1. Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected millions of people across the globe which resulted in significant health and economic impacts, and demonstrated the importance of having infrastructure available to deploy for monitoring diseases on a massive scale. Mass community testing is costly and the demand for tests can exceed the capacity of testing facilities during an infection surge [
1], not to mention that not everyone has access to testing due to economic, geographic, or social restrictions. Furthermore, test results are a lagging indicator of a pandemic’s progression, because testing is usually prompted by symptoms, which may take 2 weeks to show up after infection [
2]; furthermore, the more readily available home testing is, the less informative official clinical testing data becomes, since those not requiring medical attention will tend to also not show up in public health testing data as much as those who do require attention, creating confounding and bias in the data. Thus, delays may occur between the appearance of symptoms, testing and the reporting of test results [
3]. Finally, for COVID-19, it is estimated that as many as 45% of cases are asymptomatic [
4,
5,
6,
7]. Considering that people only seek medical attention and undergo diagnostic testing if they are symptomatic, the number of confirmed clinical cases may grossly underestimate the prevalence of the disease (and its capacity to spread).
Wastewater-based epidemiology (WWBE) is a method for monitoring presence and trends of an infectious agent that is shed in bodily secretions/excrement in communities. In WWBE for SARS-CoV-2, wastewater is collected from wastewater-treatment plants (WWTPs) and is tested by qRT-PCR for SARS-CoV-2 RNA excreted via feces, urine, and saliva. The presence and levels of viruses in wastewater samples represents a cross section of viral presence and spread in the communities served by those plants [
8,
9,
10]. WWBE has been successfully employed as a surveillance tool for diseases such as SARS, hepatitis A, and polio [
11,
12,
13]. With regard to SARS-CoV-2, viral particles are reported to be shed in feces from infected individuals even if they are asymptomatic [
14,
15,
16,
17,
18]. Recent studies have shown that WWBE is able to predict COVID-19 prevalence even earlier than clinical case data [
3,
19,
20,
21,
22], supporting the idea that WWBE can be used as an early warning system to monitor community prevalence and spread [
23,
24].
Over the last three years we have gathered SARS-CoV-2 wastewater data, which we present in this paper both to describe the programmatic data to date and to provide more about the specifics of the SARS-CoV-2 pandemic, and to identify best practices for WWBE more broadly. We present data from two complementary data gathering processes here. The primary data is wastewater samples that were collected from 44 total WWTPs across the state of Minnesota between March 2022 and December 2022. (Our team’s data gathering began in May of 2020, but the PCR assay changed in March 2022, as discussed below, so we focus on data after that time period.) The SARS-CoV-2 viral load in wastewater were obtained by qRT-PCR. These 44 WWTPs represent a broad sampling of the Minnesota population serving a total of 191 zip codes and a population of 3,825,269 people, which is approximately 67% of the total population of the state. The secondary data were from an experiment on storage and time-to-analysis conditions for qRT-PCR analysis of wastewater data gathered in the summer of 2023. This experiment was undertaken in part because of our findings from our primary data of significant heterogeneity in the data quality and predictive ability from different WWTPs.
