Transcriptional Differences in Gills Provide Insights into the Environmental Acclimatization of Wild Topmouth Gudgeon (Pseudorasbora parva) from Freshwater Invasion to Saline–Alkali Waters
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Preparation
2.3. RNA Extraction, Library Preparation, and RNA Sequencing
2.4. De Novo Assembly and Functional Annotation
2.5. Identification of Differentially Expressed Genes
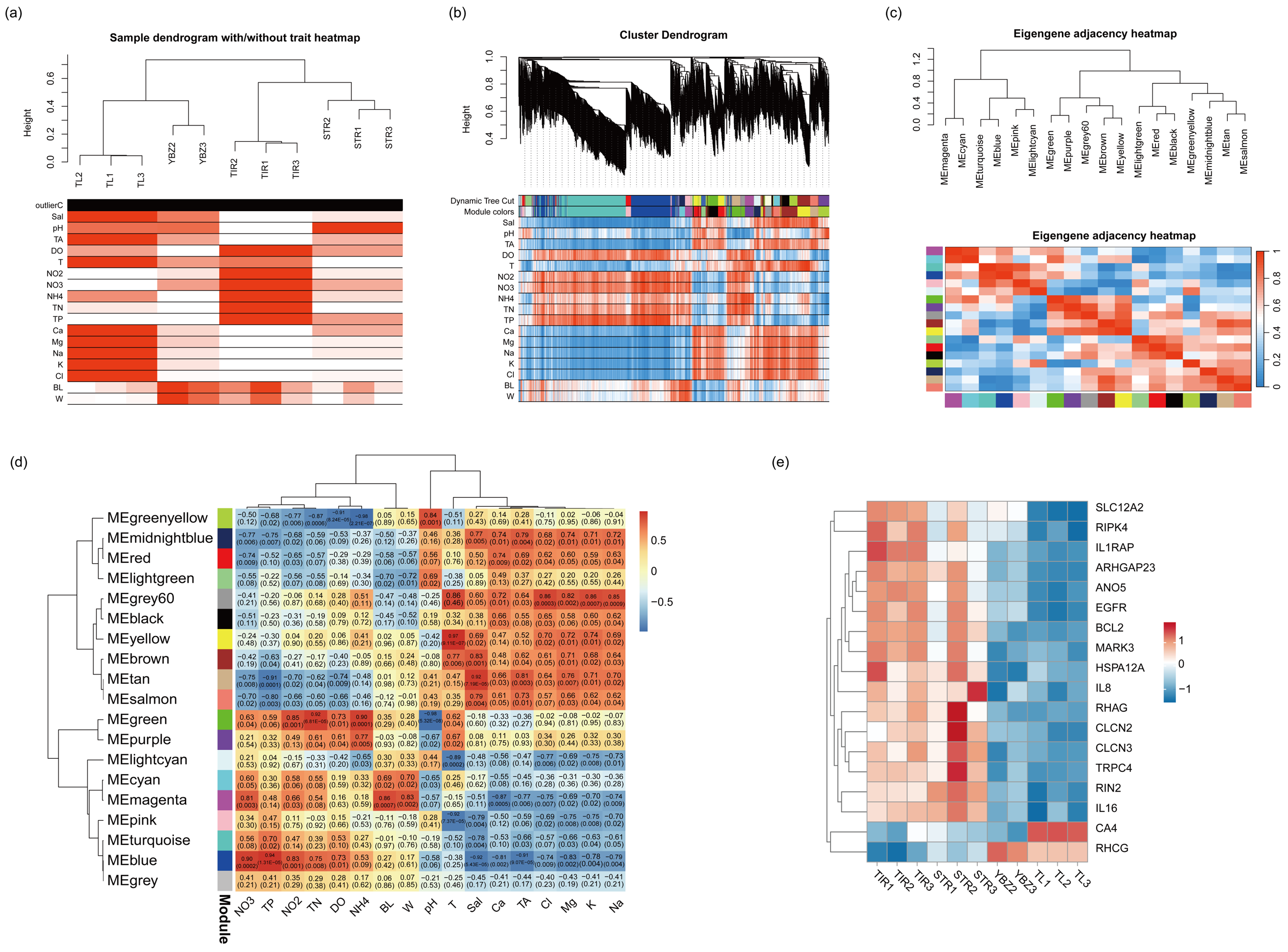
2.6. Gene Co-Expression Network Construction
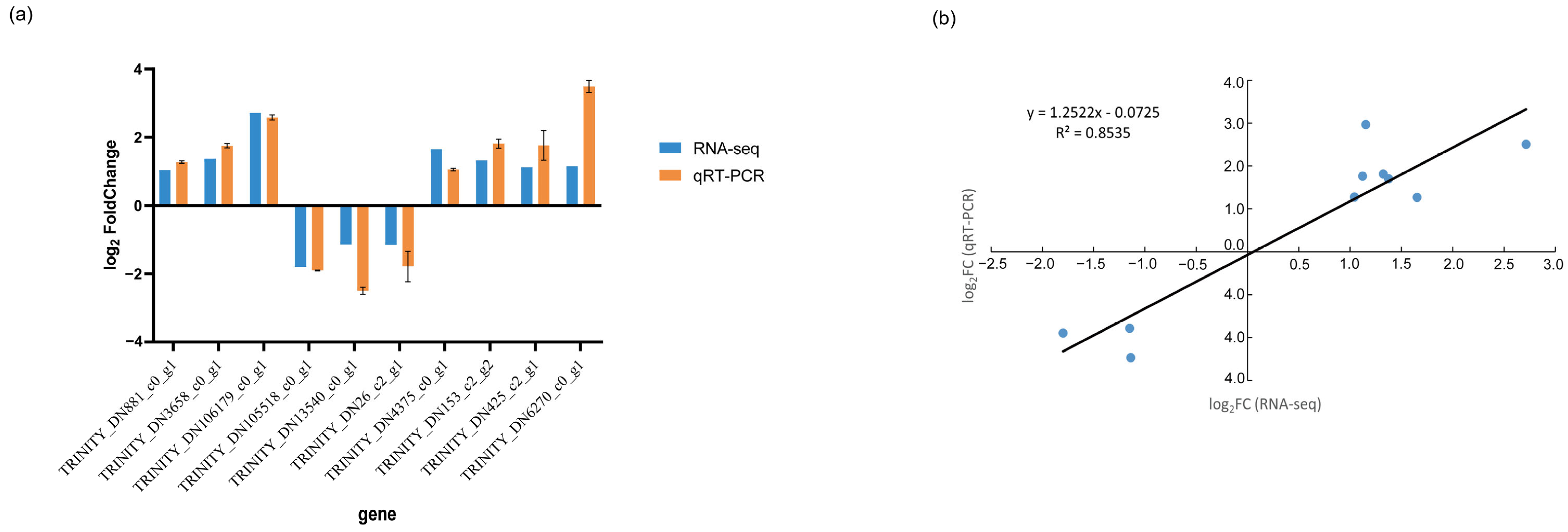
2.7. Gene Expression RT-qPCR Validation
2.8. Statistical Analysis
3. Results
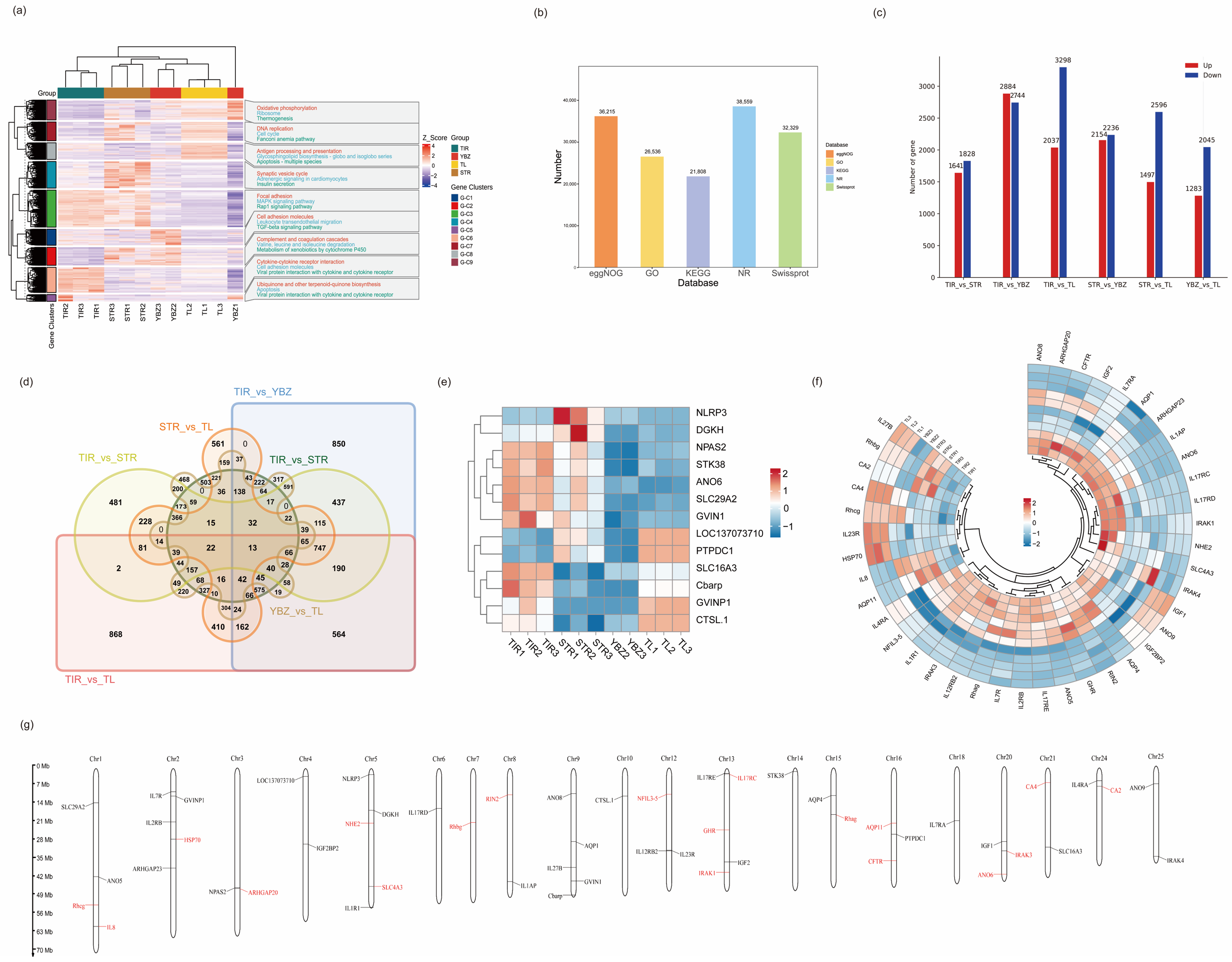
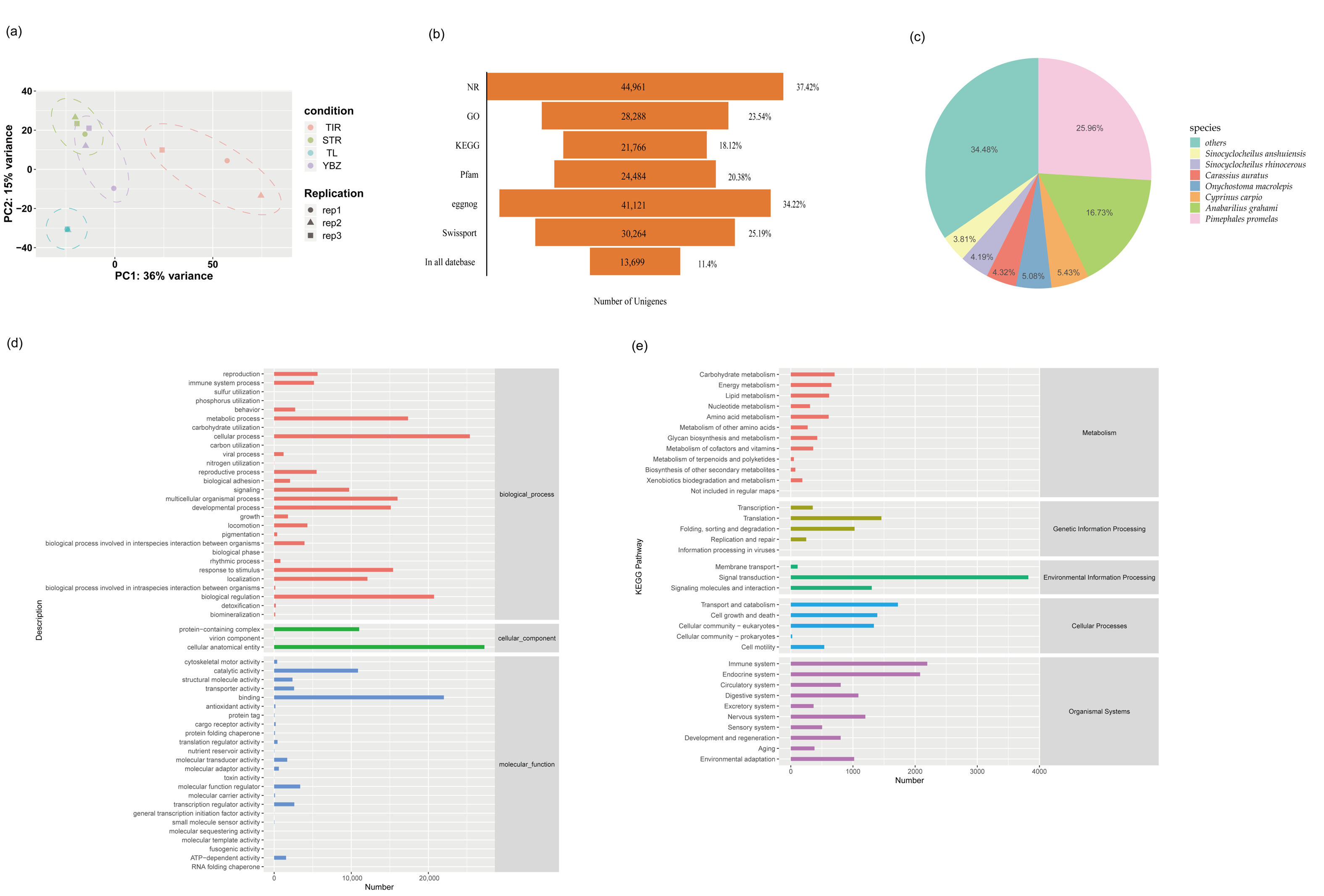
3.1. Overview of Transcriptome Profiles
3.2. Unigenes and Functional Annotation
3.3. Comparison of DEGs Under Saline Conditions
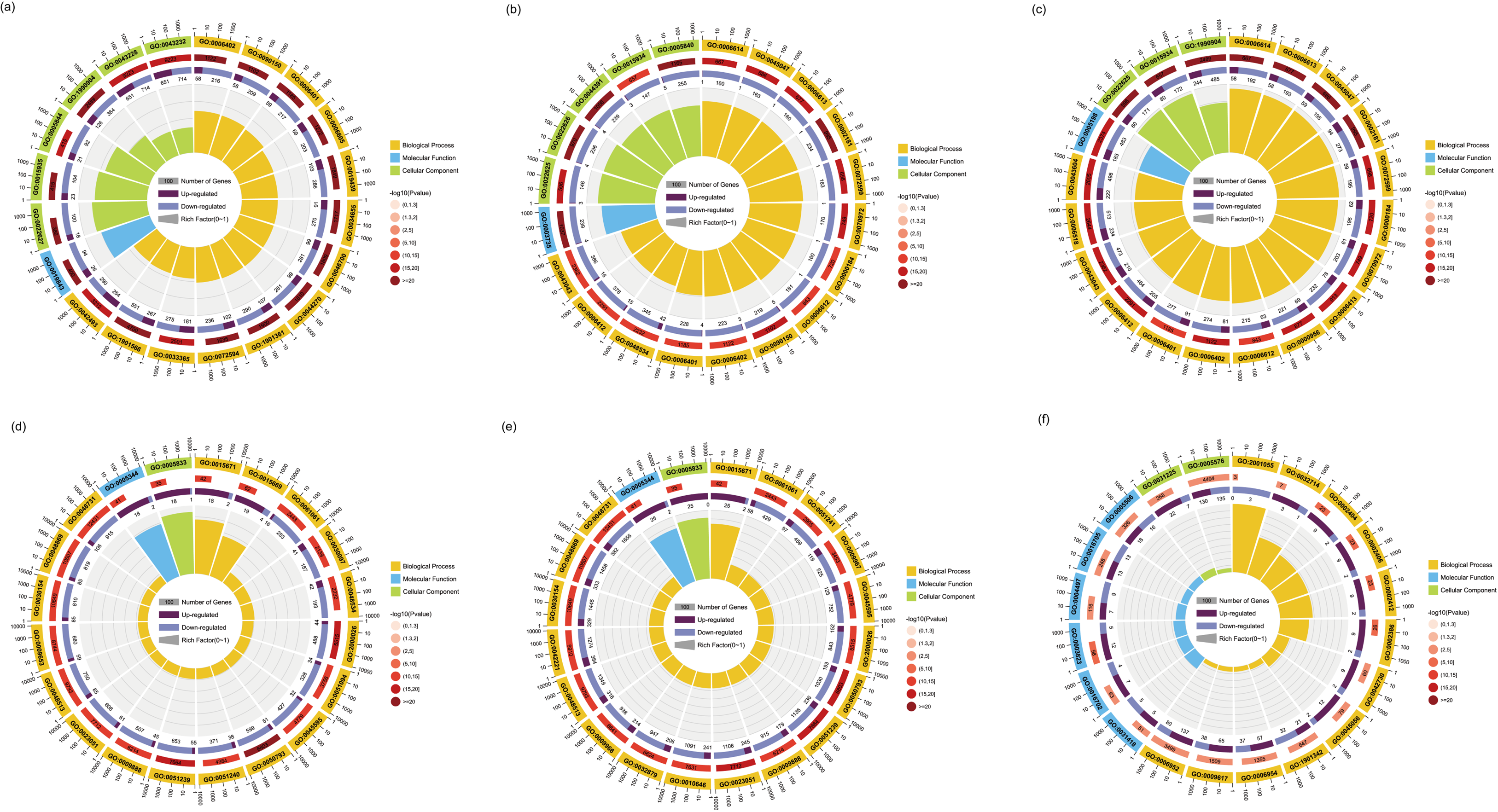
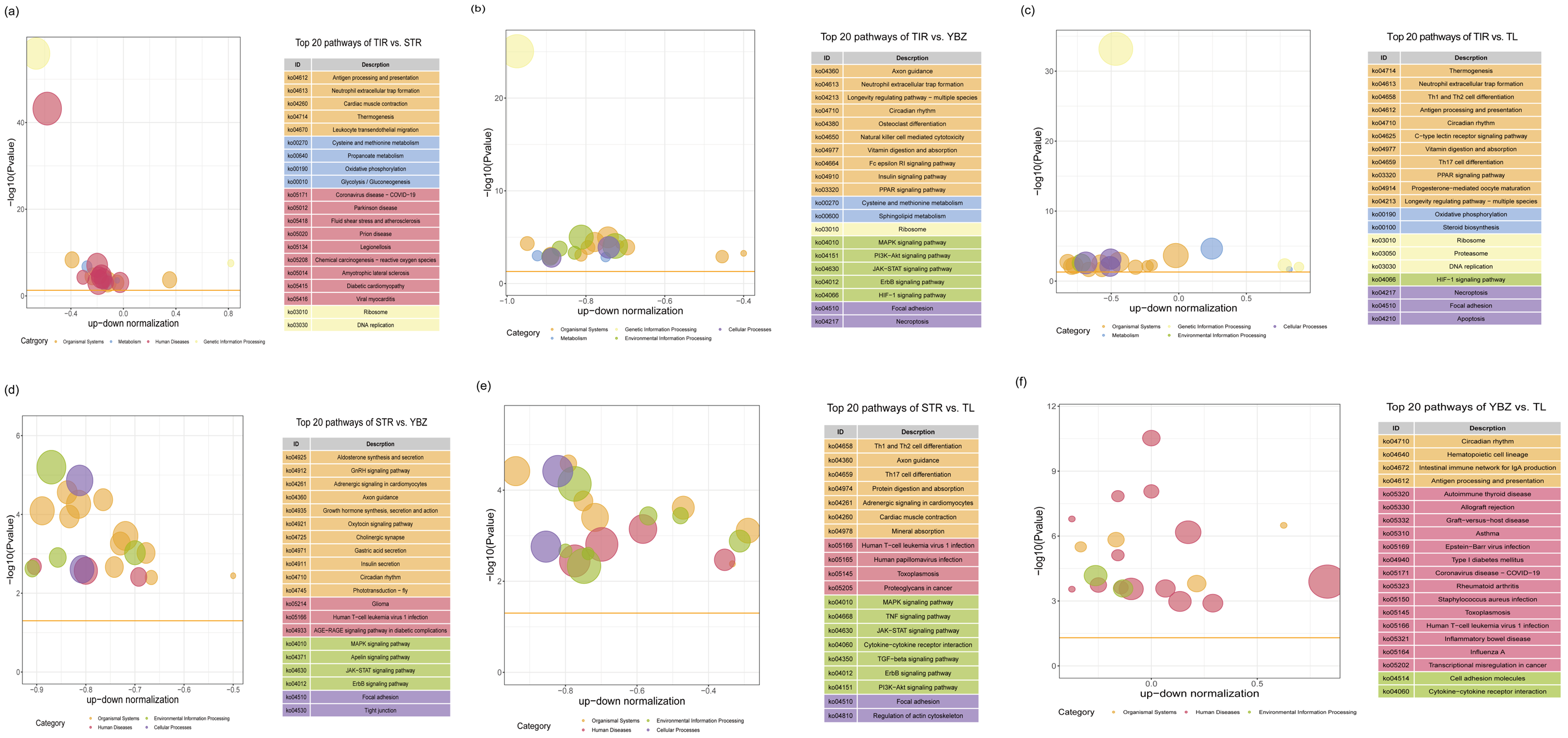
3.4. GO Terms and KEGG Pathways Enrichment Analysis of DEGs
3.5. Co-Expression Network of DEGs Under Saline–Alkali Stress
3.6. DEGs Related to Saline–Alkaline Tolerance
3.7. qRT-PCR Validation of DEGs
4. Discussion
4.1. Ion and Osmotic Regulation
4.2. Ammonia Nitrogen Excretion
4.3. Acid–Base Accommodation
4.4. Immune Response in an Invasive Environment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Reference Transcriptome Analysis Methods
Appendix A.2. Reference Genome Comparison Results
| Key DEGs | Key Biological Processes | Key Cellular Components | Key Molecular Functions | Key Pathways |
|---|---|---|---|---|
| Rhcg | Ion transport Ammonium transmembrane transport Epithelium development Regulation of pH | Apical plasma membrane Vesicle Organelle | Ammonium transmembrane transporter activity Ion transmembrane transporter activity | - |
| ano6 | Ion transport Response to stimulus Blood coagulation Cell migration Immune response | Plasma membrane Secretory granule Vesicle Membrane protein complex | Chloride channel activity Cation channel activity Phospholipid scramblase activity | Efferocytosis |
| aqp1 | Water transport Regulation of cell size Ion transport Carbohydrate transport | Plasma membrane Brush border Extracellular region | Water channel activity Ammonium transmembrane transporter activity Ion transmembrane transporter activity | Bile secretion Proximal tubule bicarbonate reclamation |
| ca2 | Ion transport Carbon dioxide transport Response to salt stress | Cytoplasm Organelle Membrane-bounded organelle | Carbonate dehydratase activity Catalytic activity | Bile secretion Pancreatic secretion |
| cftr | Ion transport immune system process Cellular response to stress | Plasma membrane Intracellular organelle Endoplasmic reticulum membrane | Anion transmembrane transporter activity ATP binding | Bile secretion Pancreatic secretion AMPK signaling pathway |
| hsp70 | Negative regulation of signal transduction Positive regulation of interleukin-8 production Negative regulation of cell death Immune response | Cytoplasm Mitochondrion Endoplasmic reticulum | Ion binding ATP hydrolysis activity Carbohydrate derivative binding | Antigen processing and presentation Endocytosis MAPK signaling pathway |
