Nitrogen Removal Mechanism and Microbial Community Changes of the MBR Bioaugmented with Two Novel Fungi Pichia kudriavzevii N7 and Candida tropicalis N9
Abstract
1. Introduction
2. Materials and Methods
2.1. Inoculum Sludge and Synthetic Wastewater
2.2. MBR Systems Description
2.3. Screening and Identification of Ammonium Transforming Strains
2.4. Construction of Compound Bioflocculant and Preparation of Inoculating Strains
2.5. Analytical Methods for Water Quality
2.6. Microbial Community Analysis
3. Results and Discussion
3.1. Strain Screening and Identification
3.2. Construction of Compound Strains
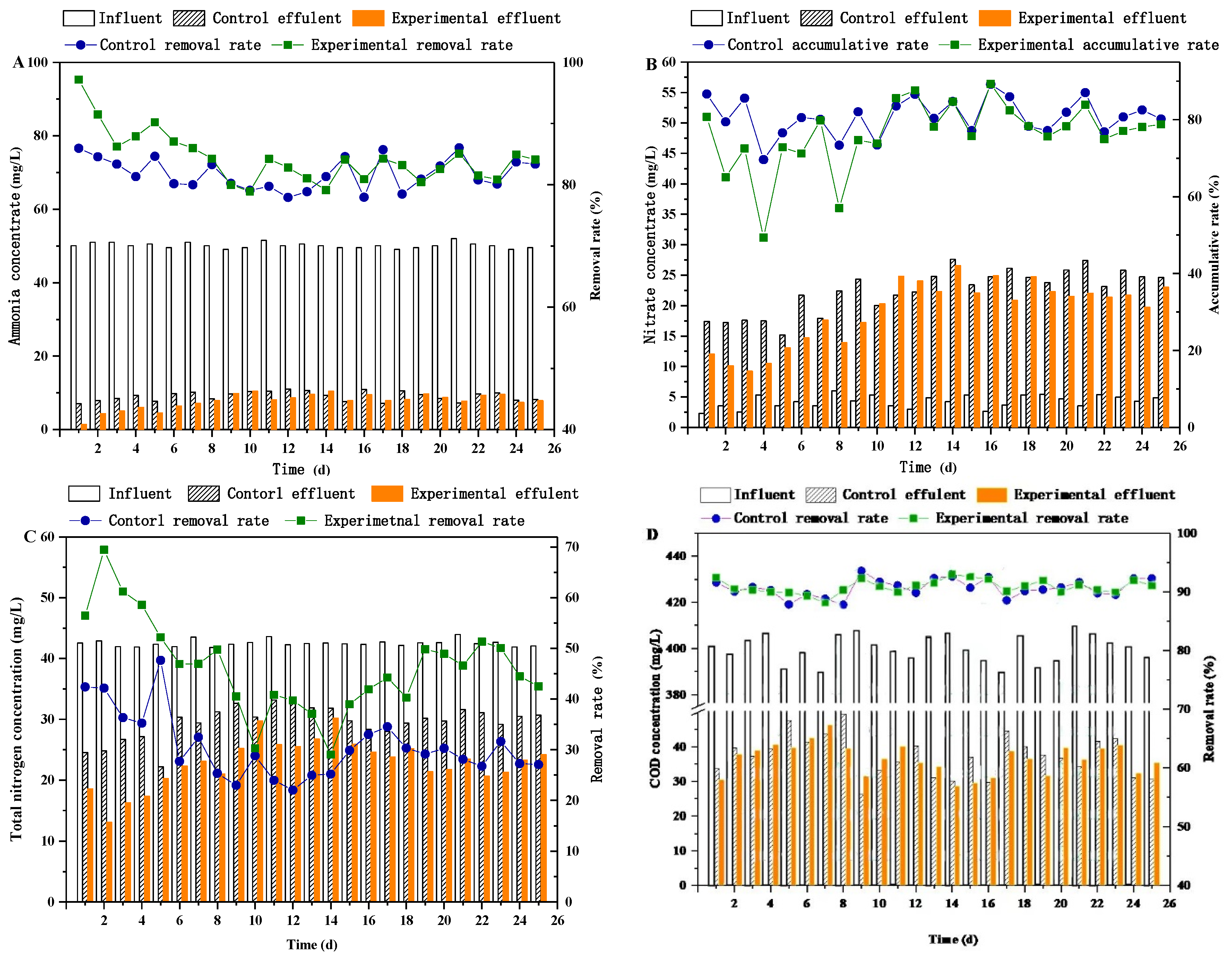
3.3. Ammonium Nitrogen
3.4. Nitrate Nitrogen
3.5. Total Nitrogen
3.6. COD