Estimating the SARS-CoV-2 RNA concentrations in wastewater (gene copies per litre) is complicated, as the dilution and fecal strength in the wastewater may vary between sampling dates due to random chance (e.g., caused by variation in a factory’s runoff). It has been recommended to multiply the viral concentration in wastewater by the flow of the sampled location (the volume of wastewater that passed through the location in a day) to obtain the viral concentrations in gene copies per day, and account for changes in sanitary sewer contributions [
25,
26]. Normalizing SARS-CoV-2 RNA concentrations by indicators of human fecal waste is also common, because feces in wastewater can have variable levels of SARS-CoV-2 depending upon the amount of water used per toilet flush or body washing [
27]. The contribution of SARS-CoV-2 from human sourced water can then be estimated by dividing the measured SARS-CoV-2 concentration by the concentration of the human waste indicator [
27]. A typically examined fecal marker is Pepper Mild Mottle Virus (PMMoV) [
19,
27,
28]. Previous studies have shown that PMMoV is the most abundant RNA virus in human feces and it is shed in large quantities in wastewater [
29,
30,
31,
32,
33]. It is also highly stable in wastewater, and its concentrations showed little seasonal variation [
30,
31]. Operating on this information, our program from 2019–2022 normalized SARS-CoV-2 RNA wastewater levels to PMMoV [
19]. However, in our ongoing surveillance work, we found that normalizing by PMMoV obfuscated the large spike in wastewater measurements during the COVID-19 Omicron wave of December 2021–January 2022, which compelled us to reexamine normalization methods. Our findings below suggest that normalization by PMMoV may increase the variation in wastewater measurements, and we do not find evidence in the current data that it substantially improves prediction.
Our study objectives in conducting these analyses were to: (1) learn how best to make use of large-scale wastewater surveillance RNA measurements, and (2) study how storage conditions affect the quality of such measurements. Our motivation for (2) was driven in part by our results in (1), in which we found nontrivial unexplained heterogeneity between WWTPs. We compare a variety of data pre-processing methods; to benchmark the effectiveness of a pre-processing method we use predictive ability of the data on clinical case counts. Rather than use statistical significance testing of model coefficients, or coefficient of determination () from linear regression for assessment of fit, we will use leave-one-out cross-validation, which is appropriate for assessing model performance when prediction is the main goal and when a variety of models and data transformations are being compared. (See the methods section for a full description).
However, we have observed nontrivial heterogeneity in the predictive ability of measurements from differing WWTPs. Researchers have started to investigate how storage conditions affect wastewater RNA and measured concentrations [
34]. In a small scale study focusing on one or a small number of WWTPs, the storage and measurement conditions can easily be controlled. As we will discuss in our results section, we observed significant heterogeneity in the relationship of the wastewater measurements with clinical COVID-19 case counts which we attribute to heterogeneous quality of data from the WWTPs. Thus, we also implemented a study on storage and time-to-analysis conditions for measurements from wastewater, to undertake efforts how much these aspects affect quality of measurements in order to define sample collection guiding principles.
Our surveillance study yields the important finding that data from different WWTPs may have very different predictive ability, even when analyzed by the same laboratory. We also conclude that parsimonious models perform the best for prediction (and the optimal model we find, for our data, is presented below). We find in our stability study that maintaining wastewater samples at 4 °C leads to less RNA degradation than freezing and thawing. The results were statistically and practically significant.
2. Materials and Methods
2.1. Viral Load Quantification
The procedure we used for quantifying viral load is as follows: 40 mL of influent was used to isolate total nucleic acids using the Enviro Total Nucleic Acid Kit for Wastewater (Promega, Madison, WI, USA) and eluted in a volume of 40 μL. Five ul was employed for quantitative RT PCR, performed in duplicate, using the TaqPath™ COVID-19 Combo Kit (ThermoFisher, Waltham, MA, USA) with copy number standards on the QuanStudio 5 RT PCR instrument in order to quantitate viral copy number. For the stability study, a large volume was obtained and then aliquoted for defined times over multiple temperatures. Four total samples from a single site were obtained and the RNA was isolated and qRT PCR was done on all samples concurrently.
