Abstract
The Rio Bravo/Grande River is a binational water resource between Mexico and the United States and supports diverse anthropogenic activities. However, limited studies on its microbiological composition focus on molecular techniques. Therefore, the aim of this study was to characterize the bacteriome and identify potentially pathogenic bacteria in surface waters of the Rio Bravo/Grande in northeastern Tamaulipas, Mexico, using the 16S rRNA gene metabarcoding technique. Surface water samples were collected from the localities of Diaz Ordaz, Reynosa, and Matamoros between 2016 and 2017. DNA extraction and sequencing were performed, focusing on the V3–V4 region of the 16S rRNA gene. A taxonomic analysis revealed the presence of 13 bacterial phyla, with Proteobacteria (40%), Firmicutes (28%), and Actinobacteria (10.2%) being the most abundant. At the genus level, Bacillus, Pseudomonas, and Acinetobacter were predominant, while 16 potentially pathogenic species, including Acinetobacter baumannii and Vibrio vulnificus, were identified. Alpha and beta diversity analyses highlighted significant differences in the bacterial diversity among the sampling sites, indicating that the river has some capacity to recover from anthropogenic and environmental disturbances. This study underscores the need for the continuous monitoring of the Rio Bravo/Grande to protect public health and maintain the water quality in the face of increasing anthropogenic pressures.
1. Introduction
The study of microbial community composition in aquatic environments is essential, as these microorganisms respond to environmental variations and influence water quality by participating in biogeochemical cycles. Identifying these communities helps assess the ecological status of rivers and their connection to pollution sources [1]. Microorganisms play a fundamental role in the biogeochemical cycles of the environment. However, the composition and dynamics of bacterial communities can be influenced by various factors, including climatic conditions, anthropogenic activities, and environmental pollution.
For example, the presence of chemical contaminants can significantly alter microbial communities in aquatic ecosystems, particularly rivers, impacting ecosystem health. A study conducted in Milwaukee, WI, USA, suggests that higher concentrations of contaminants, such as pharmaceuticals, personal care products, and agricultural runoff, can reduce microbial density and negatively affect bacterial community composition [1].
Regarding climatic conditions, it has been demonstrated that these also influence microbial dynamics. Salami et al. (2024) [2] studied the seasonal variation of bacterial communities in Lake Urmia, a hypersaline ecosystem, and found that bacterial diversity decreases in summer and winter compared to spring and autumn. This indicates that, although climatic conditions are not pollutants themselves, they play a key role in shaping bacterial populations.
On the other hand, anthropogenic activities represent one of the main sources of alteration in freshwater bodies, including rivers [2]. It has been documented that factors such as urbanization, tourism, agriculture, and industrial activities near water bodies are associated with the presence of pathogens, antimicrobial resistance genes, and modifications in bacterial community composition [3,4].
The Rio Bravo/Grande River is a binational water resource of significant importance due to its location between the United States of America and Mexico. In Mexico, the Rio Bravo/Grande flows through the states of Chihuahua, Coahuila, Nuevo León, and Tamaulipas, and its water is utilized for recreation, agriculture, livestock, and as a source of drinking water [5]. Approximately 14 million people reside in the vicinity of the Rio Bravo/Grande, with 7.2 million in the United States and 6.8 million in Mexico [6]. Moreover, the rapid industrialization of the surrounding areas, migratory flows, and the demand for livestock and agriculture have significantly impacted the surface water to the extent of affecting bacterial communities. The type of water at different points of the Rio Bravo varies according to the intensity of anthropogenic activities in the area, which causes fluctuations in the concentration of these microorganisms and, therefore, in water quality. Areas near urban and industrial centers show higher concentrations of total coliforms, highlighting the need to monitor and properly manage pollution sources to protect water resources and the health of surrounding populations. Studies have been conducted by our group on the prevalence of water quality indicator bacteria in the Rio Bravo/Grande (specifically in Reynosa), where Escherichia coli, Enterococcus spp., and Vibrio spp. have been reported, along with the presence of virulence factors, including Shiga toxin genes associated with E. coli strains and stx1 (10%) and stx2 (27%) genes, as well as the presence of multi-toxigenic Vibrio spp., which were the most prevalent in the Rio Bravo/Grande [7,8].
However, culture-based studies on bacteria do not provide a comprehensive view of the global distribution of bacteria, nor do they allow for the distinction and proportion of pathogenic bacteria. An alternative approach is metagenomics, which involves sequencing the genetic information of the majority of the bacteria present in bodies of water, as it is estimated that more than 80% of bacteria detected by molecular biology are non-cultivable. Therefore, many microorganisms are known through their Operational Taxonomic Units (OTUs), providing a broad overview of the microbial populations present in water resources [9,10]. While identifying bacteria as indicators of water quality is appropriate for estimating water body contamination, it is worth mentioning that next-generation sequencing techniques can broaden the spectrum of the bacterial populations that can be detected in water samples.
The focus of metagenomic techniques in bodies of water is to characterize microbial populations in different sources that are used for consumption or recreation [11]. The detection of bacterial communities through 16s RNA metabarcoding has been used in studies of surface waters in many countries. For example, a study conducted on rivers in India aimed to analyze beneficial microorganisms capable of living in contaminated environments, which may have potential applications in industrial settings [12]. Additionally, assessments of bacterial microbiomes can evaluate the impacts on and health of the environment, such as in the case of the collapse of the Funda Dam in Minas Gerais, Brazil, considered one of the largest environmental disasters in the history of the global mining industry, where a bacterial microbiome assessment indicated that the microbiome of rivers and lagoons continues to be severely affected [13]. On the other hand, the Tijuana River Basin, which is also of binational importance, has been recently analyzed for the identification of pathogenic microorganisms, such as bacteria, viruses, and parasites, and bacteria that are pathogenic to humans, mainly of the Bacteroides genus, were identified. Allising et al. constructed a profile of the most abundant disease-causing pathogens, identifying A. cryaerophilus as an emerging diarrheal pathogen of interest that the health system typically does not monitor [14]. The directional flow of rivers influences the structure of microbial communities, but human activities can disrupt this continuity by introducing new microorganisms and substances into the ecosystem [15].