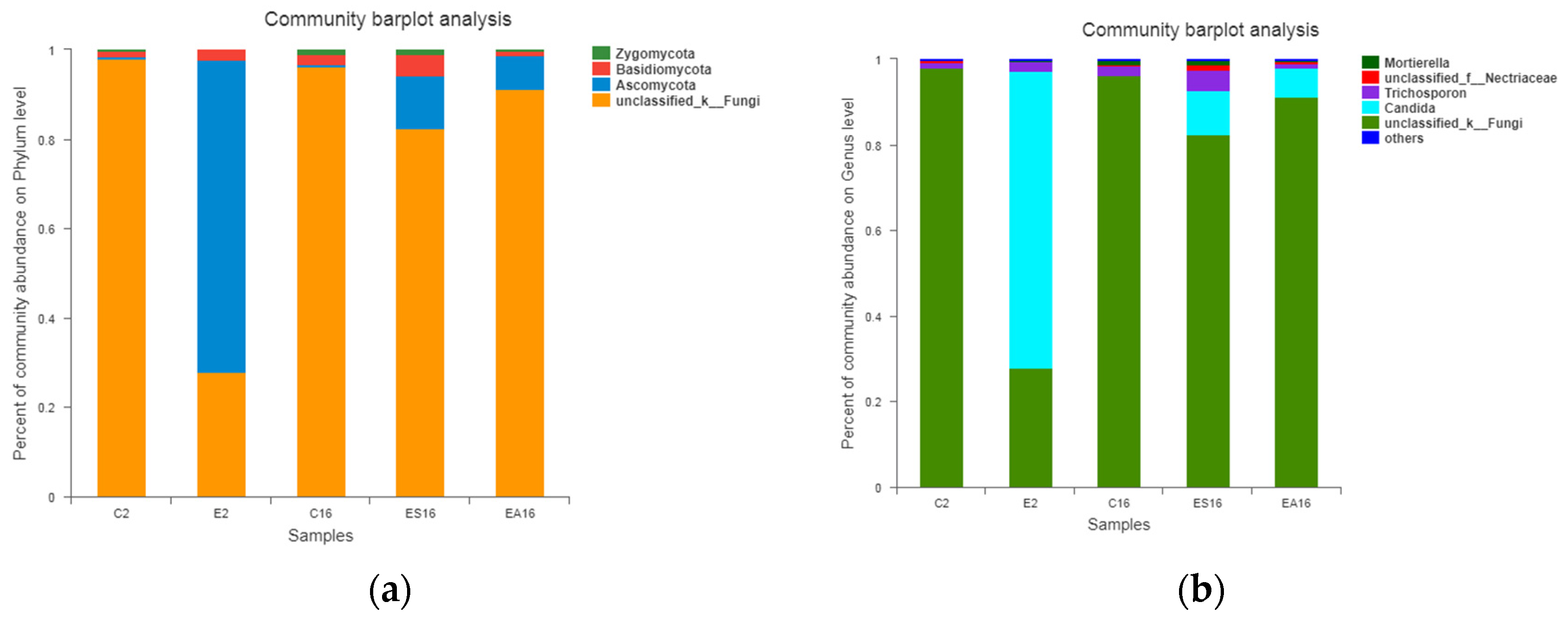
3.7. Microbial Community Analysis
3.7.1. Diversity of Fungal and Bacterial Communities in the MBR Systems
3.7.2. Microbial Community Composition in the MBR Systems
4. Conclusions
- (1)
- In the first week of the experiment, the ammonium removal rate of the MBR system inoculated with N7 and N9 increased and the effluent concentration of NO3−-N was reduced compared to the MBR system without inoculation of the compound fungi.
- (2)
- Through ITS gene sequencing, it was observed that N9 could survive in MBR-containing activated sludge for a long time, Moreover, N9 was also observed to form biofilm with other microorganisms and maintain the population advantage, demonstrating good adaptability.