2.2. Statistical Methods
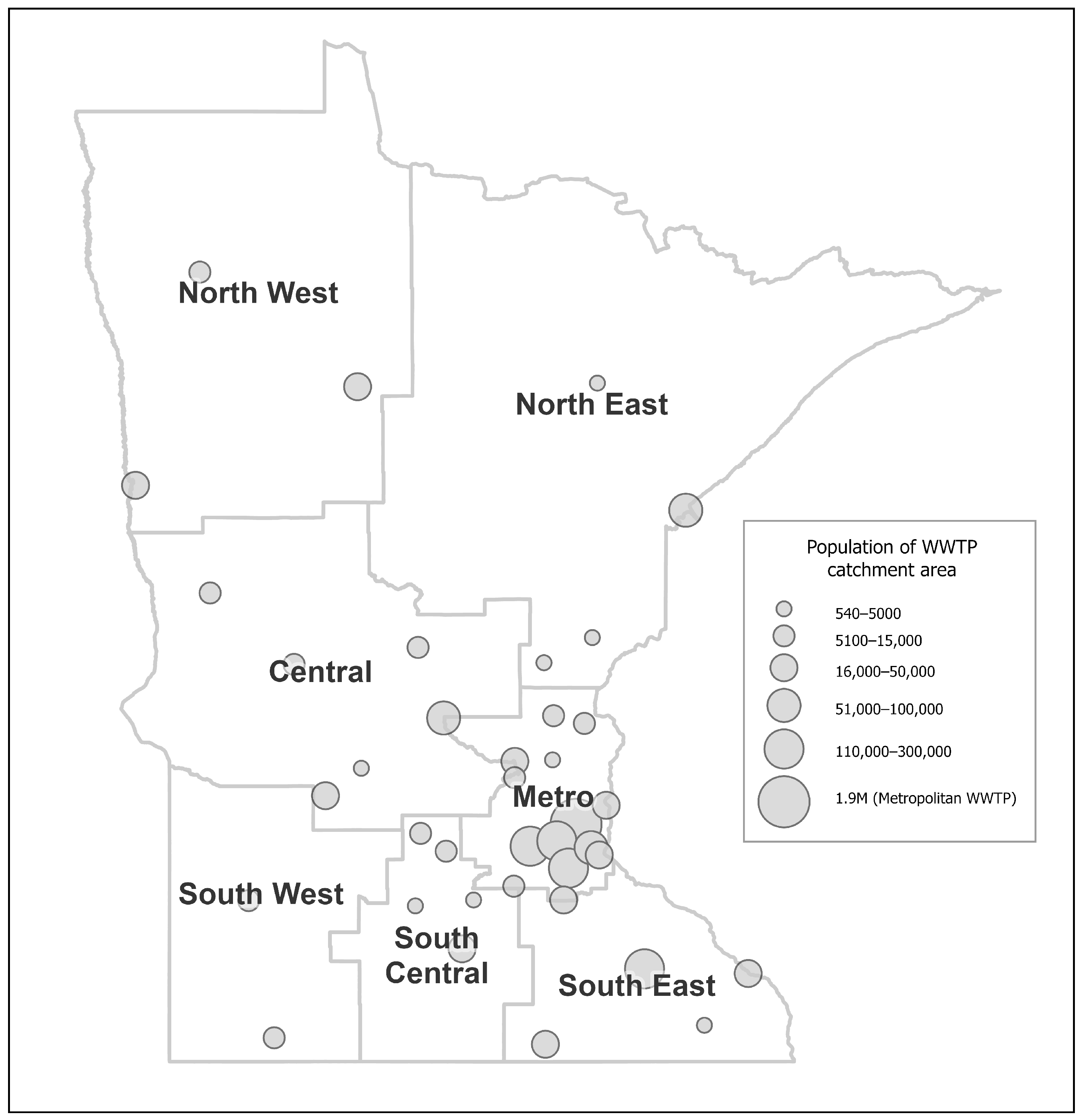
Wastewater samples were collected from a total of 44 WWTPs across the state of Minnesota (see
Figure 1 for a map of WWTP locations). From its inception to March 2022, we employed a polyethylene glycol concentration and phenol chloroform nucleic acid extraction procedure and qRT-PCR for the nucleocapsid gene in the SARS-CoV-2 genome using N1 and N2 primer:probe pairs. As the pandemic progressed so too did more sensitive and effective isolation and detection methods. Accordingly, in March 2022 we pivoted to a column-based total nucleic acid isolation method (Wizard
® Enviro Total Nucleic Acid Kit (Promega, Madison, WI, USA)) and a three gene qRT PCR method that measured the nucleocapsid (N), spike (S), and ORF1ab (O) proteins, sampled from 40 WWTPs. Our statistical findings demonstrate the latter assays are more accurate than the former assays. We present in this paper, data from the latter period (March 2022 to December 2022), using the three gene qRT PCR assay. Since the two assays are nontrivially different, they cannot be combined into a single statistical analysis. For data analysis purposes, a weekly level average of measurements was used. One sample was reported to have zero concentration of N, S and O. It was removed in further analyses.
