monitoring
3.2. Host Origin Library
3.3. Host Source Identification of E. coli from Six Runs Creek
| Categories into which isolates from the library were classified | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of Isolates | Beaver | Bird | Cattle | Deer | Dog | Lagoon | Nutria | Raccoon | Swine | Total (ni) | |||||||||||
| CC* | MC† | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | ||||
| Beaver | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |||||||||||
| Bird | 0 | 24 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 27 | |||||||||||
| Cattle | 0 | 1 | 183 | 0 | 0 | 15 | 1 | 0 | 17 | 217 | |||||||||||
| Deer | 0 | 0 | 1 | 34 | 0 | 0 | 0 | 0 | 0 | 35 | |||||||||||
| Dog | 0 | 0 | 1 | 0 | 22 | 0 | 0 | 0 | 1 | 24 | |||||||||||
| Lagoon | 0 | 0 | 19 | 0 | 0 | 246 | 0 | 0 | 59 | 324 | |||||||||||
| Nutria | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 2 | 0 | 25 | |||||||||||
| Raccoon | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 3 | |||||||||||
| Swine | 0 | 0 | 29 | 0 | 0 | 56 | 0 | 0 | 204 | 289 | |||||||||||
| Total | 4 | 0 | 24 | 1 | 183 | 51 | 34 | 0 | 22 | 0 | 246 | 72 | 23 | 2 | 2 | 2 | 204 | 78 | |||
| Total isolates (n) | 948 | ||||||||||||||||||||
| % n CC=(100)(CC)/ni) | 100.0 | 88.9 | 84.3 | 97.1 | 91.7 | 75.9 | 92.0 | 66.7 | 70.6 | 85.2‡ | |||||||||||
| %n MC=(100)(MC)/(n-ni) | 0.0 | 0.1 | 5.7 | 0.0 | 0.0 | 8.2 | 0.2 | 0.2 | 9.0 | ||||||||||||
| Categories into which isolates from the library were classified | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of Isolates | Beaver | Bird | Cattle | Deer | Dog | Lagoon | Nutria | Raccoon | Swine | Total (ni) | |||||||||||
| CC* | MC† | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | CC | MC | ||||
