Comparative Analysis of Light Microscopy and High-Throughput Sequencing for Phytoplankton Detection in Rivers Flowing into the Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Microscope Observation
2.3. Metabarcoding Analysis
3. Results
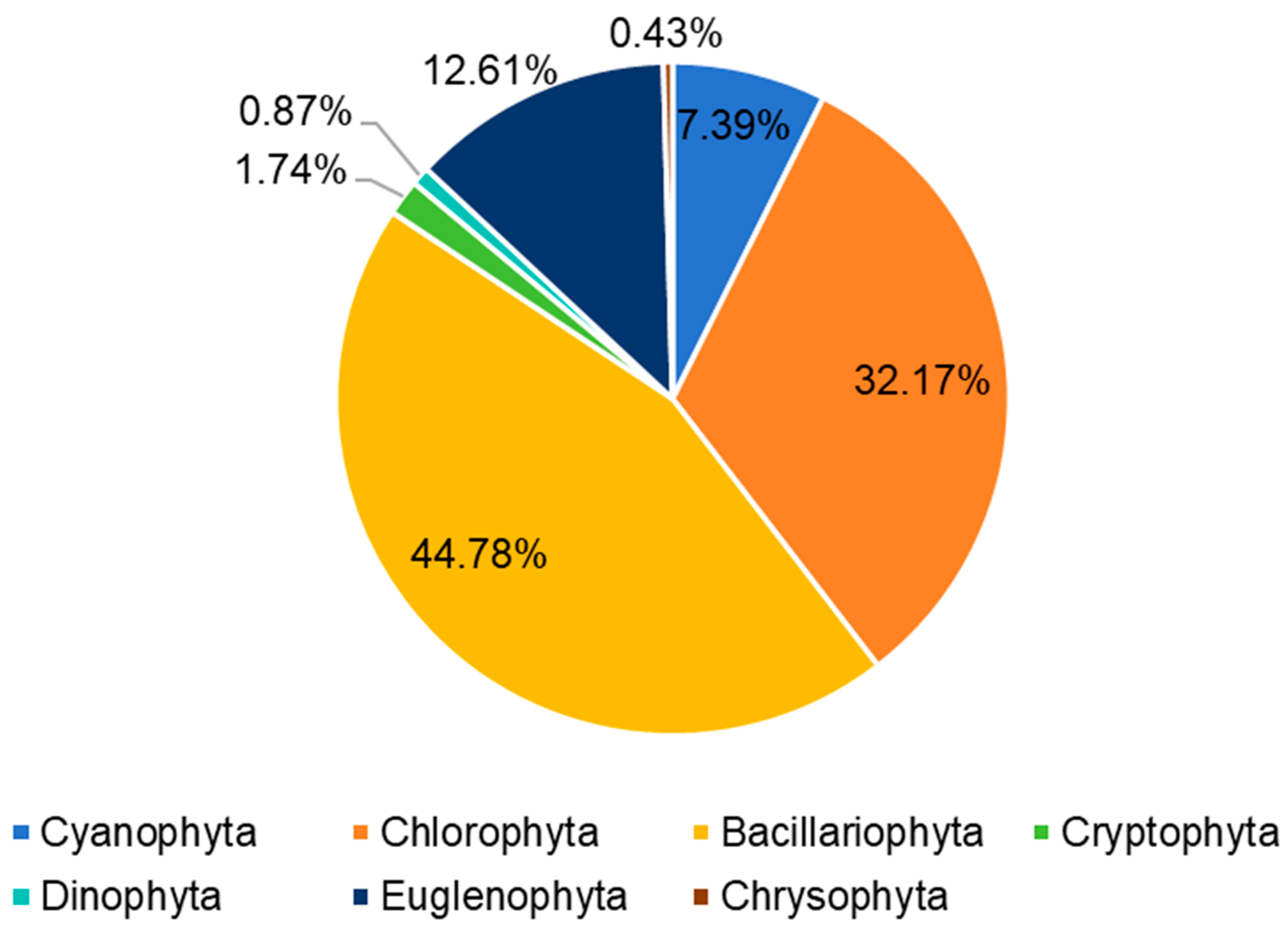
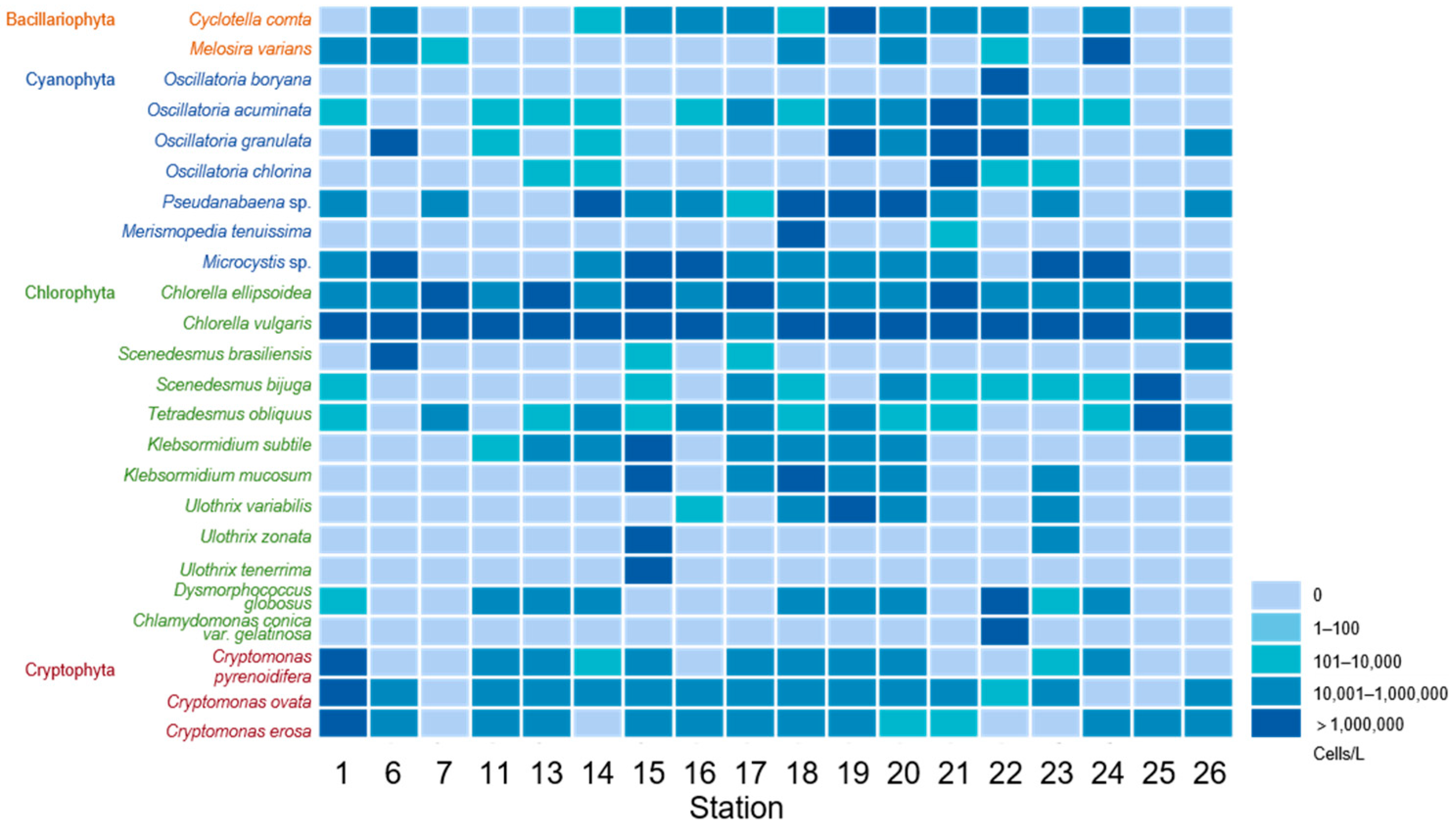
3.1. Algae Species and Abundance Observed Using Microscopy
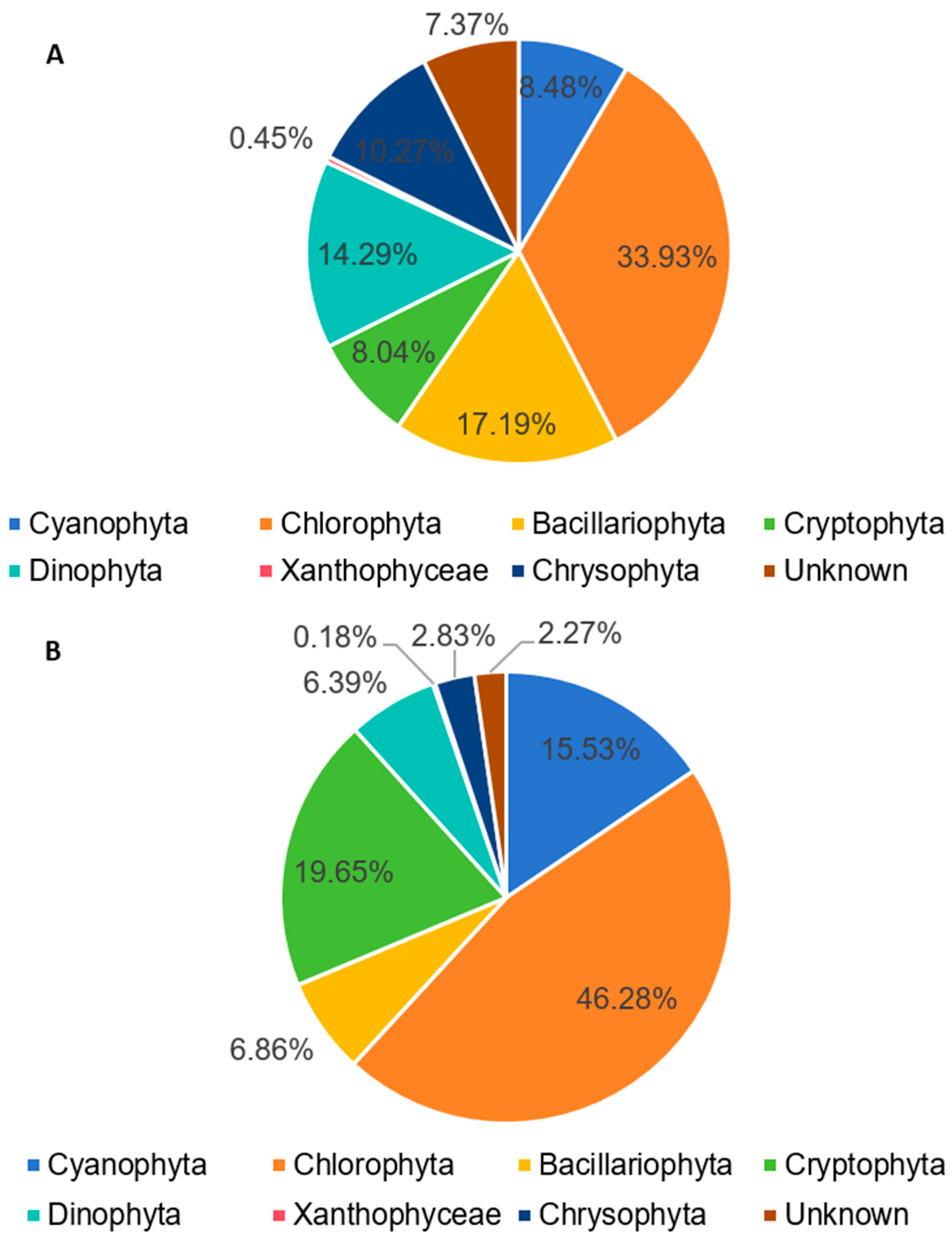
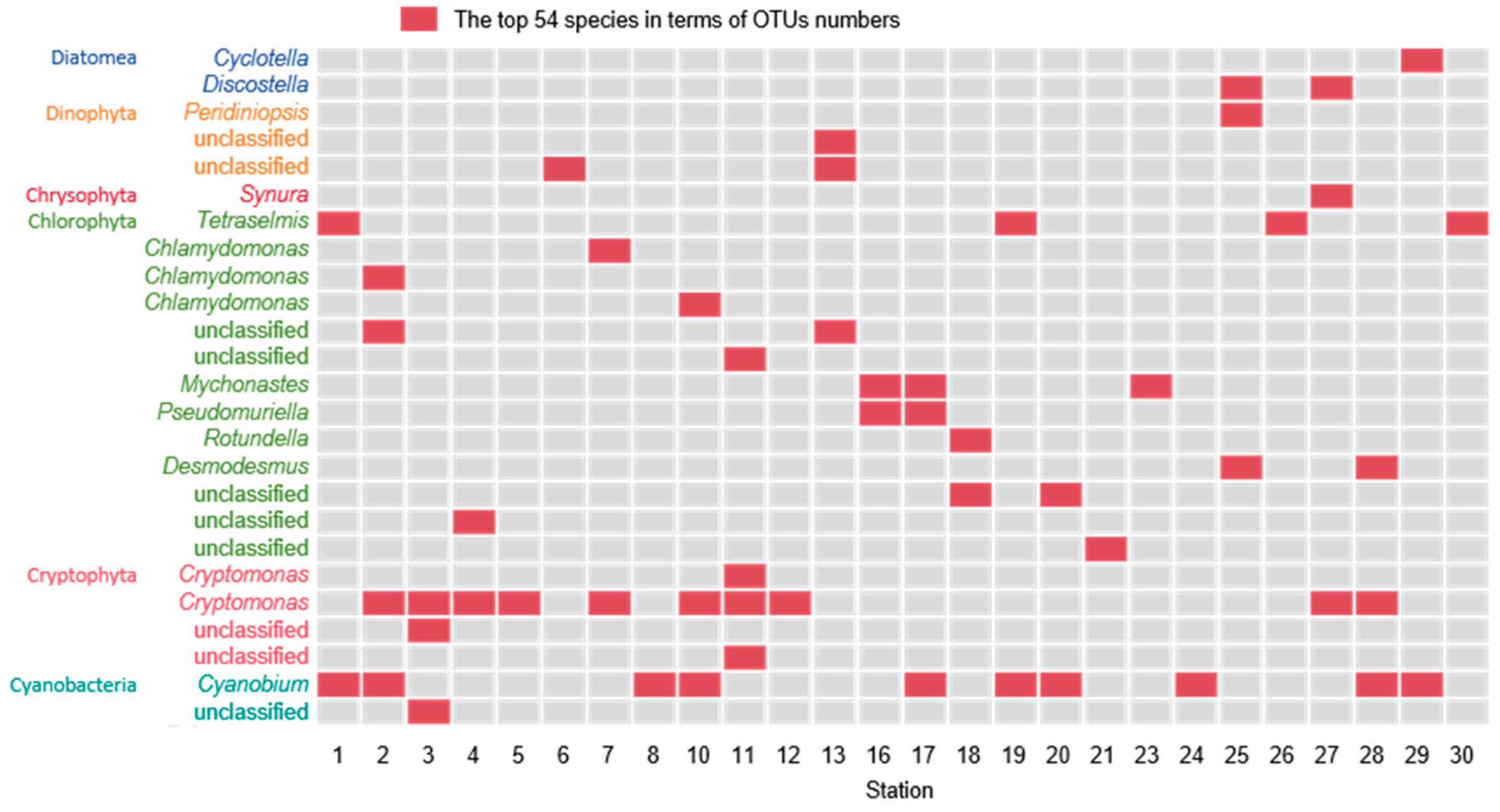
3.2. Molecular Biological Detection Result
3.3. Comparison of the Microscopy and Molecular Biological Results
4. Discussion
4.1. Identification of Phytoplankton Species
4.2. Species Abundance Counting
4.3. Phytoplankton Characteristics in the Study Area
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Litchman, E.; Klausmeier, C.A.; Dyhrman, S.T. Trait-Based Community Ecology of Phytoplankton. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 615–639. [Google Scholar] [CrossRef]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Tréguer, P.; Bowler, C.; Moriceau, B.; Dutkiewicz, S.; Gehlen, M.; Aumont, O.; Bittner, L.; Dugdale, R.; Finkel, Z.; Iudicone, D.; et al. The world ocean silica cycle. Annu. Rev. Mar. Sci. 2013, 5, 477–501. [Google Scholar] [CrossRef] [PubMed]
- Glibert, P.M. Harmful algal blooms and eutrophication: “Strategies” for nutrient uptake and growth outside the Redfield comfort zone. Chin. J. Oceanol. Limnol. 2011, 29, 724–738. [Google Scholar] [CrossRef]
- Wright, S.W.; Shearer, J.D. Rapid extraction and high-performance liquid chromatography of chlorophylls and carotenoids from marine phytoplankton. J. Chromatogr. A 1984, 294, 281–295. [Google Scholar] [CrossRef]
- Zapata, M.; Rodríguez, F.; Garrido, J.L. Separation of chlorophylls and carotenoids from marine phytoplankton—A new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 2000, 195, 29–45. [Google Scholar] [CrossRef]
- Chisholm, S.W.; Olson, R.J.; Zettler, E.R.; Goericke, R.; Waterbury, J.B.; Welschmeyer, N.A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 1988, 334, 340–343. [Google Scholar] [CrossRef]