Despite the binational importance of the Rio Bravo/Grande as a water resource, its microbiological characterization remains limited, with no sequencing-based studies reported to date in this region. Existing microbiological data—though potentially outdated—can still serve as a reference point for evaluating shifts in the bacterial composition driven by environmental, industrial, or anthropogenic influences. In this context, the present study aimed to characterize native bacterial populations and identify potential health-risk-associated bacteria in surface waters of the Rio Bravo/Grande in northeastern Tamaulipas using 16S rRNA gene metabarcoding.
2. Materials and Methods
Sampling. A total of 38 samples of surface water were collected from the Rio Bravo/Grande in northeastern Tamaulipas, in the cities of Gustavo Diaz Ordaz (DO, 6 samples), Reynosa (RY, 17 samples), and Heroica Matamoros (MT, 15 samples); the distribution is shown in Figure 1. The total length of the Rio Bravo/Grande sampling area was 208 km, and the sampling period extended from January 2016 to May 2017, before noon. One liter of surface water was collected at a distance of at least two meters from the shore at a depth of 30 cm using a sterile container [16]. The samples were transported under refrigerated conditions for analysis at the Centro de Biotecnologia Genómica of the Instituto Politécnico Nacional in Reynosa, Tamaulipas. The cumulative risk percentage was calculated by dividing the number of anthropogenic activities present at each site by the total possible (8), then expressing it as a percentage. These values were averaged across all the sampling sites to obtain the overall cumulative risk.
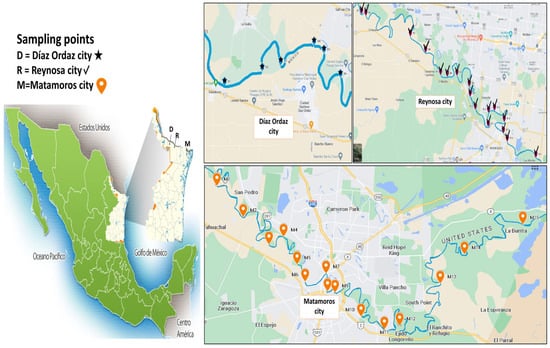
Figure 1.
The distribution of sampling points along the Rio Grande/Rio Bravo River in three cities in the northeast of Tamaulipas.
The evaluation of the microbiological quality of water using the Most Probable Number (MPN) technique. The microbiological quality was assessed based on the standards NOM-210-SSA1-2015 [17] and NOM-112-SSA1-1994 [18]. The obtained results were compared with the guidelines established in Decree No. 33903-S-MINAE—the regulation for the evaluation and classification of surface water quality [19]. According to Decree No. 33903-S-MINAE, surface water bodies are classified into five categories based on their quality and allowable uses. Class I corresponds to excellent-quality water, suitable for direct human consumption without treatment, as well as for recreational activities. Class II water is of good quality, suitable for human consumption, but requires proper treatment for potabilization. Class III water is of moderate quality, suitable for agricultural irrigation and certain industrial uses, but not safe for human consumption without complete treatment. Class IV water is of poor quality, primarily used for the irrigation of crops resistant to a low water quality and certain industrial uses; it is not suitable for human consumption or recreational activities and may contain pathogenic organisms. Finally, Class 5 water is of very poor quality, is not suitable for human consumption or industrial uses without intensive treatment, and is limited to use in the irrigation of crops tolerant to such conditions. Pathogenic organisms are commonly present in this class of water. The maximum permissible contamination limits for fecal coliforms (NMP/100 mL) are as follows: Class 1 (≤20 NMP/100 mL), Class 2 (20 to 1000 NMP/100 mL), Class 3 (1000 to 2000 NMP/100 mL), Class 4 (2000 to 5000 NMP/100 mL), and Class 5 (≥5000 NMP/100 mL).
DNA extraction. The samples were individually filtered using a 0.22 um nylon membrane (PALL Corporation, Atlanta, GA, USA). DNA extraction was carried out using the ZymoBIOMICS® DNA Miniprep kit (Zymo Research, Irvine, CA, USA), with the extraction performed on the biomass retained on the membrane, following the manufacturer’s instructions. The DNA from the samples was grouped and mixed for sequencing as follows: The samples taken from Diaz Ordaz, DO1 to DO3, were analyzed directly due to the low quantity of the samples obtained in the area. Samples DO4 to DO6 were added to a pool labeled DO4p. For the samples from the Reynosa area, four groups were formed, RY1p (including points RY1 to RY4), RY2p (points RY5 to RY8), RY3p (points RY9 to RY12), and RY4p (points RY13 to RY17). Finally, for the city of Matamoros, the samples were categorized as MT1p (including points MT1 to MT4), MT2p (points MT5 to MT8), MT3p (points MT9 to MT12), and MT4p (points MT13 to MT15). Thus, a total of 12 composite samples were sequenced and analyzed by metabarcoding: from the city of Diaz Ordaz, DO1, DO2, DO3, and DO4p; from the city of Reynosa, RY1p, RY2p, RY3p, and RY4p; and from the city of Matamoros, MT1p, MT2p, MT3p, and MT4p.