- (3)
- High throughput sequencing results illustrated that the addition of N7 and N9 caused the original community structure to be altered, enriching the fungal communities. It should not be ignored that N7 and N9 also have an important impact on the change of bacterial community structure in the MBR system.
- (4)
- Combined with the results of ITS gene sequencing and ammonium transformation characteristics, it is speculated that the compound fungi N7 and N9 inoculated every 8 days might achieve better results in practical application.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| MBR | membrane bioreactor |
| DO | dissolved oxygen |
| HRT | hydraulic retention time |
| COD | chemical oxygen demand |
| MLSS | mixed liquor suspended solids |
| ITS | internal transcribed spacer |
References
- Bernet, N.; Delgenes, N.; Akunna, J.C.; Delgenes, J.P.; Moletta, R. Combined anaerobic–aerobic sbr for the treatment of piggery wastewater. Water Res. 2000, 34, 611–619. [Google Scholar] [CrossRef]
- Zou, J.L.; Dai, Y.; Sun, T.H.; Li, Y.H.; Li, G.B.; Li, Q.Y. Effect of amended soil and hydraulic load on enhanced biological nitrogen removal in lab-scale SWIS. J. Hazard. Mater. 2009, 163, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Sarioglu, O.F.; Yasa, A.O.; Celebioglu, A.A. Efficient ammonium removal from aquatic environments by Acinetobacter calcoaceticus STB1 immobilized on an electrospun cellulose acetate nanofibrous web. Green. Chem. 2013, 15, 2566–2572. [Google Scholar] [CrossRef]
- Obaja, D.; Macé, S.; Costa, J.; Sans, C.; Mata-Alvarez, J. Nitrification, denitrification and biological phosphorus removal in piggery wastewater using a sequencing batch reactor. Bioresour. Technol. 2003, 87, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ding, Y.; Wang, Y.; Song, X.; Ambrose, R.F.; Ullman, J.L.; Winfrey, B.K.; Wang, J.; Gong, J. Treatment of rich ammonia nitrogen wastewater with polyvinyl alcohol immobilized nitrifier biofortified constructed wetlands. Ecol. Eng. 2016, 94, 7–11. [Google Scholar] [CrossRef]
- Okaiyeto, K.; Nwodo, U.U.; Mabinya, L.V.; Okoh, A.I. Characterization of a bioflocculant produced by a consortium of Halomonas sp. Okoh and Micrococcus sp. Leo. Int. J. Environ. Res. Public. Health 2013, 10, 5097–5110. [Google Scholar] [CrossRef]
- Fenn, M.E.; Poth, M.A.; Aber, J.D.; Baron, J.S.; Stottlemyer, R. Nitrogen Excess in North American Ecosystems: Predisposing Factors, Ecosystem Responses, and Management Strategies. Ecol. Appl. 1998, 8, 706–733. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, X.; Deal, B.; Han, L. Biological systems for treatment and valorization of wastewater generated from hydrothermal liquefaction of biomass and systems thinking: A review. Bioresour. Technol. 2019, 278, 329–345. [Google Scholar] [CrossRef]
- Ma, F.; Guo, J.; Zhao, L.; Chang, C.; Cui, D. Application of bioaugmentation to improve the activated sludge system into the contact oxidation system treating petrochemical wastewater. Bioresour. Technol. 2009, 100, 597–602. [Google Scholar] [CrossRef]
- Han, X.; Jin, Y.; Yu, J. Rapid formation of aerobic granular sludge by bioaugmentation technology: A review. Chem. Eng. J. 2022, 437, 134971. [Google Scholar] [CrossRef]
- Yang, R.; He, X.W.; Gan, X.; Qi, Z.D.; Zhang, X.X.; Ren, H.Q. Study on Reducing Endocrine Disrupting Toxicity of NPEO by Bioaugmentation of Bacteria Consortium. Res. Environ. Sci. 2023, 36, 345–353. [Google Scholar] [CrossRef]