| Beaver | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | |||||||||||
| Bird | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | |||||||||||
| Cattle | 0 | 0 | 136 | 0 | 0 | 1 | 1 | 0 | 0 | 138 | |||||||||||
| Deer | 0 | 0 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | 28 | |||||||||||
| Dog | 0 | 0 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 20 | |||||||||||
| Lagoon | 0 | 0 | 0 | 0 | 0 | 125 | 0 | 0 | 9 | 134 | |||||||||||
| Nutria | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 1 | 0 | 23 | |||||||||||
| Raccoon | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 5 | |||||||||||
| Swine | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 93 | 98 | |||||||||||
| Total | 4 | 0 | 19 | 0 | 136 | 1 | 28 | 0 | 20 | 0 | 125 | 6 | 22 | 3 | 3 | 1 | 93 | 9 | |||
| Total isolates(n) | 470 | ||||||||||||||||||||
| % n CC=(100CC)/(ni) | 100.0 | 100.0 | 99.3 | 100.0 | 100.0 | 95.4 | 88.0 | 75.0 | 91.2 | 94.3‡ | |||||||||||
| %n MC=(100)MC/(n-ni) | 0.0 | 0.0 | 0.3 | 0.0 | 0.0 | 1.8 | 0.7 | 0.2 | 2.4 | ||||||||||||
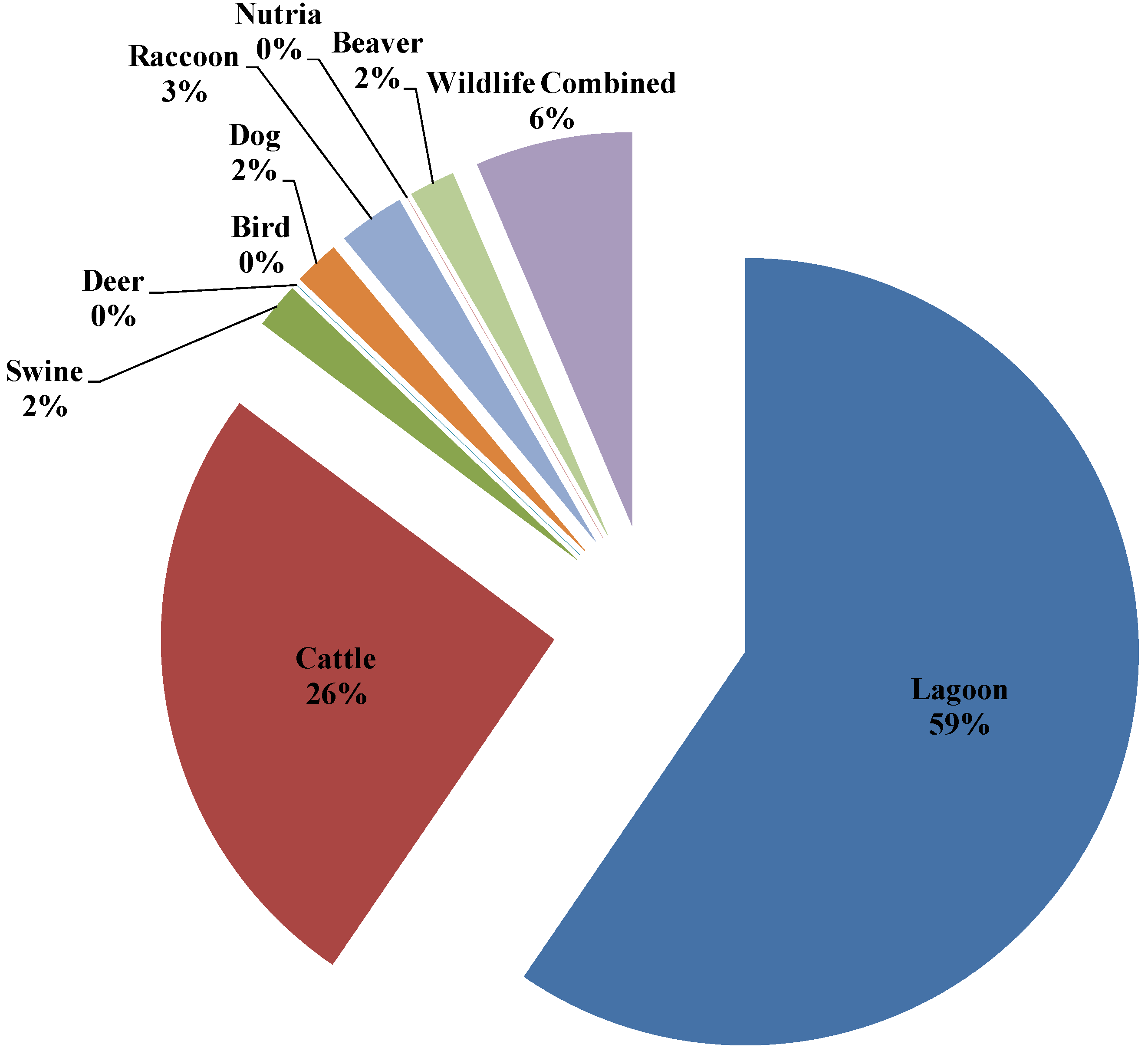
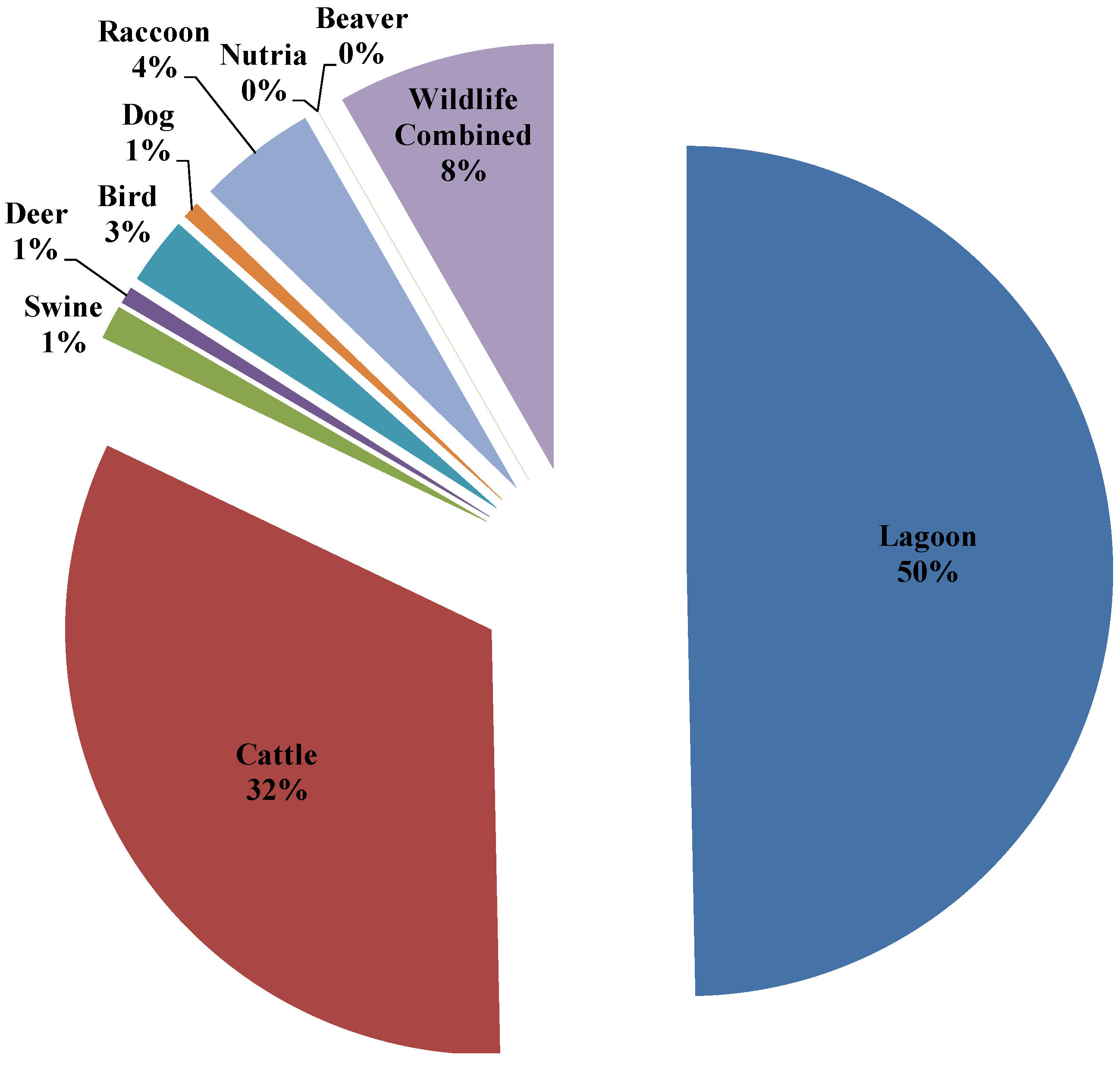
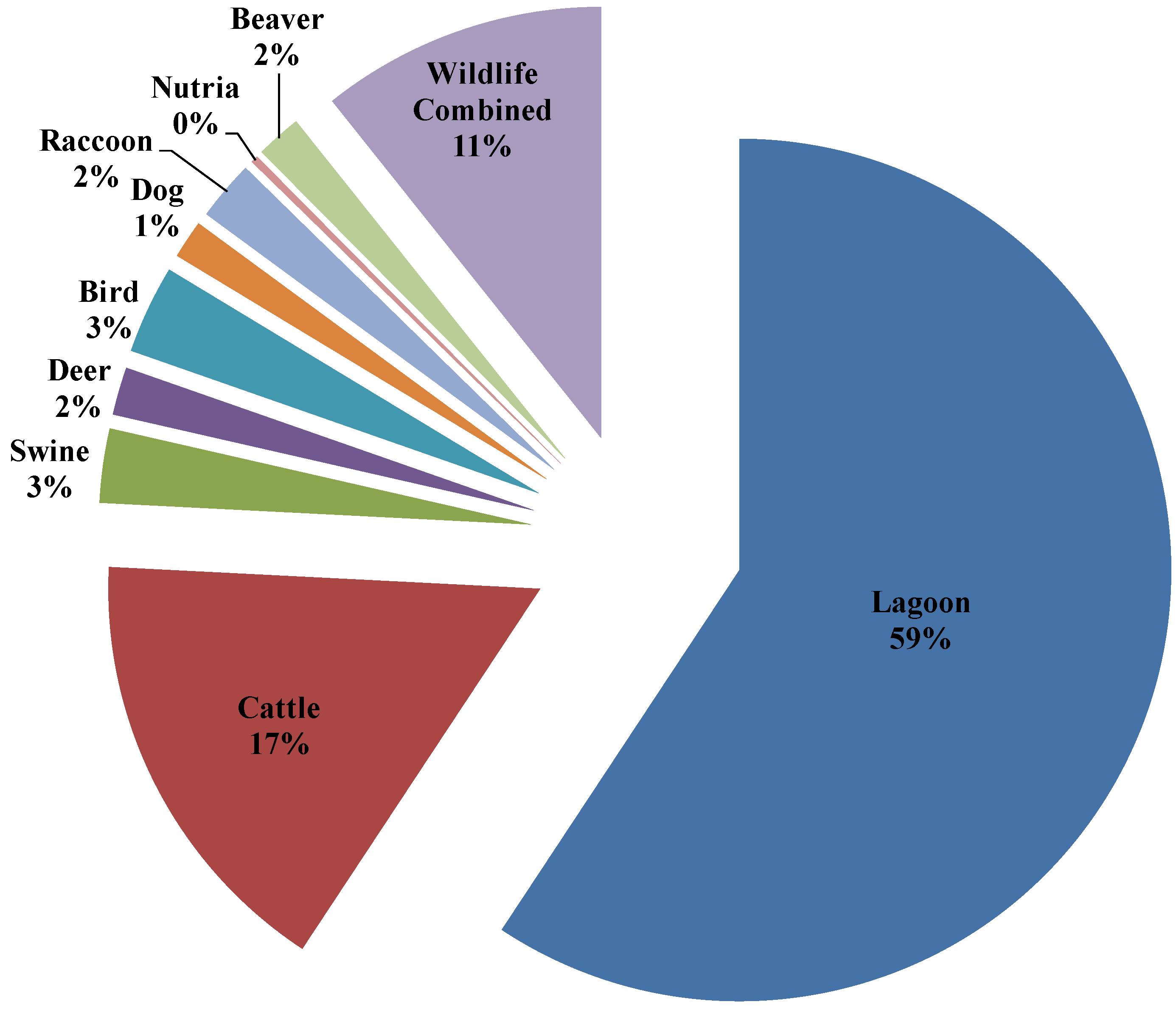
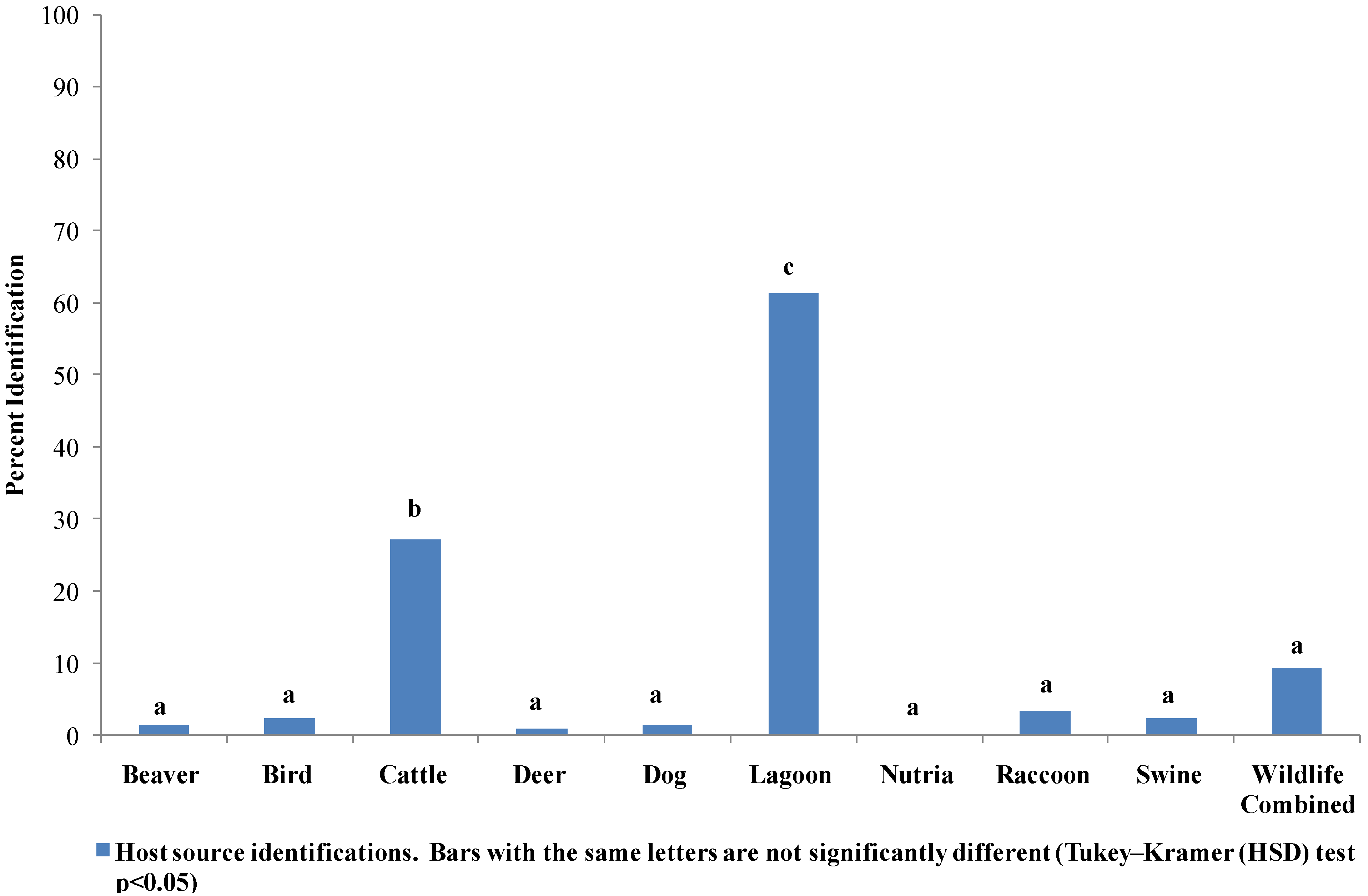
3.4. Seasonal Host Source Allocations
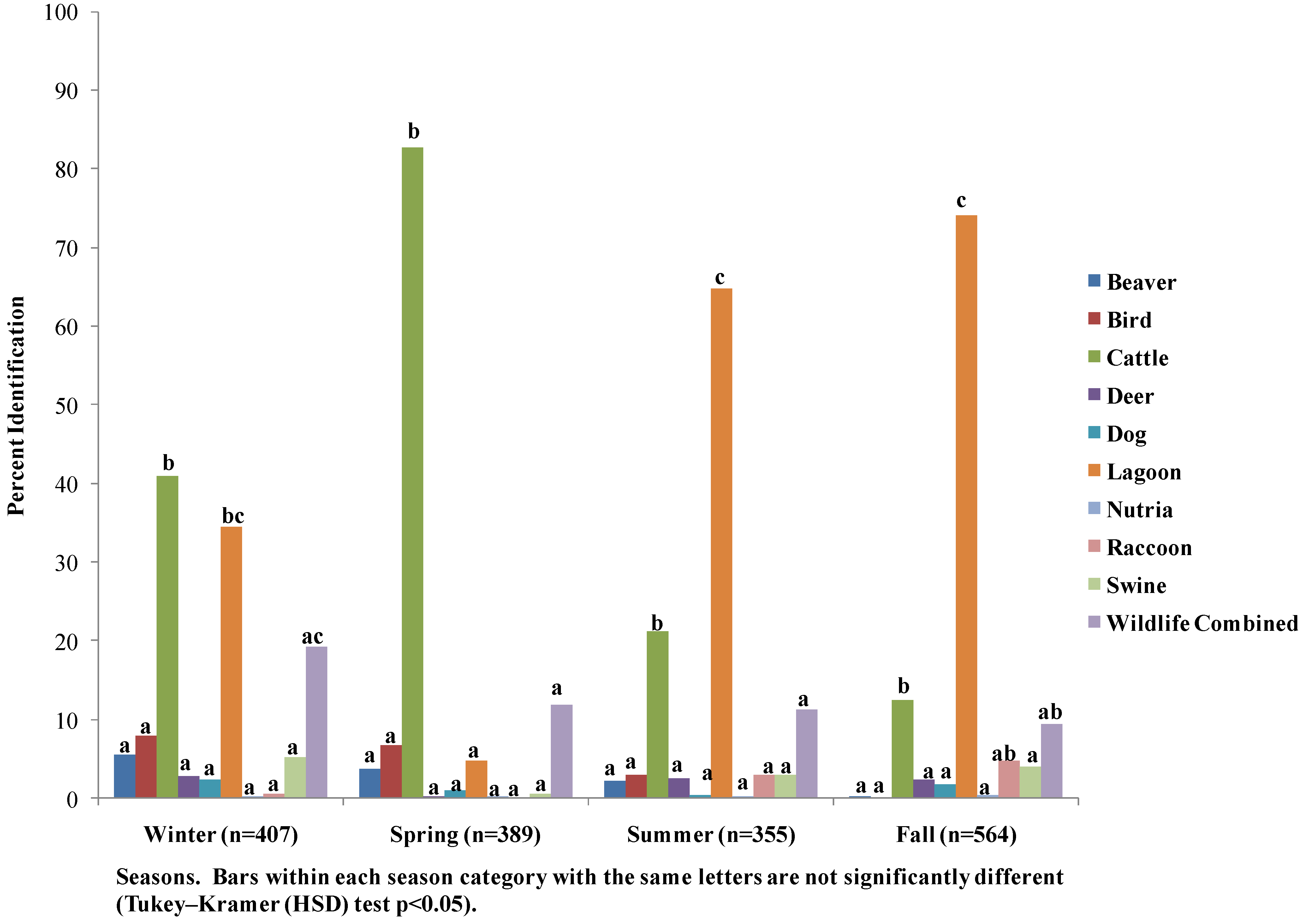
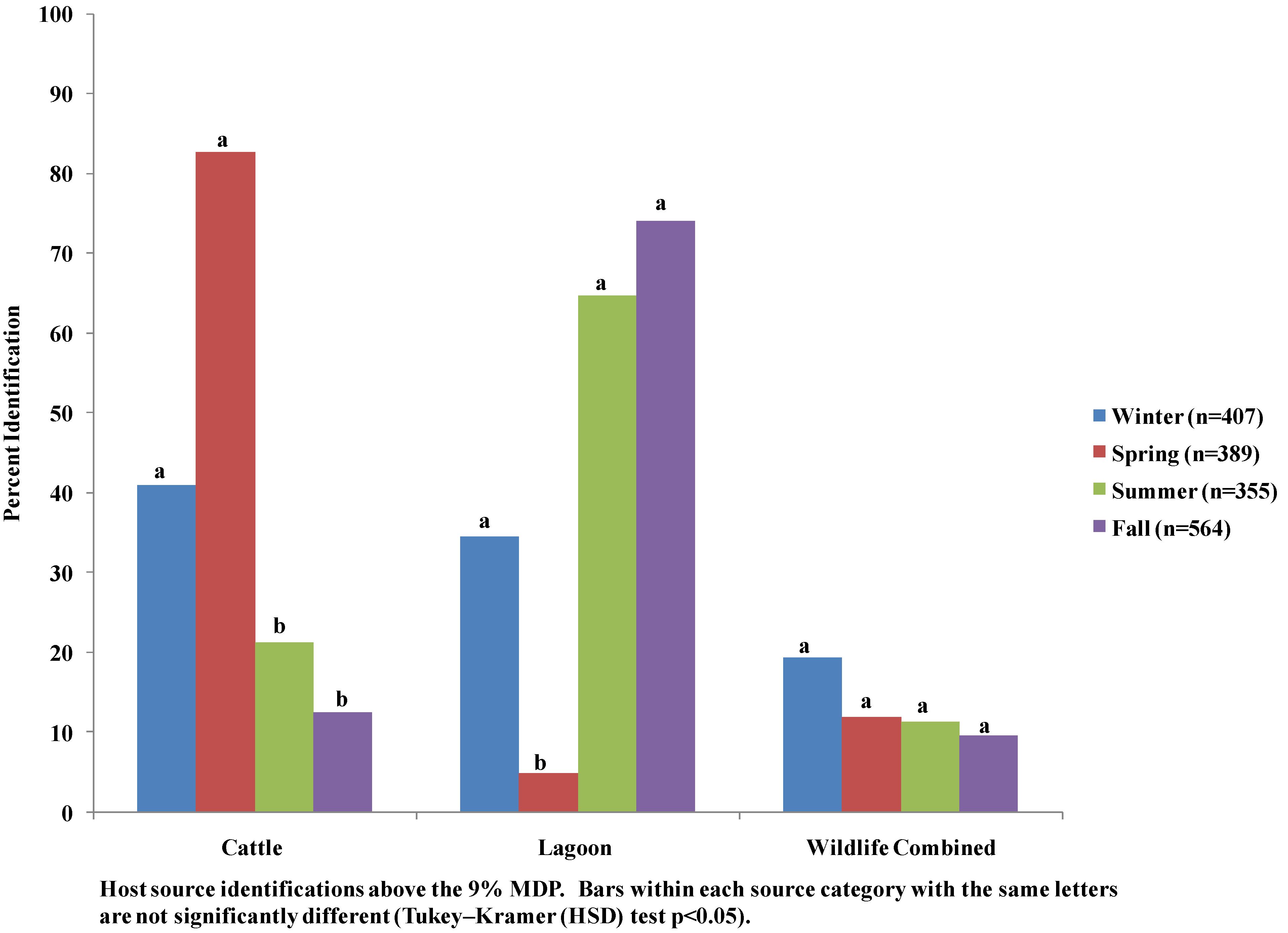
4. Discussion
5. Conclusions
Acknowledgements
References
- Simpson, J.M.; Santo Domingo, J.W; Reasoner, D.J. Microbial source tracking: State of the science. Environ. Sci. Technol. 2002, 36, 5279–5288. [Google Scholar]
- Scott, T.M.; Rose, J.B.; Jenkins, T.M.; Farrah, S.R.; Lukasik, J. Microbial source tracking: Current methodology and future directions. Appl. Environ. Microbiol. 2002, 68, 5796–5803. [Google Scholar] [CrossRef]