- Faria, D.G.; Lee, M.D.; Lee, J.B.; Lee, J.; Chang, M.; Youn, S.H.; Suh, Y.S.; Ki, J.S. Molecular diversity of phytoplankton in the East China Sea around Jeju Island (Korea), unraveled by pyrosequencing. J. Oceanogr. 2014, 70, 11–23. [Google Scholar] [CrossRef]
- Hu, X.K.; Lin, Z.R.; Zhang, Q.C.; Kong, F.Z.; Cen, J.Y.; Zeng, Y.L.; Yu, R.C. Detection of bloom-forming dinoflagellates Karenia mikimotoi and Prorocentrum donghaiense using qPCR assays. J. Appl. Phycol. 2022, 34, 1483–1496. [Google Scholar] [CrossRef]
- Andersen, P.; Throndsen, J. Estimating cell numbers. In Manual on Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Eds.; UNESCO: Paris, France, 2004. [Google Scholar]
- Utermöhl, H. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Int. Ver. Theor. Angew. Limnol. 1958, 9, 1–38. [Google Scholar] [CrossRef]
- Lohmann, H. Über das Nannoplankton und die Zentrifugierung kleinster Wasserproben zur Gewinnung desselben in lebendem Zustande. Int. Rev. Ges. Hydrobiol. 1911, 4, 1–38. [Google Scholar] [CrossRef]
- Johnson, P.W.; Sieburth, J.M. Chroococcoid cyanobacteria in the sea: A ubiquitous and diverse phototrophic biomass. Limnol. Oceanogr. 1979, 24, 928–935. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Watson, S.W.; Guillard, R.R.L.; Brand, L.E. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature 1979, 277, 293–294. [Google Scholar] [CrossRef]
- Utermöhl, H. Neue Wege in der quantitativen Erfassung des Planktons (mit besonderer Berücksichtigung des Ultraplanktons). Verh. Int. Ver. Theor. Angew. Limnol. 1931, 5, 567–596. [Google Scholar] [CrossRef]
- Sournia, A. Phytoplankton Manual; UNESCO: Paris, France, 1978. [Google Scholar]
- Hasle, G. Counting phytoplankton cells. In Phytoplankton Manual; Sournia, A., Ed.; UNESCO: Paris, France, 1978. [Google Scholar]
- Karlson, B.; Cusack, C.; Bresnan, E. Microscopic and Molecular Methods for Quantitative Phytoplankton Analysis; UNESCO: Paris, France, 2010. [Google Scholar]
- Tomas, C. Identifying Marine Phytoplankton; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Throndsen, J.; Hasle, G.; Tangen, K. Phytoplankton of Norwegian Coastal Waters; Almater Forlag AS: Oslo, Norway, 2007. [Google Scholar]