Among the remaining samples, 424 were reported to have zero virus concentration of S. The reason for the loss of signal (S gene target failure) was due to prevalence of a SARS-CoV-2 variant that acquired mutations in the spike gene that prevented the primer:probe pair from binding. Thus, the S gene was not used in any further analyses. Importantly, our analytical resolution was preserved using the N and Orf1a genes. The concentrations of a human fecal marker, PMMoV, were also determined in each wastewater sample in parallel. PMMoV concentrations below the 5th percentile were replaced with the 5th percentile value, and those above the 95th percentile were replaced with the 95th percentile value to avoid outliers. The influent flow rate was provided by the participating WWTPs. For 13 of the WWTPs, the catchment area for the WWTP was obtained from the Minnesota Pollution Control Agency (MPCA) or in some cases directly from the treatment facility. For the other WWTPs that did not have a digitized catchment area available, the city boundaries were used to approximate this area for these. Many of those were rural WWTPs where the city boundaries are likely to be a good approximation of the WWTP catchment area. Data enrichment services from Esri [
35] were used to apportion 2020 census block population to the WWTP catchment areas.
The weekly number of new infections for each WWTP service area was obtained from the Minnesota Department of Health (MDH) and aggregated to the zip code areas using the residential address for the case. Case counts were summed by zip code weekly (Sunday–Saturday) and the date beginning the week was used to associate with each case count. The case data were then aggregated by WWTP according to the zip codes previously found to be associated with each catchment area. The zip codes were spatially intersected with the sewersheds to identify all of the zip codes associated with each sewershed. Each zip code area was visually inspected to ensure that if a zip code intersected two sewersheds, that zip code would be associated only with the sewershed with the most area in the zip code. Five zip codes intersected more than one WWTP catchment area. To allow a transformation, any zero case counts were replaced with a value of one. The MDH also provided the number of people who have received at least one dose of COVID-19 vaccine, and the number of people with completed vaccine series in the areas served by each WWTP over time. The complete series could be one, two, or three doses depending on the person’s age and which vaccine they received. Being “fully vaccinated” does not include or require further booster doses in the present definition.
The wastewater samples were collected twice per week during the course of the study and were isolated on different days (Monday and Wednesday or Tuesday and Thursday) for different WWTPs. To align the wastewater samples with the clinical case counts and to avoid having to analyze data on irregular time intervals, we aggregated the wastewater measurements by week (Sunday–Saturday). For a given week with any wastewater data, we use the sample average of all the measurements of that week as the aggregated measurement. If there was no wastewater sample in a week then that week was considered missing and was not included in the analysis.
With time-aligned case counts and wastewater measurements, we ran linear regression models (the specific models will be described shortly). We fit separate models for each WWTP. Due to the relatively low number of data points for each WWTP (recall
Table 1), nonlinear or machine learning methods are not effective. To assess the model performance, a leave-one-out cross-validation (LOOCV) was used. The models were trained on
observations and validated on the remaining one observation, where
n is the sample size. The procedure was repeated
n times with each of the
n observations used exactly once for validation. The average of the
n prediction errors obtained was computed for model comparison. The evaluation metric was the root mean squared errors (RMSE) between the predicted value and the actual value. If the dependent variable was log
10-transformed, the transformation was reversed to obtain the predictions on the original scale before the computation of RMSE. This provides an unbiased approach for comparing predictive performance of different models. All statistical analyses were performed using R version 4.2.3 [
36].