- Boonnorat, J.; Techkarnjanaruk, S.; Honda, R.; Angthong, S.; Boonapatcharoen, N.; Muenmee, S.; Prachanurak, P. Use of aged sludge bioaugmentation in two-stage activated sludge system to enhance the biodegradation of toxic organic compounds in high strength wastewater. Chemosphere 2018, 202, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Hu, T.T.; Song, Y.J.; Chen, H.P.; Lv, Y.K. Heterotrophic nitrogen removal by Acinetobacter sp. Y1 isolated from coke plant wastewater. J. Biosci. Bioeng. 2015, 120, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Liu, Y.; Ai, G.; Zheng, H.; Liu, Z. The characteristics of a novel heterotrophic nitrification–aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour. Technol. 2012, 108, 35–44. [Google Scholar] [CrossRef]
- Schmalenberger, A.; Fox, A. Bacterial Mobilization of Nutrients from Biochar-Amended Soils. Adv. Appl. Microbiol. 2016, 94, 109–159. [Google Scholar] [CrossRef] [PubMed]
- Djelal, H.; Amrane, A. Biodegradation by bioaugmentation of dairy wastewater by fungal consortium on a bioreactor lab-scale and on a pilot-scale. J. Environ. Sci. 2013, 25, 1906–1912. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, T.; Song, Y.; Chen, H.; Lv, Y. Simultaneous removal of carbon and nitrogen by mycelial pellets of a heterotrophic nitrifying fungus-Penicillium sp. L1. J. Biosci. Bioeng. 2017, 123, 223–229. [Google Scholar] [CrossRef]
- Wang, W.; Cao, L.; Tan, H.; Zhang, R. Nitrogen removal from synthetic wastewater using single and mixed culture systems of denitrifying fungi, bacteria, and actinobacteria. Appl. Microbiol. Biol. 2016, 100, 9699–9707. [Google Scholar] [CrossRef]
- Espinosa-Ortiz, E.J.; Rene, E.R.; Pakshirajan, K.; Hullebusch, E.D.V.; Lens, P.N.L. Fungal pelleted reactors in wastewater treatment: Applications and perspectives. Chem. Eng. J. 2016, 283, 553–571. [Google Scholar] [CrossRef]
- Vantamuri, A.B.; Kaliwal, B.B. Decolourization and biodegradation of Navy blue HER (Reactive Blue 171) dye from Marasmiussp. BBKAV79. Biotechnology 2017, 7, 48. [Google Scholar] [CrossRef][Green Version]
- Mrozik, A.; Piotrowska-Seget, Z. Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol. Res. 2010, 165, 363–375. [Google Scholar] [CrossRef]
- Shi, S.; Fang, M.; Sun, T.; Li, A.; Zhou, J.; Qu, Y. Mineralization and Kinetics of Reactive Brilliant Red X-3B by a Combined Anaerobic–Aerobic Bioprocess Inoculated with the Coculture of Fungus and Bacterium. Appl. Biochem. Biotech. 2014, 172, 1106–1120. [Google Scholar] [CrossRef]
- Novotny, E.; Svobodová, K.I.; Benada, O.I.; Kofroňová, O.; Heissenberger, A.; Fuchs, W. Potential of combined fungal and bacterial treatment for color removal in textile wastewater. Bioresour. Technol. 2011, 102, 879–888. [Google Scholar] [CrossRef]
- Zsirai, T.; Buzatu, P.; Aerts, P. Efficacy of relaxation, backflushing, chemical cleaning and clogging removal for an immersed hollow fibre membrane bioreactor. Water Res. 2012, 46, 4499–4507. [Google Scholar] [CrossRef]
- Balci; Behzat; Ergan, B.; Erkurt, F.E. Magnetite/activated sludge hybrid process for the treatment of dye containing simulated textile wastewater. Desalin. Water Treat. 2023, 286, 101–114. [Google Scholar] [CrossRef]
- DZ/T 0064-2021; Methods for Analysis of Groundwater Quality. Ministry of Natural Re-Sources of the People’s Republic of China: Beijing, China, 2021.