| il8 | Immune system process Positive regulation of response to stimulus Inflammatory response Regulation of sodium ion transport | Vesicle Endomembrane system intracellular organelle | Signaling receptor regulator activity Cytokine activity Enzyme regulator activity | IL-17 signaling pathway) Cytokine-cytokine receptor interaction |
| il17rc | Cellular response to stimulus Interleukin-17-mediated signaling pathway Cytokine-mediated signaling | Plasma membrane Cell periphery Obsolete cell | Interleukin-17 receptor activity Signaling receptor activity Cytokine receptor activity | IL-17 signaling pathway Cytokine-cytokine receptor interaction |
References
- Banarescu, P.M. On the Relations between Hydrography and the Ranges of Freshwater Fish Species and Subspecies. Ital. J. Zool. 1998, 65, 87–93. [Google Scholar] [CrossRef]
- Copp, G.H.; Garthwaite, R.; Gozlan, R.E. Risk Identification and Assessment of Non-Native Freshwater Fishes: A Summary of Concepts and Perspectives on Protocols for the UK. J. Appl. Ichthyol. 2005, 21, 371–373. [Google Scholar] [CrossRef]
- Gavriloaie, C.; Bucur, C.; Berkesy, C. Notes Concerning the Distribution of Asian Fish Species, Pseudorasbora parva, in Europe. Aquac. Aquar. Conserv. Legis. Bioflux 2014, 7, 43–50. [Google Scholar]
- Simon, A.; Gozlan, R.E.; Robert Britton, J.; Van Oosterhout, C.; Hänfling, B. Human Induced Stepping-Stone Colonisation of an Admixed Founder Population: The Spread of Topmouth Gudgeon (Pseudorasbora parva) in Europe. Aquat. Sci. 2015, 77, 17–25. [Google Scholar] [CrossRef]
- Gozlan, R.E.; Andreou, D.; Asaeda, T.; Beyer, K.; Bouhadad, R.; Burnard, D.; Caiola, N.; Cakic, P.; Djikanovic, V.; Esmaeili, H.R.; et al. Pan-Continental Invasion of Pseudorasbora parva: Towards a Better Understanding of Freshwater Fish Invasions: Pan-Continental Invasion of Topmouth Gudgeon. Fish Fish. 2010, 11, 315–340. [Google Scholar] [CrossRef]
- Onikura, N.; Nakajima, J. Age, Growth and Habitat Use of the Topmouth Gudgeon, Pseudorasbora parva in Irrigation Ditches on Northwestern Kyushu Island, Japan. J. Appl. Ichthyol. 2013, 29, 186–192. [Google Scholar] [CrossRef]
- Chen, G.; Qiu, Y.P.; Li, L. Fish Invasions and Changes in the Fish Fauna of the Tarim Basin. Acta Ecol. Sin. 2017, 37, 700–714. [Google Scholar] [CrossRef][Green Version]
- Xu, W.; Wang, H.; Li, Y. Distribution and Aquaculture Application of Saline Alkali Water at Home and Abroad. China Fish. 2021, 7, 50–53. [Google Scholar]
- Wang, F.; Zhu, L.; Wei, Y.; Gao, P.; Liu, Y.; Zhou, K.; Sun, Z.; Lai, Q.; Yao, Z. Intestinal Ion Regulation Exhibits a Daily Rhythm in Gymnocypris przewalskii Exposed to High Saline and Alkaline Water. Sci. Rep. 2022, 12, 807. [Google Scholar] [CrossRef]
- Liu, Y.; Fang, H.; Lai, Q.; Liang, L. The Current State and Development Strategy for China’s Saline-Alkaline Fisheries. Strateg. Study Chin. Acad. Eng. 2016, 18, 74–78. [Google Scholar] [CrossRef]