- Hansen, G.; Daugbjerg, N.; Henriksen, P. Comparative Study of Gymnodinium mikimotoi and Gymnodinium aureolum, comb. nov. (=Gyrodinium aureolum) Based on Morphology, Pigment Composition, and Molecular Data. J. Phycol. 2003, 36, 394–410. [Google Scholar] [CrossRef]
- Bergholtz, T.; Daugbjerg, N.; Moestrup, Ø.; Fernández-Tejedor, M. On the identity of Karlodinium veneficum and description of Karlodinium armiger sp. nov. (Dinophyceae). J. Phycol. 2005, 42, 170–193. [Google Scholar] [CrossRef]
- Lu, D.D.; Goebel, J.; Qi, Y.Z.; Zou, J.Z.; Han, X.T.; Gao, Y.H.; Li, Y.G. Morphological and genetic study of Prorocentrum donghaiense Lu from the East China sea, and comparison with some related Prorocentrum species. Harmful Algae 2005, 4, 493–505. [Google Scholar] [CrossRef]
- Luo, Z.H.; Wang, L.; Chan, L.L.; Lu, S.H.; Gu, H.F. Karlodinium zhouanum, a New dinoflagellate Species from China, and molecular phylogeny of Karenia digitata and Karenia longicanalis (Gymnodiniales, Dinophyceae). Phycologia 2018, 57, 401–412. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Treusch, A.H.; Demir-Hilton, E.; Vergin, K.L.; Worden, A.Z.; Carlson, C.A.; Donatz, M.G.; Burton, R.M.; Giovannoni, S.J. Phytoplankton distribution patterns in the northwestern sargasso sea revealed by small subunit rRNA genes from plastids. ISME J. 2012, 6, 481–492. [Google Scholar] [CrossRef]
- Zhao, J.Y.; Geng, H.X.; Zhang, Q.C.; Li, Y.F.; Kong, F.Z.; Yan, T.; Zhou, M.J.; Yang, D.; Yuan, Y.; Yu, R.C. Green Tides in the Yellow Sea Promoted the Proliferation of Pelagophyte Aureococcus anophagefferens. Environ. Sci. Technol. 2022, 56, 3056–3064. [Google Scholar] [CrossRef] [PubMed]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Moon-van der Staay, S.Y.; De Wachter, R.; Vaulot, D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 2001, 409, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Potter, D.; Lajeunesse, T.C.; Saunders, G.W.; Anderson, R.A. Convergent evolution masks extensive biodiversity among marine coccoid picoplankton. Biodivers. Conserv. 1997, 6, 99–107. [Google Scholar] [CrossRef]
- Guillou, L.; Moon-Van Der Staay, S.Y.; Claustre, H.; Partensky, F.; Vaulot, D. Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl. Environ. Microbiol. 1999, 65, 4528–4536. [Google Scholar] [CrossRef]