- Liu, C.; Xie, J.; Song, M.; Gao, Z.; Zheng, D.; Liu, X.; Ning, G.; Cheng, X.; Bruning, H. Nitrogen removal performance and microbial community changes in subsurface wastewater infiltration systems (SWISs) at low temperature with different bioaugmentation strategies. Bioresour. Technol. 2017, 250, 603–610. [Google Scholar] [CrossRef]
- Morimura, S.; Zeng, X.; Noboru, N.; Hosono, T. Changes to the microbial communities within groundwater in response to a large crustal earthquake in Kumamoto, southern Japan. J. Hydrol. 2020, 581, 124341. [Google Scholar] [CrossRef]
- Du, P.; Du, R.; Ren, W.; Lu, Z.; Zhang, Y.; Fu, P. Variations Variations of bacteria and fungi in PM2.5 in Beijing, China. Atmos. Environ. 2018, 172, 55–64. [Google Scholar] [CrossRef]
- Kleerebezem, R.; van Loosdrecht, M.C. Mixed culture biotechnology for bioenergy production. Curr. Opin. Biotech. 2007, 18, 207–212. [Google Scholar] [CrossRef] [PubMed]
- More, T.T.; Yadav, J.S.S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Extracellular polymeric substances of bacteria and their potential environmental applications. J. Environ. Manag. 2014, 144, 1–25. [Google Scholar] [CrossRef]
- Yin, C.; Meng, F.; Chen, G.H. Spectroscopic characterization of extracellular polymeric substances from a mixed culture dominated by ammonia-oxidizing bacteria. Water Res. 2015, 68, 740–749. [Google Scholar] [CrossRef]
- Racz, L.A.; Datta, T.; Goel, R. Effect of organic carbon on ammonia oxidizing bacteria in a mixed culture. Bioresour. Technol. 2010, 101, 6454–6460. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ni, J.; Ma, T.; Liu, T.; Zheng, M. Bioaugmentation treatment of municipal wastewater with heterotrophic-aerobic nitrogen removal bacteria in a pilot-scale SBR. Bioresour. Technol. 2015, 183, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Vdovina, T.V.; Sirotkin, A.S.; Kobeleva, Y.V.; Gorshkova, E.S. Bioaugmentation of nitrifying microorganisms to increase the efficiency of the oxidation of nitrogen compounds during wastewater biofiltration. Appl. Biochem. Micro. 2020, 56(9), 948–955. [Google Scholar] [CrossRef]
- Motamedi, H.; Jafari, M. Screening Heterotrophic Ammonia Removal and Aerobic Denitrifying Bacteria from Wastewater of Ammonia Production Units of a Petrochemical Industry. Curr. Microbiol. 2020, 77, 2207–2214. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xiao, X.; Zhao, Y.; Tu, B.; Zhang, Y. Screening of efficient ammonia-nitrogen degrading bacteria and its application in livestock wastewater. Biomass Convers. Biol. 2022, 4, 1–9. [Google Scholar] [CrossRef]