Library preparation, sequencing, and bioinformatic analysis. Sequencing targeted the 16s rRNA gene using the Quick-16S™ NGS Library Prep kit (Zymo Research, Invine, CA, USA). Primers designed by Zymo Research targeting the V3–V4 region of the 16S rRNA gene were employed. A targeted amplification of sequences was performed using real-time PCR with the Quick-16S™ NGS Library Prep Kit (Zymo Research, Invine, CA, USA). A master mix was prepared consisting of 10 µL of qPCR Premix, 4 µL of the Quick-16S™ (V3–V4) primer set, and 4 µL of nuclease-free water (DNase/RNase-Free Water), for a total volume of 18 µL per reaction. This mixture was distributed into 96-well PCR plates, and 2 µL of sample DNA was added to each well. Positive controls, such as the ZymoBIOMICS™ Microbial Community DNA Standard, were included. For the amplification of the V3–V4 region of the 16S rRNA gene, primers compatible with the kit protocol were used, designed based on the Earth Microbiome Project (EMP). The primers employed were EMP 515F (forward), with the sequence GTGYCAGCMGCCGCGGTAA, and EMP 806R (reverse), with the sequence GGACTACNVGGGTWTCTAAT, both with their corresponding 5′ adapter overhangs: TCGTCGGCAGCGTCAGATGTGTATAA GAGACAG for the forward primer and G TCTCGTGGGCTCGGAGATGTGTATAA GAGACAGG for the reverse primer. The amplification conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 20 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 3 min, and finally, a plate read and hold at 4 °C.
Final PCR products were quantified using qPCR fluorescence readings and grouped based on equal molarity. The resulting pooled library was cleaned using the Select-a-Size DNA Clean & Concentrator™ kit (Zymo Research, Irvine, CA, USA), then quantified using TapeStation® (Agilent Technologies, Santa Clara, CA, USA) and Qbit® (Thermo Fisher Scientific, Waltham, WA, USA). Subsequently, the final library was sequenced on an Illumina® MiSeq™ instrument with a V3 reagent kit (600 cycles). Sequences were inferred from raw sequences using DADA [20]. Chimeric sequences were also removed using the DADA2 pipeline. For taxonomic assignment, the Uclust program from QIIME v.1.9.1 with the Zymo Research database was utilized. For the visual composition, alpha diversity, and beta diversity analyses, QIIME v.1.9.1 software was employed [21]. The taxonomic assignment and the differences between the sampling groups were determined using the LEfSe program [22] with standard program parameters. Heatmap analysis, Taxa2SV_decomcomposer, and component analysis were also conducted using internal scripts of the QIIME v1.9.1 program. The alpha diversity of the bacterial communities was assessed using the Shannon index, which was calculated with the estimate_richness function from the phyloseq package in R.
3. Results
General characteristics of water samples. Surface water from the Rio Bravo/Grande was obtained from three locations with varying anthropogenic activities, such as fishing, swimming, anthropogenic pollution, the presence of contaminants from factories, waste, agriculture, and livestock. The DO area exhibited less urbanization near the river compared to RY and MT, which were more impacted by housing, garbage, farms, and recreation activities. The maximum level of NMP/100 mL was ≥16,000 NMP/100 mL, detected in 35 sampling sites along the Rio Bravo. The minimum level of NMP/100 mL was 120 NMP/100 mL for total coliforms and 74 NMP/100 mL for fecal coliforms (corresponding to the city of MT4 and RY10). The results for total coliforms and fecal coliforms were compared based on Decree No. 33903-S-MINAE—the regulation for the evaluation and classification of the quality of surface water bodies (Table 1). The samples belonging to Class 5 also show anthropogenic activities in nearby areas, except for the sites RY10 and MT4. In the case of the samples that do not present accumulated risk percentages higher than 50% but do show high amounts of fecal and total coliforms, this can be associated with other factors unrelated to nearby anthropogenic activity. This result may be due to natural sources near the Rio Bravo, such as the runoff of organic matter from rainfall, deposits from wildlife, or natural decomposition processes in bodies of water. The RY10 sample presents an accumulated risk of 50% and MT4 of 25%; however, the coliform count is low in this area. The water sampling conditions ranged from 31 °C to 41 °C, mostly cloudy. It is important to mention this as evaporation can influence the distribution of contaminants and nutrients, affecting the microbiological quality of the water.

Table 1.
Climatic conditions, presence of coliforms, and anthropogenic activity on sampling sites.
General characteristics of sequencing data. A total of 860,108 raw sequences were obtained. Quality filtering was performed using Trimmomatic v0.39 with the PE (paired-end) option and the parameters LEADING:3, TRAILING:3, SLIDINGWINDOW:4:30, and MINLEN:30, aiming to remove adapters and low-quality bases. After filtering, the number of sequences was reduced to 331,924 ASVs. The average number of raw sequences per sampling site was 71,675, which was reduced to 27,600 ASVs per site after processing with DADA2. After chimera removal, an average of 232.5 unique sequences per sampling site were identified. Among all the sampling groups, the highest number of unique sequences was found in DO4p, with 297 sequences, and the lowest was found in RY2p, with 147 sequences.
Taxonomic assignment. The composition of bacterial diversity in the Rio Bravo/Grande. The 16s rRNA gene database from Zymo Research identified the following abundance of bacteria at the phylum level: Proteobacteria 40%, Firmicutes 28%, Cyanobacteria 9%, Bacteroidetes 4%, Verrucomicrobia 8%, Plantomicetes 1%, and Actinobacteria 10%. Bacteria belonging to a single site were identified, such as Chlamydiae (0.06%), which was detected at site DO2. The rest of the distribution, by phylum, is shown in Figure 2. The prevalence of bacterial families in the Rio Bravo/Grande were as follows: Bacillaceae (15.4%), Moraxellaceae (9.6%), Planococcaceae (8.7%), Oxalobacteraceae (6.1%), Sporichthyaceae (4.8%), Pseudomonadaceae (4.5%), Acidimicrobacteriaceae (4.4%), Rhodobacteraceae (2.6%), Sphingomonadaceae (2.3%), Comamonadaceae (1.6%), Cytophagaceae (1.4%), Caulobacterceae (1.16%), Mycobacteriaceae (1%), Alcalinogenaceae (1%), and Planctomycetaceae (1.3%), and the distribution of the rest of the families was less than 1%.