- Aillery, M.; Gollehon, N.; Johansson, R.; Key, N.; Ribaudo, M. Managing manure to improve air and water quality. Agric. Nutr. Manag. 2006, 2, 13–20. [Google Scholar]
- United States Environmental Protection Agency (USEPA). National pollutant discharge elimination system permit regulation and effluent limitation guidelines and standards for concentrated animal feeding operations (CAFOs); final rule. Federal Register 2003, 68, 7176–7274. [Google Scholar]
- Burkholder, J.; Libra, B.; Weyer, P.; Heathcote, S.; Kolpin, D.; Thorne, P.S.; Wichman, M. Impacts of waste from concentrated animal feeding operations on water quality. Environ. Health Perspect. 2007, 115, 308–312. [Google Scholar] [CrossRef]
- Carson, A.C.; Shear, B.L.; Ellersieck, M.R.; Asfaw, A. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 2001, 67, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Evenson, J.C.; Strevett, K.A. Discrininant analysis of fecal bacterial species composition for use as a phenotypic microbial source tracking method. Res. Microbiol. 2006, 157, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Dombek, P.E.; Johnson, L.K.; Zimmerley, S.T.; Sadowsky, M.J. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 2000, 66, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- McLellan, S.L.; Daniels, A.D.; Salmore, A.K. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 2003, 69, 2587–2594. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.K.; Brown, M.B.; Carruthers, E.A.; Ferguson, J.A.; Dombek, P.E.; Sadowskyl, M.J. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 2004, 70, 4478–4485. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Weir, S.C.; Howell, E.T.; Lee, H.; Trevors, J.T. Repetitive element (REP)-polymerase chain reaction (PCR) analysis of Escherichia coli isolates from recreational waters of southeastern Lake Huron. Can. J. Microbiol. 