Use of SARS-CoV-2 Concentrations
Denote the COVID-19 case count at time
t by
and denote the SARS-CoV-2 concentrations (either O or N) measured in a wastewater sample at time
t by
. In this section, we compare different linear regression models for predicting
from
. As mentioned in
Section 1, virus concentrations are commonly normalized by either flow,
or a fecal marker such as PMMoV:
However, there have been contradictory findings on whether normalization of virus concentration can improve correlations with cases [
28,
37,
38]. Moreover, it seems that no studies examined using both flow and PMMoV to normalize the virus concentration, as in
(This latter normalization has perhaps less logical coherence but to the extent that flow and PMMoV are both informative, but noisy and imperfect, it could be helpful.) We compare these three normalization approaches. It is also of interest to know whether adding lagged virus concentrations (e.g.,
and
) in the model will improve the predictive performance. Finally, it is prevalent in the wastewater literature to use a
transformation on the variables to meet assumptions for parametric analysis [
38,
39]; we compared taking the log transformation of wastewater measurements to not taking it, to validate this standard procedure. In results not shown here for the sake of brevity, we verified as well that because the N and O genes are highly correlated, it is not very important whether one or the other (or an average of the two) is used for prediction. We focus on using solely the O gene here. To summarize, we varied the following factors: 1. Normalization of the virus concentrations (unnormalized, flow, PMMoV or both), 2. The number of lagged values for the virus concentrations (0, 1 or 2), 3. Whether a
transformation was used on the dependent and independent variables. The three factors were ‘fully crossed’, resulting in a total of
conditions. The models were fit separately for each WWTP in each condition (we will show, shortly, that different model fits are needed for different WWTPs). To allow for comparisons between WWTPs with different sizes of population served, we divided the case count and virus concentrations by the population size. This ensures that the prediction errors for all WWTPs are theoretically on the same scale. Observations with missing values due to the creation of lagged variables were removed. As was mentioned above, the models were compared by the averaged-over-WWTPs LOOCV.
2.3. Storage and Time-to-Analysis Stability Methods