- Rappé, M.S.; Suzuki, M.T.; Vergin, K.L.; Giovannoni, S.J. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the pacific and Atlantic Coasts of the United States. Appl. Environ. Microbiol. 1998, 64, 294–303. [Google Scholar] [CrossRef]
- Chen, Z.F.; Zhang, Q.C.; Kong, F.Z.; Liu, Y.; Zhao, Y.; Zhou, Z.X.; Geng, H.X.; Dai, L.; Zhou, M.J.; Yu, R.C. Resolving phytoplankton taxa based on high-throughput sequencing during Brown tides in the Bohai Sea, China. Harmful Algae 2019, 84, 127–138. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, J.; Zhou, F.; Shou, L.; Chen, Q.; Tao, B.; Yan, X.; Wang, K. Controlling factors of summer phytoplankton community in the Changjiang (Yangtze River) estuary and adjacent East China sea shelf. Contin. Shelf Res. 2015, 101, 71–84. [Google Scholar] [CrossRef]
- Li, J.L.; Zheng, B.H.; Liu, L.S.; Tang, J.L. Phytoplankton community structure in the Yangtze River estuary and its relation to environmental factors. Res. Environ. Sci. 2013, 26, 403–409. [Google Scholar]
- Lv, S.; Li, X.; Wang, R.; Wang, Y.; Dong, Z.; Zhou, T.; Liu, Y.; Lin, K.; Liu, L. Autochthonous sources and drought conditions drive anomalous oxygen-consuming pollution increase in a sluice-controlled reservoir in eastern China. Sci. Total Environ. 2022, 841, 156739. [Google Scholar] [CrossRef]
- Winnepenninckx, B.; Backeljau, T.; De Wachter, R. Extraction of high-molecular-weight DNA from mollusks. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef] [PubMed]
- Lejzerowicz, F.; Esling, P.; Pillet, L.; Wilding, T.A.; Black, K.D.; Pawlowski, J. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 2015, 5, 13932. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.W.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19 (Suppl. S1), 21–31. [Google Scholar] [CrossRef]
- Wu, C.; Narale, D.D.; Cui, Z.; Wang, X.; Liu, H.; Xu, W.; Zhang, G.; Sun, J. Diversity, structure, and distribution of bacterioplankton and diazotroph communities in the Bay of Bengal during the winter monsoon. Front. Microbiol. 2022, 13, 987462. [Google Scholar] [CrossRef]