- Gao, L.; Yuan, Z.; Mao, X.; Ma, T. Salinity Levels, Trends and Drivers of Surface Water Salinization across China’s River Basins. Water Res. 2025, 281, 123556. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Guo, W.; Lai, Q.; Shi, J.; Zhou, K.; Qi, H.; Lin, T.; Li, Z.; Wang, H. Gymnocypris przewalskii Decreases Cytosolic Carbonic Anhydrase Expression to Compensate for Respiratory Alkalosis and Osmoregulation in the Saline-Alkaline Lake Qinghai. J. Comp. Physiol. B 2016, 186, 83–95. [Google Scholar] [CrossRef]
- Peng, M.; Li, Z.; Liu, X.; Niu, D.; Li, J. Inland Alkaline Brackish Water Aquaculture of Juvenile Razor Clam: Survival, Growth, Physiology and Immune Responses. Aquac. Rep. 2020, 18, 100463. [Google Scholar] [CrossRef]
- Song, L.; Zhao, Y.; Song, Y.; Zhao, L.; Ma, C.; Zhao, J. Effects of Saline-Alkaline Water on Growth Performance, Nutritional Processing, and Immunity in Nile Tilapia (Oreochromis niloticus). Aquaculture 2021, 544, 737036. [Google Scholar] [CrossRef]
- Laverty, G.; Skadhauge, E. Adaptation of Teleosts to Very High Salinity. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2012, 163, 1–6. [Google Scholar] [CrossRef]
- Gao, S.; Chang, Y.; Zhao, X.; Sun, B.; Zhang, L.; Laing, L.; Dong, Z. The effect of different bicarbonate alkalinity on the gill structure of Amur Ide (Leuciscus waleckii). Acta Hydrobiol. Sin. 2020, 44, 736–743. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, X.; Chang, Y.; Sun, B.; Liang, L.; Zhang, L.; Dong, Z. Effect of different bicarbonate alkalinities on microstructures of Kidney and Intestine in Amur ide (Leuciscus waleckii). Chin. J. Fish. 2020, 33, 24–30. [Google Scholar] [CrossRef]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef]
- Nilsen, T.O.; Ebbesson, L.O.E.; Madsen, S.S.; McCormick, S.D.; Andersson, E.; Bjornsson, B.T.; Prunet, P.; Stefansson, S.O. Differential Expression of Gill Na+,K+-ATPase Alpha- and Beta-Subunits, Na+,K+,2Cl− Cotransporter and CFTR Anion Channel in Juvenile Anadromous and Landlocked Atlantic Salmon Salmo salar. J. Exp. Biol. 2007, 210, 2885–2896. [Google Scholar] [CrossRef]
- Kunzelmann, K.; Nilius, B.; Owsianik, G.; Schreiber, R.; Ousingsawat, J.; Sirianant, L.; Wanitchakool, P.; Bevers, E.M.; Heemskerk, J.W.M. Molecular Functions of Anoctamin 6 (TMEM16F): A Chloride Channel, Cation Channel, or Phospholipid Scramblase? Pflugers Arch. 2014, 466, 407–414. [Google Scholar] [CrossRef]
- Watanabe, S.; Niida, M.; Maruyama, T.; Kaneko, T. Na+/H+ Exchanger Isoform 3 Expressed in Apical Membrane of Gill Mitochondrion-Rich Cells in Mozambique Tilapia Oreochromis mossambicus. Fish. Sci. 2008, 74, 813–821. [Google Scholar] [CrossRef]
- Larsen, P.F.; Nielsen, E.E.; Williams, T.D.; Loeschcke, V. Intraspecific Variation in Expression of Candidate Genes for Osmoregulation, Heme Biosynthesis and Stress Resistance Suggests Local Adaptation in European Flounder (Platichthys flesus). Heredity 2008, 101, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.F.; Nielsen, E.E.; Koed, A.; Thomsen, D.S.; Olsvik, P.A.; Loeschcke, V. Interpopulation Differences in Expression of Candidate Genes for Salinity Tolerance in Winter Migrating Anadromous Brown Trout (Salmo trutta L.). BMC Genet. 2008, 9, 12. [Google Scholar] [CrossRef]