2009, 55, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Parveen, S.; Portier, K.M.; Robinson, K.; Edmiston, L.; Tamplin, M.L. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 1999, 65, 3142–3147. [Google Scholar] [PubMed]
- Leung, K.T.; Mackereth, R.; Tien, Y.C.; Topp, E. A comparison of AFLP and ERIC-PCR analyses for discriminating Escherichia coli from cattle, pig and human sources. FEMS Microbiol. Ecol. 2004, 47, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Myoda, S.P.; Carson, C.A.; Fuhrmann, J.J.; Hahm, B.K.; Hartel, P.G.; Yampara-Lquise, H.; Johnson, L.; Kuntz, R.L.; Nakatsu, C.H.; Sadowsky, M.J.; Samadpour, M. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 2003, 1, 167–180. [Google Scholar] [PubMed]
- Buchan, A.; Alber, M.; Hodson, R.E. Strain-specific differentiation of environmental Escherichia coli isolates via denaturing gradient gel electrophoresis (DGGE) analysis of the 16S-23S intergenic spacer region. FEMS Microbiol. Ecol. 2001, 35, 313–321. [Google Scholar] [PubMed][Green Version]
- Field, K.G; Bernhard, A.E; Brodeur, T.J. Molecular approaches to microbiological monitoring: Fecal Source Detection. Environ. Monit. Assess. 2003, 81, 313–32. [Google Scholar] [CrossRef] [PubMed]
- Khatib, L.A.; Tsai, Y.L.; Olson, B.H. A biomarker for the identification of swine fecal pollution in water, using the STII toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 63, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, C.; Crozier, J.B.; Mentz, K.A.; Booth, A.M.; Graves, A.K.; Nelson, N.J.; Reneau, R.B., Jr. Carbon source utilization profiles as a method to identify sources of faecal pollution in water. J. Appl. Microbiol. 