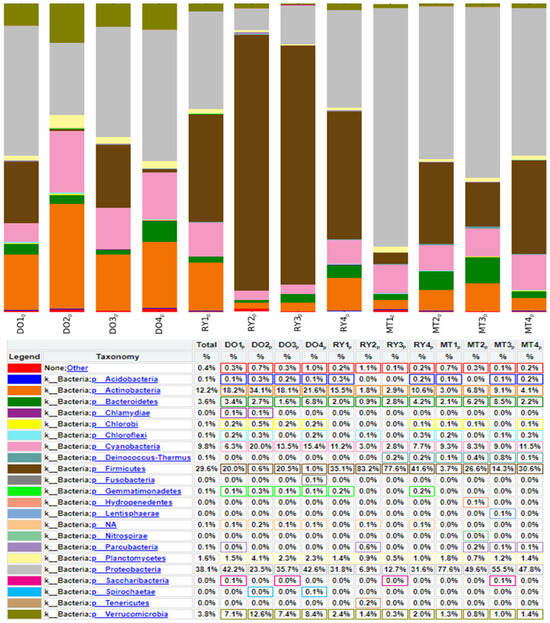
Figure 2.
Distribution of bacteria by phylum in surface water samples from the Rio Bravo. k: class. p: phylum. %: the percentage of sequences obtained representing the abundance of detected bacteria.
The DO sites showed a higher abundance of Acidimicrobacteriaceae (11.59%) and Rhodobacteraceae (4%). The detection of these families in water bodies may indicate the presence of active microbial communities that play important ecological roles, such as the degradation of organic matter, biogeochemical cycles, and the promotion of plant growth. In Reynosa sites, the family Bacillaceae (33.31%) and Planococcaceae (21.83%) were more abundant. Finally, the highest abundances of Moraxellaceae (15.97%), Oxalobacteraceae (12.06%), and Pseudomonadaceae (9.72%) were observed in Matamoros sites. At the genus level, the most abundant were Bacillus (13.9%), Pseudomonas (9.5%), Acinetobacter (9.4%), Cyanobacterium (8%), Synechococcus (5%), Exiguobacterium (3.9%), Sphingomonas (1.6%), Planomicrobium (1.6%), Brevumdimonas (1.6%), Fictibacillus (1.5%), and Mycobacterium (1.1%). Figure 3 shows the distribution of bacteria at the genus and species levels. In Diaz Ordaz city, Synechococcus and Planomicrobium were detected, which are characteristic of surface waters and natural environments, but the percentage of these genera decreased from 10.5% to 0.7% for Synechoccus and from 1.9% to 0% for Planomicrobium downstream, where the waters merged with surface water sites in the Reynosa area. We also detected the genera Pseudomonas (4.5%), Exiguobacterium (11.6%), Bacillus (13.9%), and Acinetobacter (16.1%) in all three sampling sites along the Rio Bravo/Grande River.
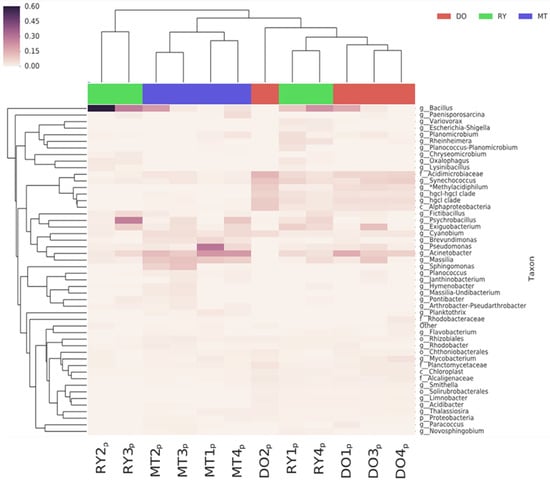
Figure 3.
Heat map showing abundance of genera in analyzed sample pools.
The genus Bacillus was the most prevalent, reaching up to 13.9%, predominantly in the Reynosa areas (RY2p 60.3%), compared to (DO116.6%) and MT (MT2: 20.6%). Brevumdimonas (DO: 1.5%; MT: 6.6%) and Sphingomonas (DO: 1.5%; MT: 6.6%) were mostly detected in Matamoros and Diaz Ordaz. The Zymo Research database repository contains algorithms for the identification of pathogenic bacteria (Table 2), where the following were identified: Acinetobacter calcoaceticus, Acinetobacter baumanii complex, and Peptoclostridium sp., were prevalent in DO sites; in RY sites, A. baumanii, S. agalactiae, Vibrio vulnificus, and Shigella sp., were found; and for MT, the predominant species was Acinetobacter iwoffi.

Table 2.
Human pathogens detected.
Analysis of α-diversity and β-diversity. In the α-diversity analysis, the highest number of ASVs of bacterial species was observed at points DO4p, followed by DO1p and RY4p; similar ASV richness was observed at sites DO2p, DO3p, RY1p, MT2p, MT3p, and MT4p. The sites with the lowest numbers of ASVs were MT1p, RY1p, and RY3p (Figure 4). The Shannon index obtained was as follows.: DO1p 3.701817, DO2p 3.209975, DO3P 3.552543, DO4 4.230499, RY1p 3.804948, RY2p 3.345132, RY3p 3.331037, RY4p 3.847665, MT1p 3.393235, MT2p 3.984850, MT3p 3.918910, and MT4p 4.010361.
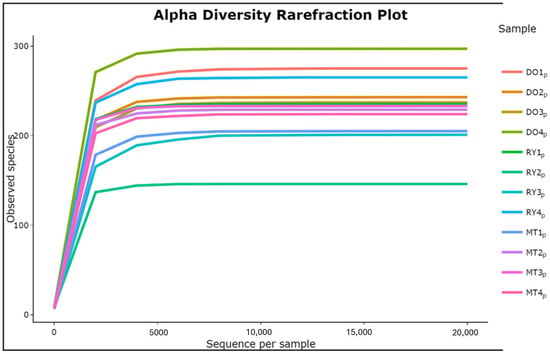
Figure 4.
Rarefaction curves for alpha diversity showing the detected ASVs in the sample pools.
The β-diversity analysis of the sampling sites is shown in Figure 5, where three groups are observed. The first groups include sites DO2p-DO4p-DO3p, which showed the highest abundance of ASVs, followed by the group MTp (all sites), which exhibited a similar species distribution. MT3p-MT4p and MT2p were similar in both species composition and abundance distribution. Sites RYp (all sites) did not cluster together; these sites showed lower richness and reduced species distribution.