- Liang, D.H.; Hu, Y.; Liang, D.; Chenga, J.; Chena, Y. Bioaugmentation of moving bed biofilm reactor (mbbr) with achromobacter jl9 for enhanced sulfamethoxazole (smx) degradation in aquaculture wastewater. Ecotox Environ. Saf. 2020, 207, 111258. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.; Yang, K.; Shu, Y.; Xiao, B.; Wu, X. Efficacy of auto-aggregating aerobic denitrifiers with coaggregation traits for bioaugmentation performance in biofilm-formation and nitrogen-removal. Bioresour. Technol. 2021, 337, 125391. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, S.; Geng, N.; Wu, Z.; Xiong, W.; Su, H. Artificially constructing mixed bacteria system for bioaugmentation of nitrogen removal from saline wastewater at low temperature. J. Environ. Manag. 2022, 324, 116351. [Google Scholar] [CrossRef]
- Bouchez, T.; Patureau, D.; Dabert, P.; Juretschko, S.; Doré, J.; Delgenès, P.; Moletta, R.; Wagner, M. Ecological study of a bioaugmentation failure. Environ. Microbiol. 2010, 2, 179–190. [Google Scholar] [CrossRef]
- Yang, M.; Lu, D.; Qin, B.; Liu, Q.; Zhao, Y.; Liu, H.; Ma, J. Highly efficient nitrogen removal of a coldness-resistant and low nutrient needed bacterium, Janthinobacterium sp. M-11. Bioresour. Technol. 2015, 256, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Luo, X.; Li, X.; Huang, D.; Huang, Q.; Zhang, X.X.; Chen, W. Bioaugmentation of woodchip bioreactors by pseudomonas nicosulfuronedens d1-1 with functional species enrichment. Bioresour. Technol. 2023, 385, 129309. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Li, Z.; Sun, Q.; Xu, Y.; Ye, Q. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 2015, 200, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Patureau, D.; Bernet, N.; Moletta, R. Combined nitrification and denitrification in a single aerated reactor using the aerobic denitrifier Commonas sp. strain SGLY2. Water Res. 1997, 31, 1363–1370. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, Y.; Ma, Y.; Du, J.; Pang, L.; Zhang, H. Ammonia removal and microbial characteristics of partial nitrification in biofilm and activated sludge treating low strength sewage at low temperature. Ecol. Eng. 2016, 93, 104–111. [Google Scholar] [CrossRef]
- Wang, X.; Tian, Y.; Zhao, X.; Peng, S.; Wu, Q.; Yan, L. Effects of aeration position on organics, nitrogen and phosphorus removal in combined oxidation pond-constructed wetland systems. Bioresour. Technol. 2015, 198, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, J.; Bai, S.; Ma, F.; Wang, L. Microbial population dynamics in response to bioaugmentation in a constructed wetland system under 10 °C. Bioresour. Technol. 2016, 205, 166–173. [Google Scholar] [CrossRef]
- Ou, D.; Li, H.; Li, W.; Wu, X.; Wang, Y.; Liu, Y. Salt-tolerance aerobic granular sludge: Formation and microbial community characteristics. Bioresour. Technol. 2018, 249, 132–138. [Google Scholar] [CrossRef]