- Deane, E.E.; Luk, J.C.Y.; Woo, N.Y.S. Aquaporin 1a Expression in Gill, Intestine, and Kidney of the Euryhaline Silver Sea Bream. Front. Physio. 2011, 2, 39. [Google Scholar] [CrossRef]
- Bao, X.N.; Mu, C.K.; Zhang, C.; Wang, Y.F.; Song, W.W.; Li, R.H.; Wang, C.L. mRNA Expression Profiles of Heat Shock Proteins of Wild and Salinity-Tolerant Swimming Crabs, Portunus trituberculatus, Subjected to Low Salinity Stress. Genet. Mol. Res. 2014, 13, 6837–6847. [Google Scholar] [CrossRef]
- Lee, C.E.; Petersen, C.H. Genotype-by-environment Interaction for Salinity Tolerance in the Freshwater-invading Copepod Eurytemora affinis. Physiol. Biochem. Zool. 2002, 75, 335–344. [Google Scholar] [CrossRef]
- DeLong, E.F.; Karl, D.M. Genomic Perspectives in Microbial Oceanography. Nature 2005, 437, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, G.; Xu, P. Gills Full-Length Transcriptomic Analysis of Osmoregulatory Adaptive Responses to Salinity Stress in Coilia nasus. Ecotoxicol. Environ. Saf. 2021, 226, 112848. [Google Scholar] [CrossRef]
- Root, L.; Campo, A.; MacNiven, L.; Con, P.; Cnaani, A.; Kültz, D. Nonlinear Effects of Environmental Salinity on the Gill Transcriptome versus Proteome of Oreochromis niloticus Modulate Epithelial Cell Turnover. Genomics 2021, 113, 3235–3249. [Google Scholar] [CrossRef]
- Li, X.; Liu, S.; Wang, Y.; Lu, W.; Zhang, Q.; Cheng, J. Genomic and Transcriptomic Landscape and Evolutionary Dynamics of Heat Shock Proteins in Spotted Sea Bass (Lateolabrax maculatus) under Salinity Change and Alkalinity Stress. Biology 2022, 11, 353. [Google Scholar] [CrossRef]
- Zhou, B.; Qi, D.; Liu, S.; Qi, H.; Wang, Y.; Zhao, K.; Tian, F. Physiological, Morphological and Transcriptomic Responses of Tibetan Naked Carps (Gymnocypris przewalskii) to Salinity Variations. Comp. Biochem. Physiol. D Genom. Proteom. 2022, 42, 100982. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, Q.; Zhao, H.; Chen, C. Transcriptome Profiles Revealed High- and Low-Salinity Water Altered Gill Homeostasis in Half-Smooth Tongue Sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. D-Genom. Proteom. 2022, 42, 100989. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, J.; Leung, C.T.; Lin, X.; Chan, T.F.; Tse, W.K.F.; Lai, K.P. Transcriptomic Analysis in Marine Medaka Gill Reveals That the Hypo-Osmotic Stress Could Alter the Immune Response via the IL17 Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 12417. [Google Scholar] [CrossRef] [PubMed]
- Chutia, P.; Das, M.; Saha, N. Transcriptome Analysis of Gills Reveals Novel Insights into the Molecular Response of Stinging Catfish (Heteropneustes fossilis) to Environmental Hypertonicity. Gene 2023, 851, 147044. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Qin, R.; Peng, Y.; Huang, Y.; Zhu, C.; Li, G.; Jiang, D.; Shi, H. Divergent Molecular Responses of Greater Amberjack (Seriola dumerili) to Acute Salinity Stress Revealed by Comparative Transcriptome Analysis. Front. Mar. Sci. 2023, 10, 1185015. [Google Scholar] [CrossRef]
- Mohindra, V.; Chowdhury, L.M.; Chauhan, N.; Paul, A.; Singh, R.K.; Kushwaha, B.; Maurya, R.K.; Lal, K.K.; Jena, J.K. Transcriptome Analysis Revealed Osmoregulation Related Regulatory Networks and Hub Genes in the Gills of Hilsa Shad, Tenualosa ilisha, during the Migratory Osmotic Stress. Mar. Biotechnol. 2023, 25, 161–173. [Google Scholar] [CrossRef]