2003, 94, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Katouli, M. Phenotypic variations of enterococci in surface waters: analysis of biochemical fingerprinting data from multi-catchments. J. Appl. Microbiol. 2008, 105, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.K.; Hagedorn, C.; Brooks, A.; Hagedorn, R.L.; Martin, E. Microbial source tracking in a rural watershed dominated by cattle. Water Res. 2007, 41, 3729–3739. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Whitlock, J.; Withington, V. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: Use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 2000, 66, 3698–3704. [Google Scholar] [CrossRef] [PubMed]
- Harwood, V.J.; Wiggins, B.; Hagedorn, C.; Ellender, R.D.; Gooch, J.; Kern, J.; Samadpour, M.; Chapman, A.; Robinson, B.J.; Thompson, B.C. Phenotypic library-based microbial source tracking methods: Efficacy in the California collaborative study. J. Water Health 2003, 1, 153–166. [Google Scholar] [PubMed]
- Wiggins, B.A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 1996, 62, 3997–4002. [Google Scholar] [PubMed]
- Hagedorn, C.; Robinson, S.L.; Filtz, J.R.; Grubbs, S.M.; Angier, T.A.; Reneau, R.B. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 1999, 65, 5522–5531. [Google Scholar] [PubMed]
- Carroll, S.; Hargreaves, M.; Goonetilleke, A. Sourcing faecal pollution from onsite wastewater treatment systems in surface waters using antibiotic resistance analysis. J. Appl. Microbiol. 2005, 99, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Orosz-Coghlan, P.A.; Rusin, P.A.; Karpiscak, M.M.; Gerba, C.P. Microbial source tracking of E. coli in constructed wetlands. Water Environ. Res. 2006, 78, 227–32. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.P.; Dawes, L.; Hargreaves, M.; Goonetilleke, A. Faecal pollution source identification in an urbanizing catchment using antibiotic resistance profiling, discriminant analysis and partial least squares regression. Water Res. 2009, 43, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.; Price, B.; Ware, A. Alternative estimate of source distribution in microbial source tracking using posterior probabilities. Water Res. 2010, 44, 2629–2637. [Google Scholar] [CrossRef]
- Booth, A.M.; Hagedorn, C.; Graves, A.K.; Hagedorn, S.C.; Mentz, K.H. Sources of fecal pollution in Virginia’s Blackwater river. J. Environ. Eng. 2003, 129, 547–552. [Google Scholar] [CrossRef]
- Choi, S.; Chu, W.; Brown, J.; Becker, S.J.; Harwood, V.J.; Jiang, S.C. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 2003, 46, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.K.; Hagedorn, C.; Teetor, A.; Mahal, M.; Booth, A.M.; Reneau, R.B., Jr. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 2002, 31, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, J.E.; Jones, D.T.; Harwood, V.J. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 2002, 36, 4273–4282. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, B.A.; Cash, P.W.; Creamer, W.S.; Dart, S.E.; Garcia, P.P.; Gerecke, T.M.; Han, J.; Henry, B.L; Hoover, K.B.; Johnson, E.L.; Jones, K.C.; McCarthy, J.G.; McDonough, J.A.; Mercer, S.A.; Noto, M.J.; Park, H.; Phillips, M.S.; Purner, S.M.; Smith, B.M.; Stevens, E.N.; Varner, A.K. Use of antibiotic resistance analysis for representativeness testing of multiwatershed libraries. Appl. Environ. Microbiol. 2003, 69, 3399–3405. [Google Scholar] [CrossRef]
- Stewart, J.R.; Ellender, R.D; Gooch, J.A.; Jiang, S.; Myoda, S.P.; Weisberg, S.B. Recommendations for microbial source tracking: lessons learned from a methods comparison study. J. Water Health 2003, 1, 225–231. [Google Scholar] [PubMed]