We conducted a ‘stability study’ about stability of RNA in wastewater under different conditions. We used a large volume of WW from a single site that was partitioned into samples that were stored at varying conditions (4 °C, −20 °C, and −80 °C) for 2, 5, 7, or 14 days. Nucleic acid for each sample (n ) was then analyzed by qRT PCR concurrently (for a total of 4 × 4 × 5 = 80 data points).
3. Results
3.1. Description of Surveillance Data
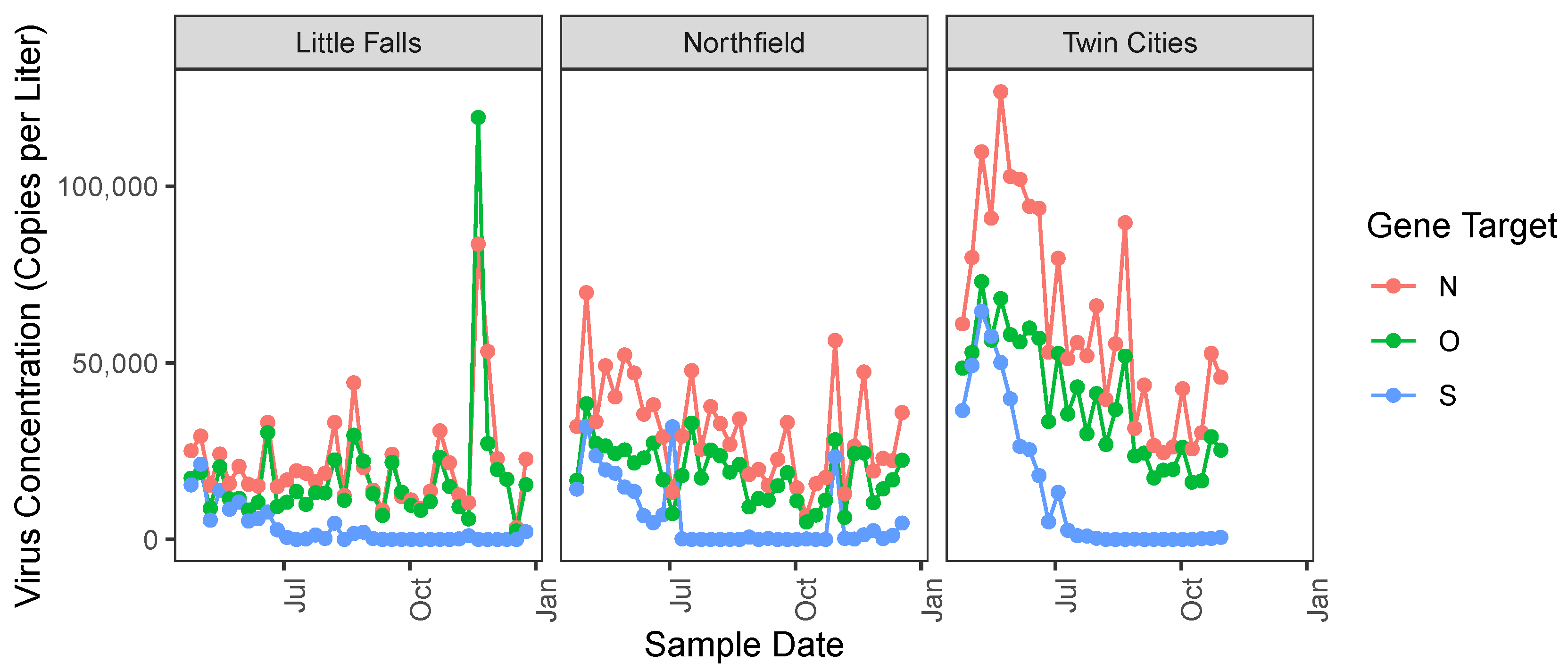
Figure 2 displays the concentrations of N, S and O in three WWTPs, Little Falls, Northfield and Twin Cities, which are chosen as representatives of smaller, midsized, and large population sizes, respectively (see
Table 1 below for tabulated populations).
Table 1 provides some descriptions of the participating WWTPs, including the sampling period, the number of weeks where the WWTP was included in the study (because we had clinical case count data and had or could impute wastewater measurements), the size of the population served, as well as the sample mean of the weekly case count.
3.2. Use of SARS-CoV-2 Concentrations
Table 2 displays the (rescaled, for ease of presentation) RMSE (root mean squared prediction error) of the cross-validated RMSE of the linear regression models under different conditions across WWTPs. Lower indicates more accurate prediction. (Recall that the case count variable was divided by the corresponding catchment area population size). Overall the results in
Table 2 suggest that taking logs is highly important for performance; this validates the standard practice of taking log transformations not just for parameter estimation but for prediction. We find that the normalization method is not as important especially when logs are taken. When the raw data were used, normalizing by flow alone slightly reduced the RMSE, whereas involving PMMoV in the normalization led to an increase in RMSE. Recall that PMMoV is defined by truncating the highest and lowest quantile values to mitigate the high variability in its measurement; but this was not enough to remove the high level of variability totally. Our interpretation, focusing especially on the non-logged models, is that PMMoV introduces extra variability which causes degradation. For instance, in our data when we normalize by PMMoV, the spike in wastewater RNA levels from the omicron surge that occurred in December 2021 in Minnesota does not appear, because the variability of the PMMoV measurements obfuscates it. Although in some contexts and for some WWTPs it seems that using PMMoV is helpful, our interpretation is that its high level of variability prevent it from being used reliably in normalization, at least at this time. On the other hand, our findings validate the use of flow in that even without a log transformation, normalization by flow is seen to achieve nearly optimal predictive performance.