- Mori, H.; Maruyama, F.; Kato, H.; Toyoda, A.; Dozono, A.; Ohtsubo, Y.; Nagata, Y.; Fujiyama, A.; Tsuda, M.; Kurokawa, K. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Fish, J.A.; Chai, B.; Wang, Q.; Sun, Y.; Brown, C.T.; Tiedje, J.M.; Cole, J.R. FunGene: The functional gene pipeline and repository. Front. Microbiol. 2013, 4, 291. [Google Scholar] [CrossRef]
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, U.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010, 188, 223–241. [Google Scholar] [CrossRef]
- Fraga, S.; Penna, A.; Bianconi, I.; Paz, B.; Zapata, M. Coolia canariensis sp. nov. (Dinophyceae), a New Nontoxic Epiphytic Benthic dinoflagellate from the Canary Islands. J. Phycol. 2008, 44, 1060–1070. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Won, N.I.; Baek, S.H. Comparison of HPLC pigment analysis and microscopy in phytoplankton assessment in the Seomjin River estuary, Korea. Sustainability 2020, 12, 6880. [Google Scholar] [CrossRef]
- Kavanagh, S.; Brennan, C.; O’Connor, L.; Moran, S.; Salas, R.; Lyons, J.; Silke, J.; Maher, M. Real-time PCR detection of Dinophysis species in Irish coastal waters. Mar Biotechnol. 2010, 12, 534–542. [Google Scholar] [CrossRef]
- Barkallah, M.; Elleuch, J.; Smith, K.F.; Chaari, S.; Ben Neila, I.B.; Fendri, I.; Michaud, P.; Abdelkafi, S. Development and application of a real-time PCR assay for the sensitive detection of diarrheic toxin producer Prorocentrum lima. J. Microbiol. Methods 2020, 178, 106081. [Google Scholar] [CrossRef]
- Yadav, D.P.; Jalal, A.S.; Garlapati, D.; Hossain, K.; Goyal, A.; Pant, G.; Learning-Based, D. Deep learning-based ResNeXt model in phycological studies for future. Algal Res. 2020, 50, 102018. [Google Scholar] [CrossRef]
- Muylaert, K.; Sabbe, K.; Vyverman, W. Spatial and temporal dynamics of phytoplankton communities in a freshwater tidal estuary (Schelde, Belgium). Estuar. Coast. Shelf Sci. 2000, 50, 673–687. [Google Scholar] [CrossRef]
- Schuchardt, B.; Schirmer, M. Phytoplankton maxima in the tidal freshwater reaches of two coastal plain estuaries. Estuar. Coast. Shelf Sci. 1991, 32, 187–206. [Google Scholar] [CrossRef]