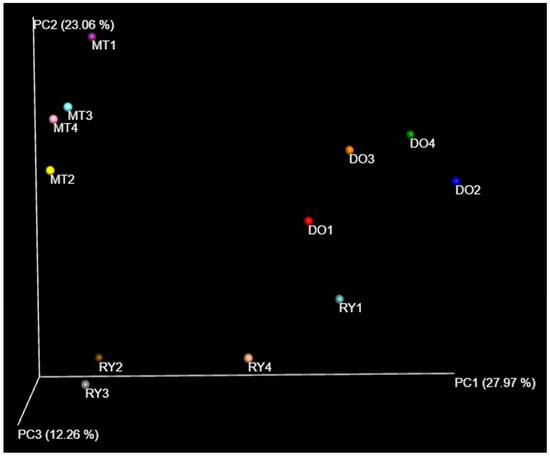
Figure 5.
The PCoA analysis of the water sample pools, by the Bray Curtis method. Colors indicate water samples.
4. Discussion
The Rio Bravo/Grande River is a body of water that has a direct impact both economically and on public health, as it is used for various activities, such as the irrigation of crops and recreational activities including swimming, sport fishing, and boating. In addition, the ability to use water from the Rio Bravo/Grande is determined by its processing for water purification, which is carried out for the homes of the inhabitants of the northern region of Tamaulipas. Surface waters, such as rivers, contain nutrients that are incorporated into biogeochemical cycles. However, problems relating to microbiological water quality associated with anthropogenic contaminants have disturbed native microbial communities in rivers to the extent of causing a decrease in native species [23]. In our study, we identified that the presence of high levels of fecal coliforms is a danger to the population that uses the Rio Bravo for primary contact, and we found the presence of contaminants such as dead animals near the area, pollution from garbage, and wastewater discharge near the river. These pollutants can be the source of potential pathogens that could affect the quality of the river, as well as that of the surrounding population (Table 2). This issue is not unique to the Rio Bravo/Grande River, as similar findings have been reported in other highly contaminated water bodies. For instance, the Ganges River suffers from severe pollution due to wastewater, industrial waste, and plastic debris. Religious rituals and cremations also contribute to its deterioration. This has led to the growth of antibiotic-resistant bacteria, affecting public health. Despite cleanup efforts, pollution persists due to population growth and poor waste management [24]. Another important factor that could have contributed to the growth of total and fecal coliforms is climatic conditions, as temperature plays a significant role in bacterial proliferation [2]. Summer temperatures in Tamaulipas average between 31 °C and 39 °C. High temperatures promote the growth of pathogenic and extremophilic bacteria. Additionally, the excessive presence of nitrates due to environmental pollution can lead to a eutrophic environment, increasing the levels of cyanobacteria, which could be harmful to aquatic life [25]. In this study, no physicochemical parameters were measured, except for those shown in Table 1, which represents a limitation in our research. However, anthropogenic factors can have a significant impact on the bacterial diversity of the Rio Bravo, and the results obtained show the potential influence of these factors on the bacterial variability in the region that should be confirmed by a more directed analysis approach. We identified lower ASV richness at RY sites (Shannon index of 3.331037) and DO sites (Shannon index of 4.230499), with an abundance of the phyla Proteobacteria and Firmicutes; the increase in the presence of these bacteria may be due to Reynosa being the area most affected by anthropogenic activity, leading to a greater exposure to contamination. This is similar to findings by [11], in Lima, Peru, where they indicate that the presence of phyla such as Bacteroidota, Campylobacterota, Fusobacteriota, and Proteobacteria are associated with freshwater and domestic sewage [11].
Similar results were obtained by [26] in China, from the Weihe River, indicating that there is not much difference in the characterization of surface waters by phylum; hence, it is preferable to identify potential pathogens. In our study, we observed the alteration (increase and decrease) of Proteobacteria along the Rio Bravo/Grande River (Figure 2). This is because Proteobacteria are a typical population found in freshwater environments and constitute a significant phylum of bacteria found across a wide range of ecosystems, including soils, freshwater, saltwater, and sediments. Proteobacteria have been regarded as useful indicators of environmental health in water systems [27]. Some Proteobacteria are known to thrive in contaminated waters or those with high nutrient loads, making them potential indicators of water quality and the presence of anthropogenic impacts. Monitoring the levels and diversity of Proteobacteria could help assess the overall health of the river and identify potential public health risks [27].
The families Bacillaceae and Moraxellaceae were the most abundant in this study and were present in all the samples along the sampled portion of the Rio Bravo/Grande River. The Bacillaceae family is widely distributed in different natural environments but also thrives in niches with anthropogenic activity as these microorganisms have the ability to form spores, allowing them to withstand extreme environmental conditions, such as high temperatures and drought, enabling them to persist for extended periods [28], making the Rio Bravo/Grande River a perfect niche for these microorganisms. It is worth noting that high temperatures and frequent droughts in the Rio Bravo/Grande region favor the survival of spore-forming bacteria, such as those from the Bacillaceae family, due to their ability to form spores that are resistant to extreme conditions. Thermal fluctuations and prolonged drought periods create an ideal environment for the sporulation of these bacteria [29]. Additionally, anthropogenic pollution, such as wastewater discharge and agricultural activity along the river, alters aquatic ecosystems, providing additional niches where these bacteria can persist and proliferate [17]. The anthropogenic pollution present at the sites may be associated with the proliferation of these bacteria, as nutrients and contaminants of human origin can promote their growth and spread. Considering that in our study the highest temperature recorded was 41 °C, these conditions likely contributed to the survival and proliferation of spore-forming bacteria in the region.
The Moraxellaceae family is widely distributed in various environments and holds clinical significance. Among the genera associated with human pathogens, we can find Acinetobacter, a bacterium that is commonly found in aquifer environments and soils; however, it has also been isolated from hospital areas. Its significance lies in the fact that it has shown resistance to multiple antibiotics [30,31].