- Blondeau-Bidet, E.; Tine, M.; Gonzalez, A.-A.; Guinand, B.; Lorin-Nebel, C. Coping with Salinity Extremes: Gill Transcriptome Profiling in the Black-Chinned Tilapia (Sarotherodon melanotheron). Sci. Total Environ. 2024, 929, 172620. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An Automatic Genome Annotation and Pathway Reconstruction Server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R Package for Identifying Differentially Expressed Genes from RNA-Seq Data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. Gene Set Enrichment Analysis with topGO. Bioconductor Improv 2009, 27, 776. [Google Scholar]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.; Huang, L. ImageGP: An Easy-to-use Data Visualization Web Serverfor Scientific Researchers. iMeta 2022, 1, e5. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Li, B.; Zeng, X.; Liu, S.; Zhuang, Z. Screening and Evaluating Reference Genes for Quantitative Real-Time PCR in Striped Jack (Pseudocaranx dentex). Prog. Fish. Sci. 2023, 44, 107–115. [Google Scholar] [CrossRef]
- Sun, H.; Lu, X.; Tong, G.; Yin, J.; Xue, S.; Zhang, L.; Han, Y. Screening of Reference Genes for Real-Time Quantitative PCR in Stewart’s Naked High-Asian-Carp Oxygymnocypris stewarti. J. Dalian Ocean. Univ. 2019, 34, 370–375. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare Tools: A Zero-code Interactive Online Platform for Biological Data Analysis and Visualization. Imeta 2024, 3, e228. [Google Scholar] [CrossRef]
- Fridman, S. Ontogeny of the Osmoregulatory Capacity of Teleosts and the Role of Ionocytes. Front. Mar. Sci. 2020, 7, 709. [Google Scholar] [CrossRef]
- Suzuki, J.; Umeda, M.; Sims, P.J.; Nagata, S. Calcium-Dependent Phospholipid Scrambling by TMEM16F. Nature 2010, 468, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Sirianant, L.; Ousingsawat, J.; Wanitchakool, P.; Schreiber, R.; Kunzelmann, K. Cellular Volume Regulation by Anoctamin 6: Ca2+, Phospholipase A2 and Osmosensing. Pflüg. Arch.—Eur. J. Physiol. 2016, 468, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Simões, F.; Ousingsawat, J.; Wanitchakool, P.; Fonseca, A.; Cabrita, I.; Benedetto, R.; Schreiber, R.; Kunzelmann, K. CFTR Supports Cell Death through ROS-Dependent Activation of TMEM16F (Anoctamin 6). Pflüg. Arch. Eur. J. Physiol. 2018, 470, 305–314. [Google Scholar] [CrossRef]
- Gonen, T.; Walz, T. The Structure of Aquaporins. Q. Rev. Biophys. 2006, 39, 361–396. [Google Scholar] [CrossRef]
- Ishibashi, K.; Sasaki, S.; Fushimi, K.; Uchida, S.; Kuwahara, M.; Saito, H.; Furukawa, T.; Nakajima, K.; Yamaguchi, Y.; Gojobori, T. Molecular Cloning and Expression of a Member of the Aquaporin Family with Permeability to Glycerol and Urea in Addition to Water Expressed at the Basolateral Membrane of Kidney Collecting Duct Cells. Proc. Natl. Acad. Sci. USA 1994, 91, 6269–6273. [Google Scholar] [CrossRef]