- Horner, R. A Taxonomic Guide to Some Common Marine Phytoplankton; Biopress: Bristol, UK, 2002. [Google Scholar]
- Throndsen, J.; Hasle, G.R.; Tangen, K. Norsk Kystplanktonflora; Almater Forlag AS: Oslo, Norway, 2003. [Google Scholar]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef]
- Miyata, K.; Inoue, Y.; Amano, Y.; Nishioka, T.; Nagaike, T.; Kawaguchi, T.; Morita, O.; Yamane, M.; Honda, H. Comparative environmental RNA and DNA metabarcoding analysis of river algae and arthropods for ecological surveys and water quality assessment. Sci. Rep. 2022, 12, 19828. [Google Scholar] [CrossRef]
- Bombin, S.; Wysor, B.; Lopez-Bautista, J.M. Assessment of littoral algal diversity from the northern Gulf of Mexico using environmental DNA metabarcoding. J. Phycol. 2021, 57, 269–278. [Google Scholar] [CrossRef]
- Wu, W.; Liu, H. Disentangling protist communities identified from DNA and RNA surveys in the Pearl River–South China sea Continuum during the wet and dry seasons. Mol. Ecol. 2018, 27, 4627–4640. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Chen, Y. Advances in the study of biodiversity of phytoplankton and red tide species in China (II): The East China Sea. Oceanol. Limnol. Sin 2021, 52, 363–384. [Google Scholar]
- Wanigatunge, R.P.; Magana-Arachchi, D.N.; Chandrasekharan, N.V.; Kulasooriya, S.A. Genetic diversity and molecular phylogeny of cyanobacteria from Sri Lanka based on 16S rRNA gene. Environ. Eng. Res. 2014, 19, 317–329. [Google Scholar] [CrossRef]
- Bilbao, J.; Pavloudi, C.; Blanco-Rayón, E.; Franco, J.; Madariaga, I.; Seoane, S. Phytoplankton community composition in relation to environmental variability in the Urdaibai estuary (SE Bay of Biscay): Microscopy and eDNA metabarcoding. Mar. Environ. Res. 2023, 191, 106175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, Q.; Gibson, K.; Chen, N. Metabarcoding dissection of harmful algal bloom species in the East China sea off Southern Zhejiang Province in late spring. Mar. Pollut. Bull. 2021, 169, 112586. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xu, Q.; Gibson, K.; Liu, S.; Chen, N. Metabarcoding analysis of harmful algal bloom species in the Changjiang estuary, China. Sci. Total Environ. 2021, 782, 146823. [Google Scholar] [CrossRef]