- Liang, J.; Wang, Q.; Li, Q.X.; Jiang, L.; Kong, J.; Ke, M.; Arslan, M.; El-Din, M.G.; Chen, C. Aerobic sludge granulation in shale gas flowback water treatment: Assessment of the bacterial community dynamics and modeling of bioreactor performance using artificial neural network. Bioresour. Technol. 2020, 313, 123687. [Google Scholar] [CrossRef]
- Mcfrederick, Q.S.; Mueller, U.G.; James, R.R. Interactions between fungi and bacteria influence microbial community structure in the Megachile rotundata larval gut. Proc. Soc. B-Biol. Sci. 2014, 281, 20132653. [Google Scholar] [CrossRef]
- Zheng, G.; Liu, S.; Yang, F.; Han, B.; Furukawa, K. Characterization of functional microbial community in a membrane-aerated biofilm reactor operated for completely autotrophic nitrogen removal. Bioresour. Technol. 2008, 99, 2749–2756. [Google Scholar] [CrossRef]
- Niu, L.; Li, Y.; Xu, L.; Wang, P.; Wang, L. Ignored fungal community in activated sludge wastewater treatment plants: Diversity and altitudinal characteristics. Environ. Sci. Pollut. Res. 2016, 24, 1–9. [Google Scholar] [CrossRef]
- Felczak, A.; Bernat, P.A.; Alska, S.R.; Lisowska, K. Quinoline biodegradation by filamentous fungus Cunninghamella elegans and adaptive modifications of the fungal membrane composition. Environ. Sci. Pollut. Res. 2016, 23, 8872–8880. [Google Scholar] [CrossRef]
- Merlin, C.; Devers, M.; Crouzet, O. Characterization of chlordecone-tolerant fungal populations isolated from long-term polluted tropical volcanic soil in the French West Indies. Environ. Sci. Pollut. Res. 2014, 21, 4914–4927. [Google Scholar] [CrossRef]
- Cho, O.; Matsukura, M.; Sugita, T. Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 2015, 39, 87–88. [Google Scholar] [CrossRef]
- Evans, T.N.; Seviour, R.J. Estimating Biodiversity of Fungi in Activated Sludge Communities Using Culture-Independent Methods. Microb. Ecol. 2012, 63, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Le, N.C.; Mai, C.T.N.; Minh, N.N.; Ha, H.P.; Lien, D.T.; Tuan, D.V.; Dong, V.Q.; Ike, M.; Uyen, D.T.T. Degradation of sec-hexylbenzene and its metabolites by a biofilm-forming yeast Trichosporon asahii B1 isolated from oil-contaminated sediments in Quangninh coastal zone, Vietnam. J. Environ. Sci. Health A 2016, 51, 267–275. [Google Scholar] [CrossRef]
- Liu, Y.-R.; Zheng, Y.-M.; He, J.-Z. Toxicity of profenofos to the springtail, Folsomia candida, and ammonia-oxidizers in two agricultural soils. Ecotoxicology 2012, 21, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Yu, J.S.; Hui, J.L.; Si, D.L.; Ming, J.S.; Wei, X.S. Multistage A-O Activated Sludge Process for Paraformaldehyde Wastewater Treatment and Microbial Community Structure Analysis. J. Chem. 2016, 2016, 2746715. [Google Scholar] [CrossRef]