- Kim, Y.K.; Lee, S.Y.; Kim, B.S.; Kim, D.S.; Nam, Y.K. Isolation and mRNA expression analysis of aquaporin isoforms in marine medaka Oryzias dancena, a euryhaline teleost. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef]
- Wang, M.; Yang, J.; Xu, P.; Xu, G.; Xu, D.; You, Y.; Liu, K.; Duan, J.; Zhou, Y.; Fang, D.; et al. Molecular Cloning and Expression Analysis of Aquaporin-1 from the Coilia nasus under High-Salinity Conditions. J. Fish. Sci. China 2017, 24, 449–458. [Google Scholar] [CrossRef]
- Aoki, M.; Kaneko, T.; Katoh, F.; Hasegawa, S.; Tsutsui, N.; Aida, K. Intestinal Water Absorption through Aquaporin 1 Expressed in the Apical Membrane of Mucosal Epithelial Cells in Seawater-Adapted Japanese Eel. J. Exp. Biol. 2003, 206, 3495–3505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Z.; Ma, A.; Cui, W.; Qu, J. Response of Aquaporin (AQP1, AQP3) and Ion Channel Protein (CFTR, NHE1) of Turbot (Scophthalmus maximus) to Low-Salinity Stress. Progrees Fish. Sci. 2024, 41, 41–49. [Google Scholar] [CrossRef]
- Gan, Y.; Zhao, J.; Thammaratsuntorn, J.; Li, C. cDNA Cloning of Aquaporin 3 in Sarotherodon melanothern and its tissue expression patterns under salinity stresses. Chin. J. Zool. 2014, 49, 560–569. [Google Scholar] [CrossRef]
- Giffard-Mena, I.; Boulo, V.; Aujoulat, F.; Fowden, H.; Castille, R.; Charmantier, G.; Cramb, G. Aquaporin Molecular Characterization in the Sea-Bass (Dicentrarchus labrax): The Effect of Salinity on AQP1 and AQP3 Expression. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 148, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Deane, E.E.; Woo, N.Y.S. Tissue Distribution, Effects of Salinity Acclimation, and Ontogeny of Aquaporin 3 in the Marine Teleost, Silver Sea Bream (Sparus sarba). Mar. Biotechnol. 2006, 8, 663–671. [Google Scholar] [CrossRef]
- Ellis, L.V.; Bollinger, R.J.; Weber, H.M.; Madsen, S.S.; Tipsmark, C.K. Differential Expression and Localization of Branchial AQP1 and AQP3 in Japanese Medaka (Oryzias latipes). Cells 2019, 8, 422. [Google Scholar] [CrossRef]
- Cutler, C.P.; Martinez, A.-S.; Cramb, G. The Role of Aquaporin 3 in Teleost Fish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 148, 82–91. [Google Scholar] [CrossRef]
- Huang, M.; Wu, M.-X.; Zhang, L.-J.; Mi, D.; Zhang, Y.-L. Effects of Carbonate Alkalinity on Branchial Gene Expression in the Large-Scale Loach (Paramisgurnus dabryanus). Front. Mar. Sci. 2022, 9, 983615. [Google Scholar] [CrossRef]
- Wilkie, M.P.; Wright, P.A.; Iwama, G.K.; Wood, C.M. The Physiological Adaptations of the Lahontan Cutthroat Trout (Oncorhynchus clarki henshawi) Following Transfer from Well Water to the Highly Alkaline Waters of Pyramid Lake, Nevada (pH 9.4). Physiol. Zool. 1994, 67, 355–380. [Google Scholar] [CrossRef]
- Wilkie, M.P.; Wood, C.M. The Adaptations of Fish to Extremely Alkaline Environments. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1996, 113, 665–673. [Google Scholar] [CrossRef]
- Randall, D.J.; Wood, C.M.; Perry, S.F.; Bergman, H.; Maloiy, G.M.O.; Mommsen, T.P.; Wright, P.A. Urea Excretion as a Strategy for Survival in a Fish Living in a Very Alkaline Environment. Nature 1989, 337, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, M.P. Ammonia Excretion and Urea Handling by Fish Gills: Present Understanding and Future Research Challenges. J. Exp. Zool. 2002, 293, 284–301. [Google Scholar] [CrossRef] [PubMed]