The Comamonadaceae family (1.6%) was less abundant than reported by [23] in the Nile River in Cairo, where a higher abundance of Comamonadaceae was identified. This family of bacteria is common in surface waters and plays an important role in denitrifying bacteria in aquatic environments, thereby reducing the nitrogen levels in the water, which helps prevent problems such as eutrophication and excessive algae growth since they are aerobic degraders of aromatic compounds [32]. The presence of these bacteria could contribute to the Rio Bravo River’s (upstream to downstream) recovery from the presence of anthropogenic activities. Although they are effective to some extent, the process is usually slow and influenced by other factors.
At the genus level, Bacillus was the most abundant in all the samples. These microorganisms can be found in water, soil, animal intestines, and human intestines. They are also considered the most abundant Gram-positive bacteria in surface water, along with Micrococcus spp. and Corynebacterium spp. [33]. Although Micrococcus spp. and Corynebacterium spp. are commonly considered Gram-positive bacteria present in surface waters, they were not detected in our metagenomic analysis. This absence could be due to their presence at concentrations below the detection threshold or an actual decrease in their abundance, possibly related to the specific environmental conditions of the Rio Bravo/Grande River. Surface waters with higher anthropogenic impacts may harbor bacteria that are pathogenic to humans. However, in this study, we observed a distribution of potential pathogens in all the sampling points, including areas with a lower environmental impact. This could be because some bacteria listed in Table 2 may inhabit different environmental niches but harbor virulence factors that could make them potential human pathogens. These genera have also been identified in the surface waters used in rural areas in Haiti: Acinetobacter, Aeromonas, Bacillus, Clostridium, Enterobacter, Escherichia, Klebsiella, Peptoclostridium, and Shigella y Yersinia [34]. We also detected the presence of Mycobacterium spp., Vibrio sp., Legionella sp., and Leptospira sp. in the Rio Bravo/Grande River; 16 potential pathogenic species were identified (Table 2). DO presented 14 out of 16 pathogenic species, RY had 10 out of 14, and MT had 7 out of 14. In the case of the MT sampling sites, these areas flow into the Gulf of Mexico, so conditions such as salinity and pH may vary, favoring the resident bacterial communities of the river [17]. However, one of the limitations of our study is that the physicochemical parameters were not measured, which we consider a limiting factor. These conditions may influence the abundance and composition of the bacterial communities. Therefore, without measuring these parameters, it is difficult to determine the dynamics of these bacteria. Nevertheless, it is worth mentioning that the focus of the study was on describing the bacterial diversity of the Rio Bravo/Grande river.
In the analyses of α and β diversity, more than 300 unique species were observed from an evaluation of 20,000 sequences used for this analysis, with the DO areas showing moderate richness due to a lower anthropogenic impact. The genera found in this zone were Planomicrobuym and Synechococcus, which have been reported as photosynthetic cocci present in surface waters and are among the most abundant in natural areas, mainly freshwater environments [35]. It has been reported that the presence of the genera Sphingomonas, Sphingobium, and Novosphingobium from the family Sphingomonadaceae, as well as members of Comamonadaceae, have the ability to degrade a wide range of aromatic compounds [25]. This has positive implications for environmental conservation and could explain why the DO (Sphingomonas 1.6%) zones are not as severely impacted compared to the RY zones.
In the RY sites, Pyschrobacillus was the most abundant genus. These microorganisms have been poorly studied, but they are attributed to the capability of degrading contaminants [36,37]. This bacterium is not commonly found in freshwater as it is an extremophile and grows under conditions below 15 °C. Notwithstanding, in a study published by Rodriguez in 2020 in Spain, they identified the presence of a new species named Psychrobacillus vulpis sp., isolated from a red fox in Spain [38]. This suggests that this bacterium may likely be associated with the presence of fox feces near the Rio Bravo/Grande river.
Although the number of total and fecal coliforms is high, it has been described that coliforms are not always an accurate indicator of fecal contamination, as some bacteria within these groups originate from natural environments (mainly soil and water). This demonstrates the impact on the microbiological quality in the surface waters of the Rio Bravo/Grande River, confirming that the presence of microbiological indicators with potential pathogenic genes significantly affects the surface water of the Rio Bravo/Grande River.
5. Conclusions
A wide bacterial diversity was identified in the Rio Bravo/Grande River, with Bacillaceae (15.4%) and Moraxellaceae (9.6%) as the predominant families. An upstream pattern was observed, with more potential pathogens in the upper river and fewer near the mouth, suggesting recent contamination and a possible natural recovery capacity. Despite this, the persistent presence of pathogens remains a public health concern, especially for those relying on the river for water. Total and fecal coliforms were found in high concentrations, particularly upstream, indicating microbial contamination, although their presence may also come from natural sources. According to Decree No. 33903-S-MINAE, these waters fall under Class 5, indicating high levels of pollution and unsuitability for direct human use. This is the first metagenomic study of the Rio Bravo/Grande River, revealing human pathogenic bacteria in its surface waters and indicating that the river may act as a reservoir for such organisms. However, no physicochemical analyses were performed, limiting a full understanding of contamination sources. The presence of biodegrading bacteria suggests a potential for slow bioremediation. Ongoing monitoring is essential to assess contamination trends and guide preventive actions. In conclusion, it is vital for local authorities and communities to adopt integrated water management strategies that address both microbial and physicochemical factors, ensuring public health and sustainable access to water.