- Stoeckel, D.M.; Mathes, M.V.; Hyer, K.E.; Hagedorn, C.; Kator, H.; Lukasik, J.; O’Brien, T.L.; Fenger, T.W.; Samadpour, M.; Strickler, K.M.; Wiggins, B.A. Comparison of seven protocols to identify fecal contamination sources using E. coli. Environ. Sci. Technol. 2004, 38, 6109–6117. [Google Scholar] [CrossRef]
- Olivas, Y.; Faulkner, B.R. Fecal source tracking by antibiotic resistance analysis on a watershed exhibiting low resistance. Environ. Monit. Assess. 2008, 139, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Price, B.; Venso, E.; Frana, M.; Greenberg, J.; Ware, A. A comparison of ARA and DNA data for microbial source tracking based on source-classification models developed using classification trees. Water Res. 2007, 41, 3575–3584. [Google Scholar] [CrossRef] [PubMed]
- USDA. Soil Survey of Sampson County, North Carolina; USDA-SCS: Washington DC, USA, 1985; Map no. 4. [Google Scholar]
- Mathew, A.G.; Upchurch, W.G.; Chattin, S.E. Incidence of antibiotic resistance in fecal Eschericia coli isolated from commercial swine farms. J. Animal Sci. 1998, 76, 429–434. [Google Scholar]
- Ritter, K.J.; Carruthers, E.; Carson, C.A.; Ellender, R.D.; Harwood, V.J.; Kingsley, K.; Nakatsu, C.; Sadowsky, M.; Shear, B.; West, B.; Whitlock, J.E.; Wiggins, B.A.; Wilbur, J.D. Assessment of statistical methods used in library-based approaches to microbial source tracking. J. Water Health 2003, 1, 209–223. [Google Scholar] [PubMed]
- North Carolina Department of Environment and Natural Resources. In Classifications and Water Quality Standards Applicable to Surface Water and Wetlands of North Carolina; Division of Water Quality: Raleigh, NC, USA, 1997; p. 41.