- Nakada, T.; Hoshijima, K.; Esaki, M.; Nagayoshi, S.; Kawakami, K.; Hirose, S. Localization of Ammonia Transporter Rhcg1 in Mitochondrion-Rich Cells of Yolk Sac, Gill, and Kidney of Zebrafish and Its Ionic Strength-Dependent Expression. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2007, 293, R1743–R1753. [Google Scholar] [CrossRef] [PubMed]
- Sashaw, J.; Nawata, M.; Thompson, S.; Wood, C.M.; Wright, P.A. Rhesus Glycoprotein and Urea Transporter Genes in Rainbow Trout Embryos Are Upregulated in Response to Alkaline Water (pH 9.7) but Not Elevated Water Ammonia. Aquat. Toxicol. 2010, 96, 308–313. [Google Scholar] [CrossRef]
- Chang, Y.-M.; Tang, R.; Dou, X.-J.; Tao, R.; Sun, X.-W.; Liang, L.-Q. Transcriptome and Expression Profiling Analysis of Leuciscus waleckii: An Exploration of the Alkali-Adapted Mechanisms of a Freshwater Teleost. Mol. BioSyst. 2014, 10, 491–504. [Google Scholar] [CrossRef]
- He, Q.; Chang, Y.; Su, B.; Sun, B.; Sun, X.; Liang, L. Effects of carbonate alkalinities on oxygen consumption, ammonia excretion and ammonia excretion gene expression in Leuciscus waleckii Dybowski. J. Shanghai Ocean. Univ. 2016, 25, 551–558. [Google Scholar] [CrossRef]
- Wang, Y.S.; Gonzalez, R.J.; Patrick, M.L.; Grosell, M.; Zhang, C.; Feng, Q.; Du, J.; Walsh, P.J.; Wood, C.M. Unusual Physiology of Scale-Less Carp, Gymnocypris przewalskii, in Lake Qinghai: A High Altitude Alkaline Saline Lake. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2003, 134, 409–421. [Google Scholar] [CrossRef]
- Chang, Y.; Liang, L. Advances of research of physiological and molecular mechanisms related to alkali-saline adaptation for fish species inhabiting alkali-saline water. J. Fish. China 2021, 45, 798–812. [Google Scholar] [CrossRef]
- Wu, S.C.; Horng, J.L.; Liu, S.T.; Hwang, P.P.; Wen, Z.H.; Lin, C.S.; Lin, L.Y. Ammonium-Dependent Sodium Uptake in Mitochondrion-Rich Cells of Medaka (Oryzias latipes) Larvae. Am. J. Physiol. Cell Physiol. 2010, 298, C237–C250. [Google Scholar] [CrossRef]
- Hsu, H.-H.; Lin, L.-Y.; Tseng, Y.-C.; Horng, J.-L.; Hwang, P.-P. A New Model for Fish Ion Regulation: Identification of Ionocytes in Freshwater- and Seawater-Acclimated Medaka (Oryzias latipes). Cell Tissue Res. 2014, 357, 225–243. [Google Scholar] [CrossRef]
- Liu, S.-T.; Horng, J.-L.; Chen, P.-Y.; Hwang, P.-P.; Lin, L.-Y. Salt Secretion Is Linked to Acid-Base Regulation of Ionocytes in Seawater-Acclimated Medaka: New Insights into the Salt-Secreting Mechanism. Sci. Rep. 2016, 6, 31433. [Google Scholar] [CrossRef] [PubMed]
- Sender, S.; Böttcher, K.; Cetin, Y.; Gros, G. Carbonic Anhydrase in the Gills of Seawater-and Freshwater-Acclimated Flounders Platichthys flesus: Purification, Characterization, and Immunohistochemical Localization. J. Histochem. Cytochem. 1999, 47, 43–50. [Google Scholar] [CrossRef]