- Huang, H.; Xu, Q.; Gibson, K.; Chen, Y.; Chen, N. Molecular characterization of harmful algal blooms in the Bohai Sea using metabarcoding analysis. Harmful Algae 2021, 106, 102066. [Google Scholar] [CrossRef]
- Liu, S.; Gibson, K.; Cui, Z.; Chen, Y.; Sun, X.; Chen, N. Metabarcoding analysis of harmful algal species in Jiaozhou Bay. Harmful Algae 2020, 92, 101772. [Google Scholar] [CrossRef]
- Chen, N.S.; Zhang, M.J.; Liu, S.M.; Cui, Z.M. Diversity of HAB species in coastal regions of China. Oceanol. Limnol. Sin. 2023, 54, 599–624. [Google Scholar]
- Welschmeyer, N.A. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and Pheopigments. Limnol. Oceanogr. 1994, 39, 1985–1992. [Google Scholar] [CrossRef]
- Asai, R.; Matsukawa, R.; Ikebukuro, K.; Karube, I. Highly Sensitive chemiluminescence Flow-Injection Detection of the Red Tide Phytoplankton Heterosigma carterae. Anal. Chim. Acta 1999, 390, 237–244. [Google Scholar] [CrossRef]
- Cai, Q.; Li, R.; Zhen, Y.; Mi, T.; Yu, Z. Detection of two Prorocentrum species using sandwich hybridization integrated with nuclease protection assay. Harmful Algae 2006, 5, 300–309. [Google Scholar] [CrossRef]
- Gran-Stadniczeñko, S.; Egge, E.; Hostyeva, V.; Logares, R.; Eikrem, W.; Edvardsen, B. Protist diversity and seasonal dynamics in Skagerrak plankton communities as revealed by metabarcoding and microscopy. J. Eukaryot. Microbiol. 2019, 66, 494–513. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Chang, Z.; Li, J.; Chen, Z.; Yang, L.; Luo, Q. Phytoplankton Community Structure and Diversity in the Indoor Industrial Aquaculture System for litopenaeus vannamei Revealed by High-Throughput Sequencing and Morphological Identification. Aquac. Res. 2019, 50, 2563–2576. [Google Scholar] [CrossRef]
- Lim, S.L.; Chu, W.L.; Phang, S.M. Use of Chlorella vulgaris for bioremediation of textile wastewater. Bioresour. Technol. 2010, 101, 7314–7322. [Google Scholar] [CrossRef]
- Cunha, A.E.P.; Sátiro, J.R.; Escobar, B.P.; Simões, R.M.S. Chlorella vulgaris growth, pigment and lipid accumulation: Effect of progressive light and hydrogen peroxide exposure. J. Chem. Technol. Biotechnol. 2023, 98, 442–450. [Google Scholar] [CrossRef]