Author Contributions
Methodology, R.R.-C., M.G.A.-A., A.V.M.-V., W.L.C.-P., G.R., S.F.-D., R.F.-M. and E.A.-C.; Resources, A.V.M.-V.; Writing—original draft, R.R.-C.; Writing—review & editing, V.B.-G.; Supervision, M.G.A.-A. and V.B.-G.; Funding acquisition, G.R. and V.B.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data available under request to the authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Muhie, S.; Gautam, A.; Mylroie, J.; Sowe, B.; Campbell, R.; Perkins, E.J.; Hammamieh, R.; Garcia-Reyero, N. Effects of environmental chemical pollutants on microbiome diversity: Insights from shotgun metagenomics. Toxics 2025, 13, 142. [Google Scholar] [CrossRef] [PubMed]
- Salami, R.; Saidi, A.; Hejazi, M.A.; Panahi, B.; Hamid, R. Seasonal dynamics of the bacterial community in Lake Urmia, a hypersaline ecosystem. Biology 2025, 14, 75. [Google Scholar] [CrossRef]
- Furtak, K.; Gawryjołek, K.; Marzec-Grządziel, A.; Niedźwiecki, J. The Influence of human agricultural activities on the quality of selected fluvisols from the Vistula River Valley, Poland—Preliminary research. Agronomy 2024, 14, 480. [Google Scholar] [CrossRef]
- Di Cesare, A.; Mammola, S.; Sabatino, R.; Fontaneto, D.; Eckert, E.M.; Rogora, M.; Tonsi, T.; Corno, G. Where do the antibiotic resistance genes come from? A modulated analysis of sources and loads of resistances in Lake Maggiore. FEMS Microbiol. Ecol. 2024, 100, fiae025. [Google Scholar] [CrossRef]
- Virgin, T.L.; Sonthiphand, P.; Coyotzi, S.; Hall, M.W.; Venkiteswaran, J.J.; Elgood, R.J.; Schiff, S.L.; Neufeld, J.D. Microbial communities change along the 300 km length of the Grand River for extreme high- and low-flow regimes. Can. J. Microbiol. 2024, 70, 289–302. [Google Scholar] [CrossRef]
- Fuentes, M.D.; Gutierrez, S.; Sahagun, D.; Gomez, J.; Mendoza, J.; Ellis, C.C.; Bauer, S.; Blattner, J.; Lee, W.-Y.; Alvarez, M.; et al. Assessment of antibiotic levels, multi-drug resistant bacteria and genetic biomarkers in the waters of the Rio Grande River between the United States-Mexico border. J. Health Pollut. 2019, 9, 190912. [Google Scholar] [CrossRef]
- Esteve-Gassent, M.D.; Pérez de León, A.A.; Romero-Salas, D.; Feria-Arroyo, T.P.; Patino, R.; Castro-Arellano, I.; Gordillo-Pérez, G.; Auclair, A.; Goolsby, J.; Rodriguez-Vivas, R.I.; et al. Pathogenic landscape of transboundary zoonotic diseases in the Mexico-US border along the Rio Grande. Front. Public Health 2014, 2, 177. [Google Scholar] [CrossRef]
- Requena-Castro, R.; Aguilera-Arreola, M.G.; Martínez-Vázquez, A.V.; Bocanegra-García, V. Prevalencia de genes de virulencia de Escherichia coli en aguas superficiales del Río Bravo en la ciudad de Reynosa, Tamaulipas. Mex. J. Biotechnol. 2018, 3, 87–93. [Google Scholar] [CrossRef]
- Guardiola-Avila, I.; Martínez-Vázquez, V.; Requena-Castro, R.; Juárez-Rendón, K.; Aguilera-Arreola, M.; Rivera, G.; Bocanegra-García, V. Isolation and identification of Vibrio species in the Rio Bravo/Grande and water bodies from Reynosa, Tamaulipas. Lett. Appl. Microbiol. 2018, 67, 190–196. [Google Scholar] [CrossRef]
- Pédron, J.; Guyon, L.; Lecomte, A.; Blottière, L.; Chandeysson, C.; Rochelle-Newall, E.; Raynaud, X.; Berge, O.; Barny, M.-A. Comparison of environmental and culture-derived bacterial communities through 16S metabarcoding: A powerful tool to assess media selectivity and detect rare taxa. Microorganisms 2020, 8, 1129. [Google Scholar] [CrossRef]
- Romero, P.E.; Calla-Quispe, E.; Castillo-Vilcahuaman, C.; Yokoo, M.; Fuentes-Rivera, H.L.; Ramirez, J.L.; Ampuero, A.; Ibáñez, A.J.; Wong, P. From the Andes to the desert: 16S rRNA metabarcoding characterization of aquatic bacterial communities in the Rimac river, the main source of water for Lima, Peru. PLoS ONE 2021, 16, e0250401. [Google Scholar] [CrossRef] [PubMed]
- Rusiñol, M.; Martínez-Puchol, S.; Timoneda, N.; Fernández-Cassi, X.; Pérez-Cataluña, A.; Fernández-Bravo, A.; Moreno-Mesonero, L.; Moreno, Y.; Alonso, J.L.; Figueras, M.J.; et al. Metagenomic analysis of viruses, bacteria and protozoa in irrigation water. Int. J. Hydrog. Environ. Health 2020, 224, 113440. [Google Scholar] [CrossRef]
- Behera, B.K.; Patra, B.; Chakraborty, H.J.; Sahu, P.; Rout, A.K.; Sarkar, D.J.; Parida, P.K.; Raman, R.K.; Rao, A.R.; Rai, A.; et al. Metagenome analysis from the sediment of river Ganga and Yamuna: In search of beneficial microbiome. PLoS ONE 2020, 15, e0239594. [Google Scholar] [CrossRef]
- de Almeida, P.I.N.; de Jesus, H.E.; Pereira, P.H.F.; Vieira, C.E.D.; Bianchini, A.; Martins, C.D.M.G.; dos Santos, H.F. The microbial profile of rivers and lagoons three years after the impact of the world’s largest mining disaster (Fundão dam, Brazil). Environ. Res. 2023, 216, 114710. [Google Scholar] [CrossRef]
- Allsing, N.; Kelley, S.T.; Fox, A.N.; Sant, K.E. Metagenomic analysis of microbial contamination in the U.S. portion of the Tijuana River Watershed. Int. J. Environ. Res. Public Health 2022, 20, 600. [Google Scholar] [CrossRef]
- NMX-AA-003-SCFI-2019; Análisis de agua–Muestreo de aguas Naturales, Residuales y Potables—Guía para la Preservación y Manejo de Muestras. Secretaría de Economía: Jalisco, Mexico; Dirección General de Normas: Mexico City, Mexico, 2019.