- Fincher, L.M.; Parker, C.D.; Chauret, C.P. Surface Water Quality: Occurrence and Antibiotic Resistance of Escherichia coli O157:H7 in a Watershed in North-Central Indiana. J. Environ. Qual. 2009, 38, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Burnes, B.S. Antibiotic resistance analysis of fecal coliforms to determine fecal pollution sources in a mixed-use watershed. Environ. Monit. Assess. 2003, 85, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, B.A.; Andrews, R.W.; Conway, R.A.; Corr, C.L.; Dobratz, E.J.; Dougherty, D.P.; Eppard, J.R.; Knupp, S.R.; Limjoco, M.C.; Mettenburg, J.M.; Rinehardt, J.M.; Sonsino, J.; Torrijos, R.L.; Zimmerman, M.E. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 1999, 65, 3483–3486. [Google Scholar] [PubMed]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liwimbi, L.; Graves, A.K.; Israel, D.W.; Heugten, E.v.; Robinson, B.; Cahoon, C.W.; Lubbers, J.F. Microbial Source Tracking in a Watershed Dominated by Swine. Water 2010, 2, 587-604. https://doi.org/10.3390/w2030587
Liwimbi L, Graves AK, Israel DW, Heugten Ev, Robinson B, Cahoon CW, Lubbers JF. Microbial Source Tracking in a Watershed Dominated by Swine. Water. 2010; 2(3):587-604. https://doi.org/10.3390/w2030587
Chicago/Turabian StyleLiwimbi, Lloyd, Alexandria K. Graves, Daniel W. Israel, Eric van Heugten, Bradford Robinson, Charles W. Cahoon, and Joice F. Lubbers. 2010. "Microbial Source Tracking in a Watershed Dominated by Swine" Water 2, no. 3: 587-604. https://doi.org/10.3390/w2030587
APA StyleLiwimbi, L., Graves, A. K., Israel, D. W., Heugten, E. v., Robinson, B., Cahoon, C. W., & Lubbers, J. F. (2010). Microbial Source Tracking in a Watershed Dominated by Swine. Water, 2(3), 587-604. https://doi.org/10.3390/w2030587