- Luo, X.; Yuan, X.; Wang, S.; Sun, F.; Hou, Z.; Hu, Q.; Zhai, L.; Cui, Z.; Zou, Y. Methane production and characteristics of the microbial community in the co-digestion of spent mushroom substrate with dairy manure. Bioresour. Technol. 2018, 250, 611–620. [Google Scholar] [CrossRef] [PubMed]
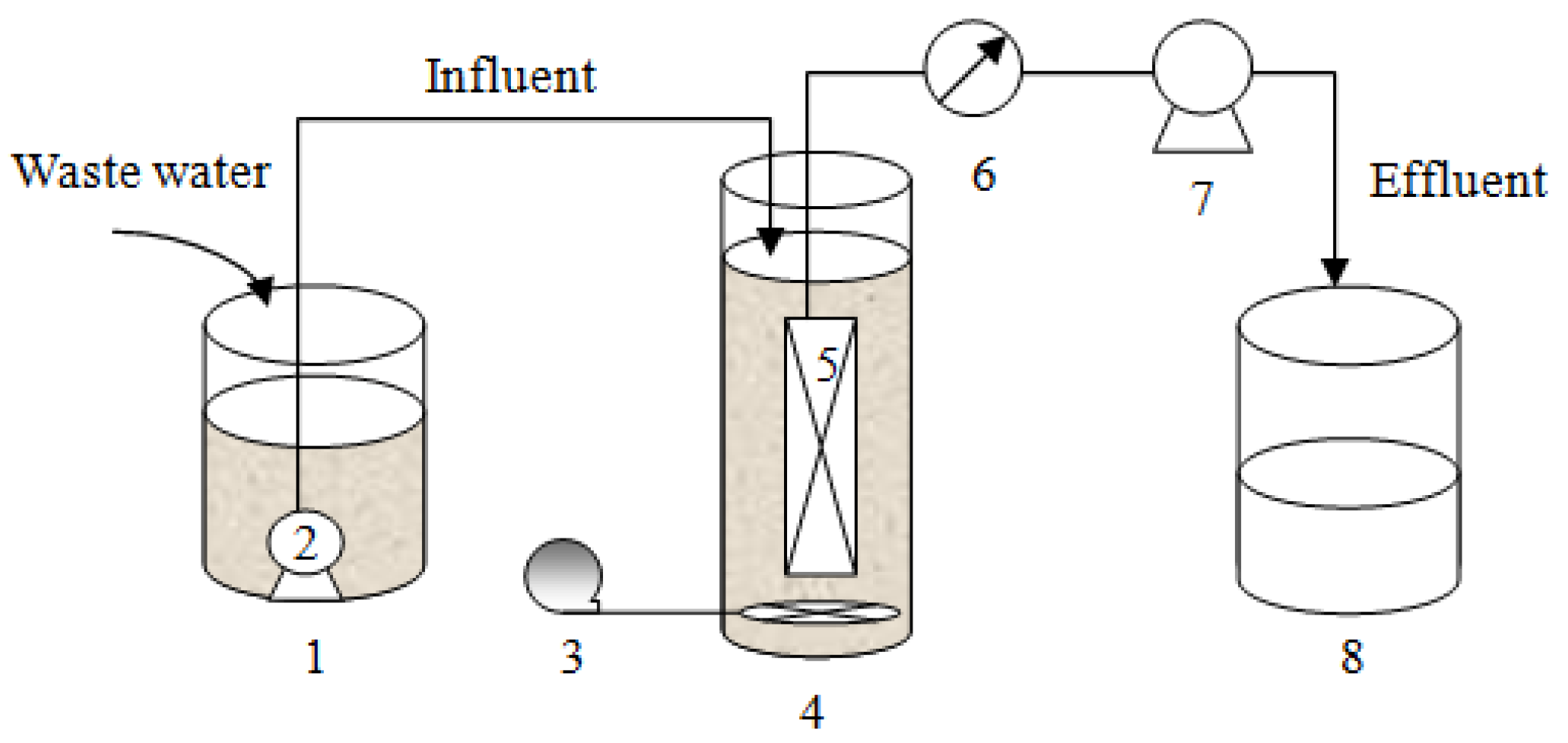


| Parameter | Units | Value |
|---|---|---|
| DO | mg·L−1 | 3–4 |
| HRT | h | 12 |
| COD | mg·L−1 | 400 |
| NH4+-N | mg·L−1 | 50 |
| MLSS | mg·L−1 | 4500 |
| Temperature | °C | 30 |
| Strains | Ammonium Nitrogen Removal Rate (%) | Compound Strains (N9:N7) | Ammonium Nitrogen Removal Rate (%) |
|---|---|---|---|
| N6T a | 75.2 ± 0.8 ** | 1:1 | 87.9 ± 1.6 ** |
| N9 b | 82.1 ± 1.3 ** | 1:2 | 86.3 ± 0.9 ** |
| N7 c | 80.3 ± 0.1 ** | 1:3 | 83.9 ± 1.5 |
| N6T + N9 | 80.8 ± 0.8 | 3:1 | 90.2 ± 1.6 ** |
| N6T + N7 | 79.7 ± 1.7 | 2:1 | 94.0 ± 0.6 ** |
| N9 + N7 | 88.1 ± 0.4 ** | ||
| N6T + N9 + N7 | 82.7 ± 1.6 ** |
| Fungi | Chao | Ace | Shannon | Simpson | Coverage |
| C2 | 63.91 | 67.56 | 1.07 | 0.63 | 0.99 |
| E2 | 68.67 | 73.12 | 1.09 | 0.52 | 0.99 |
| C16 | 97.50 | 125.30 | 1.37 | 0.51 | 0.99 |
| ES16 | 71.86 | 72.72 | 2.09 | 0.27 | 0.99 |
| EA16 | 62.50 | 62.95 | 1.66 | 0.41 | 0.99 |
| Bacteria | Chao | Ace | Shannon | Simpson | Coverage |
| C2 | 313.55 | 312.12 | 3.38 | 0.10 | 0.99 |
| E2 | 344.63 | 342.90 | 3.76 | 0.06 | 0.99 |
| C16 | 331.89 | 324.40 | 3.83 | 0.05 | 0.99 |
| ES16 | 320.75 | 316.18 | 3.40 | 0.10 | 0.99 |
| EA16 | 319.32 | 319.71 | 3.56 | 0.07 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Gui, Q.; Zheng, W.; Zhang, Y.; Wang, K. Nitrogen Removal Mechanism and Microbial Community Changes of the MBR Bioaugmented with Two Novel Fungi Pichia kudriavzevii N7 and Candida tropicalis N9. Water 2024, 16, 757. https://doi.org/10.3390/w16050757
Ma M, Gui Q, Zheng W, Zhang Y, Wang K. Nitrogen Removal Mechanism and Microbial Community Changes of the MBR Bioaugmented with Two Novel Fungi Pichia kudriavzevii N7 and Candida tropicalis N9. Water. 2024; 16(5):757. https://doi.org/10.3390/w16050757
Chicago/Turabian StyleMa, Minglei, Qiang Gui, Weisheng Zheng, Yingjie Zhang, and Kai Wang. 2024. "Nitrogen Removal Mechanism and Microbial Community Changes of the MBR Bioaugmented with Two Novel Fungi Pichia kudriavzevii N7 and Candida tropicalis N9" Water 16, no. 5: 757. https://doi.org/10.3390/w16050757
APA StyleMa, M., Gui, Q., Zheng, W., Zhang, Y., & Wang, K. (2024). Nitrogen Removal Mechanism and Microbial Community Changes of the MBR Bioaugmented with Two Novel Fungi Pichia kudriavzevii N7 and Candida tropicalis N9. Water, 16(5), 757. https://doi.org/10.3390/w16050757