- Tan, Q.; Wang, X.; Zheng, L.; Wu, H.; Xing, Y.; Tian, Q.; Zhang, Y. Anthropogenic pressure induced discontinuities of microbial communities along the river. J. Environ. Manag. 2025, 373, 123764. [Google Scholar] [CrossRef]
- NORMA MEXICANA NMX-AA-42-2015; Análisis de Agua-Enumeración de Organismos Coliformes Totales, Organismos Coliformes Fecales (Termotolerantes) y Escherichia coli, Método del Número más Probable en Tubos Múltiples. Diario Oficial de la Federación: Mexico City, Mexico, 2015.
- NORMA OFICIAL MEXICANA NOM-112-SSA1-1994; Bienes y Servicios. Determinación de Bacterias Coliformes. Técnica del Número más Probable. Diario Oficial de la Federación: Mexico City, Mexico, 1994.
- Decreto Ejecutivo N° 33903-S; Reglamento para la Evaluación y Clasificación de la Calidad de Cuerpos de Agua Superficiales. Diario Oficial La Gaceta: San José, Costa Rica, 2007.
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Rout, A.K.; Tripathy, P.S.; Dixit, S.; Behera, D.U.; Behera, B.; Das, B.K.; Behera, B.K. Unveiling the microbiome landscape: A metagenomic study of bacterial diversity, antibiotic resistance, and virulence factors in the sediments of the River Ganga, India. Antibiotics 2023, 12, 1735. [Google Scholar] [CrossRef]
- Sodhi, K.K.; Kumar, M.; Singh, D.K. Assessing the bacterial diversity and functional profiles of the River Yamuna using Illumina MiSeq sequencing. Arch. Microbiol. 2021, 203, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.; Arroyo, R.; Espinosa-Martos, I.; Fernández, L.; Rodríguez, J.M. Identification of emerging human mastitis pathogens by MALDI-TOF and assessment of their antibiotic resistance patterns. Front. Microbiol. 2017, 8, 1258. [Google Scholar] [CrossRef]
- Wang, X.; Gu, J.; Gao, H.; Qian, X.; Li, H. Abundances of clinically relevant antibiotic resistance genes and bacterial community diversity in the Weihe River, China. Int. J. Environ. Res. Public Health 2018, 15, 708. [Google Scholar] [CrossRef]
- Chen, X.; Wei, W.; Wang, J.; Li, H.; Sun, J.; Ma, R.; Jiao, N.; Zhang, R. Tide driven microbial dynamics through virus-host interactions in the estuarine ecosystem. Water Res. 2019, 160, 118–129. [Google Scholar] [CrossRef]
- Wu, G.; Chen, J.; Shi, X.; Kim, J.; Xia, J.; Zhang, L. Impacts of global climate warming on meteorological and hydrological droughts and their propagations. Earth’s Future 2022, 10, e2021EF002542. [Google Scholar] [CrossRef]
- Mandic-Mulec, I.; Stefanic, P.; van Elsas, J.D. Ecology of bacillaceae. In The Bacterial Spore: From Molecules to Systems; ASM Press: Washington, DC, USA, 2016; pp. 59–85. [Google Scholar] [CrossRef]
- Lennert, K.J.; Borsodi, A.K.; Anda, D.; Krett, G.; Kós, P.B.; Engloner, A.I. The effect of urbanization on planktonic and biofilm bacterial communities in different water bodies of the Danube River in Hungary. Sci. Rep. 2024, 14, 23881. [Google Scholar] [CrossRef]
- Song, J.; Choo, Y.-J.; Cho, J.-C. Perlucidibaca piscinae gen. nov.; sp. nov.; a freshwater bacterium belonging to the family Moraxellaceae. Int. J. Syst. Evol. Microbiol. 2008, 58, 97–102. [Google Scholar] [CrossRef]
- Eraqi, W.A.; ElRakaiby, M.T.; Megahed, S.A.; Yousef, N.H.; Elshahed, M.S.; Yassin, A.S. The nile river microbiome reveals a remarkably stable community between wet and dry seasons, and sampling sites, in a Large Urban Metropolis (Cairo, Egypt). OMICS A J. Integr. Biol. 2018, 22, 553–564. [Google Scholar] [CrossRef]
- Willems, A. The Prokaryotes: The Family Comamonadaceae, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Dvořák, P.; Casamatta, D.A.; Poulíčková, A.; Hašler, P.; Ondřej, V.; Sanges, R. Synechococcus: 3 billion years of global dominance. Mol. Ecol. 2014, 23, 5538–5551. [Google Scholar] [CrossRef]
- Rodríguez, M.; Reina, J.C.; Béjar, V.; Llamas, I. Psychrobacillus vulpis sp. nov.; a new species isolated from faeces of a red fox in Spain. Int. J. Syst. Evol. Microbiol. 2020, 70, 882–888. [Google Scholar] [CrossRef]
- Mukherjee, N.; Bartelli, D.; Patra, C.; Chauhan, B.V.; Dowd, S.E.; Banerjee, P. Microbial Diversity of Source and Point-of-Use Water in Rural Haiti—A Pyrosequencing-Based Metagenomic Survey. PLoS ONE 2016, 11, e0167353. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthi, S.; Ruckmani, A.; Pukall, R.; Chakrabarti, T. Psychrobacillus gen. nov. and proposal for reclassification of Bacillus insolitus Larkin & Stokes, 1967, B. psychrotolerans Abd-El Rahman et al.; 2002 and B. psychrodurans Abd-El Rahman et al.; 2002 as Psychrobacillus insolitus comb. nov.; Psychrobacillus psychrotolerans comb. nov. and Psychrobacillus psychrodurans comb. nov. Syst. Appl. Microbiol. 2010, 33, 367–373. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).