- Wilson, J.M.; Randall, D.J.; Donowitz, M.; Vogl, A.W.; Ip, A.K.-Y. Immunolocalization of Ion-Transport Proteins to Branchial Epithelium Mitochondria-Rich Cells in the Mudskipper (Periophthalmodon schlosseri). J. Exp. Biol. 2000, 203, 2297–2310. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.H.; Lee, T.H. The Novel Correlation of Carbonic Anhydrase II and Anion Exchanger 1 in Gills of the Spotted Green Pufferfish, Tetraodon nigrovirids. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2007, 307, 411–418. [Google Scholar] [CrossRef]
- Gu, J.; Dai, S.; Liu, H.; Cao, Q.; Yin, S.; Lai, K.P.; Tse, W.K.F.; Wong, C.K.C.; Shi, H. Identification of Immune-Related Genes in Gill Cells of Japanese Eels (Anguilla japonica) in Adaptation to Water Salinity Changes. Fish Shellfish Immunol. 2018, 73, 288–296. [Google Scholar] [CrossRef]
- Bekeredjian-Ding, I.; Inamura, S.; Giese, T.; Moll, H.; Endres, S.; Sing, A.; Zähringer, U.; Hartmann, G. Staphylococcus aureus Protein A Triggers T Cell-Independent B Cell Proliferation by Sensitizing B Cells for TLR2 Ligands. J. Immunol. 2007, 178, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.W.; Gaffen, S.L. IL-17RC: A Partner in IL-17 Signaling and Beyond. Semin. Immunopathol. 2010, 32, 33–42. [Google Scholar] [CrossRef]
- Houde, A.L.S.; Akbarzadeh, A.; Günther, O.P.; Li, S.; Patterson, D.A.; Farrell, A.P.; Hinch, S.G.; Miller, K.M. Salmonid Gene Expression Biomarkers Indicative of Physiological Responses to Changes in Salinity and Temperature, but Not Dissolved Oxygen. J. Exp. Biol. 2019, 222, jeb198036. [Google Scholar] [CrossRef]
- Makrinos, D.L.; Bowden, T.J. Natural Environmental Impacts on Teleost Immune Function. Fish Shellfish Immunol. 2016, 53, 50–57. [Google Scholar] [CrossRef]
- Jing, L.B. Analysis of the Characteristics and Changing Trends of Surface Water Chemical Compositions in the Main Stream of the Taizi River in Liaoyang City. Ground Water 2024, 46, 102–104. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Duan, Y.; Liu, X.; Huo, B.; Liu, J.; Tang, R.; Li, D. Transcriptional Differences in Gills Provide Insights into the Environmental Acclimatization of Wild Topmouth Gudgeon (Pseudorasbora parva) from Freshwater Invasion to Saline–Alkali Waters. Water 2025, 17, 1794. https://doi.org/10.3390/w17121794
Liu L, Duan Y, Liu X, Huo B, Liu J, Tang R, Li D. Transcriptional Differences in Gills Provide Insights into the Environmental Acclimatization of Wild Topmouth Gudgeon (Pseudorasbora parva) from Freshwater Invasion to Saline–Alkali Waters. Water. 2025; 17(12):1794. https://doi.org/10.3390/w17121794
Chicago/Turabian StyleLiu, Lu, Yuanshuai Duan, Xuan Liu, Bin Huo, Jieya Liu, Rong Tang, and Dapeng Li. 2025. "Transcriptional Differences in Gills Provide Insights into the Environmental Acclimatization of Wild Topmouth Gudgeon (Pseudorasbora parva) from Freshwater Invasion to Saline–Alkali Waters" Water 17, no. 12: 1794. https://doi.org/10.3390/w17121794
APA StyleLiu, L., Duan, Y., Liu, X., Huo, B., Liu, J., Tang, R., & Li, D. (2025). Transcriptional Differences in Gills Provide Insights into the Environmental Acclimatization of Wild Topmouth Gudgeon (Pseudorasbora parva) from Freshwater Invasion to Saline–Alkali Waters. Water, 17(12), 1794. https://doi.org/10.3390/w17121794