- Zurawell, R.W.; Chen, H.; Burke, J.M.; Prepas, E.E. Hepatotoxic cyanobacteria: A review of the biological importance of microcystins in freshwater environments. J. Toxicol. Environ. Health Part B Crit. Rev. 2005, 8, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef]
- Sarkar, S.; Alhasson, F.; Kimono, D.; Albadrani, M.; Seth, R.K.; Xiao, S.; Porter, D.E.; Scott, G.I.; Brooks, B.; Nagarkatti, M.; et al. Microcystin exposure worsens nonalcoholic fatty liver disease associated ectopic glomerular toxicity via NOX-2-MIR21 axis. Environ. Toxicol. Pharmacol. 2020, 82, 103563. [Google Scholar] [CrossRef]

| Microscopy | Molecular Biological | |
|---|---|---|
| Phyla number | 7 | 7 |
| Identified genera number | 73 | 38 |
| Species number | 230 | 432 OTUs |
| Identified species number | 230 | 148 |
| Cell number counting | Yes | Relative abundance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Liu, Y.; Wang, R.; Dong, Z.; Lin, K.; Lv, S.; Liu, Y.; Lu, W.; Liu, L. Comparative Analysis of Light Microscopy and High-Throughput Sequencing for Phytoplankton Detection in Rivers Flowing into the Sea. Water 2025, 17, 1559. https://doi.org/10.3390/w17111559
Hu X, Liu Y, Wang R, Dong Z, Lin K, Lv S, Liu Y, Lu W, Liu L. Comparative Analysis of Light Microscopy and High-Throughput Sequencing for Phytoplankton Detection in Rivers Flowing into the Sea. Water. 2025; 17(11):1559. https://doi.org/10.3390/w17111559
Chicago/Turabian StyleHu, Xiaokun, Yunlong Liu, Rui Wang, Zhaojun Dong, Kuixuan Lin, Shucong Lv, Yuchen Liu, Wenze Lu, and Lusan Liu. 2025. "Comparative Analysis of Light Microscopy and High-Throughput Sequencing for Phytoplankton Detection in Rivers Flowing into the Sea" Water 17, no. 11: 1559. https://doi.org/10.3390/w17111559
APA StyleHu, X., Liu, Y., Wang, R., Dong, Z., Lin, K., Lv, S., Liu, Y., Lu, W., & Liu, L. (2025). Comparative Analysis of Light Microscopy and High-Throughput Sequencing for Phytoplankton Detection in Rivers Flowing into the Sea. Water, 17(11), 1559. https://doi.org/10.3390/w17111559