Including extra lags generally worsened the predictive performance, perhaps surprisingly. These results are in the context of only having a few dozen observations per WWTP, but we argue this is common given that over a longer time horizon the parameters and prediction models are likely to change in the face of evolving virus and immunity landscape. It is important to note that different models yielded different best fits for individual WWTPs; here we are presenting results averaged over all WWTPs. Results are affected both by high performing and low performing WWTPs. It is worth noting that in some of the ‘higher performing’ WWTPs, predictive performance was improved by using extra lags. For instance, in the EB, EP, HT, MH, MK, and TC WWTPs, including lagged variables improves performance according to LOOCV.
Overall, the model with the lowest RMSE over all WWTPs is the one where virus concentrations are normalized by flow, with
transformation, and without any lags:
where
’s are the regression coefficients, and
is the residual. Although it is not clear from our results if the normalization by flow is truly important, this is the model to which we explore various extensions in the upcoming sections.
3.2.1. Heterogeneity in Fit and Individual WWTP Validation
It would be ideal if it were possible to develop one model using the pooled data from all WWTPs. Unfortunately our results suggest this is not possible; we compared the estimated error variance and the estimated regression coefficients of
from the model (
1) across WWTPs. We found significantly different regression coefficients and variance estimates, see
Table 3 (giving estimated regression coefficients and
p-values of
for each WWTP) and
Table 4 (giving estimated
values for each WWTP). We found that the estimated coefficients ranged from −0.06 to 0.66 (with summary statistics of mean = 0.28, SD = 0.23). The ratio of the largest to smallest variance estimate was larger than 10. This indicates that separate fits are needed for separate WWTPs. More broadly, it suggests the important finding that in any large-scale surveillance plan, measurements from different WWTPs (even when analyzed by the same lab) may not be directly comparable and may require different interpretation.
3.2.2. Use of Vaccination Rate
We examine how best to incorporate vaccination data in our linear regression models. Let
be the percentage of people who have received at least one doses of vaccine out of the population served by a WWTP, and
be the percentage of fully vaccinated people. The models to be investigated were built upon the model in Equation (
1). In addition to the normalized virus concentration
, the models also considered the main effects of
and/or
, as well as two- or three-way interactions between
,
and
. The models are listed in
Table 5, along with their means and standard deviations of the cross-validated RMSE across WWTPs. The model that includes the interaction between
and
,
minimizes the mean RMSE, but the differences are not large (and the results were not sensitive to whether
or
was used). The clearest result is that including too many variables leads to worse predictions. However, in the next subsection we see perhaps surprisingly that including vaccination may add more variability than it removes (especially when predicting two weeks ahead).
3.2.3. Prediction Accuracy over Different Forecast Horizons
We assess the performance of predictions over different forecast horizons (same week, one week ahead and two weeks ahead). The model in Equation (
2) was fitted separately for each WWTP, with
and
being the dependent variable. The means and standard deviations of the cross-validated RMSE are reported in the first three rows of
Table 6. At the current noise level, we do not see a degradation in ability to predict one week ahead versus predicting the current week’s case count. The predictions for two weeks ahead are substantially worse than the predictions for the first week. For comparisons, we fitted the model in Equation (
1) again with different forecast horizons (
Table 6).
Table 7 gives results for models that exclude vaccinatino information. Interestingly, when predicting two weeks ahead, the simplest model (excluding vaccination information) does much better in terms of both mean and variance, which shows the value of parsimonious models.
3.3. Storage and Time-to-Analysis Stability Study Findings
Although one might expect a priori for measurements from different WWTPs that are studied in the same lab to have broadly similar characteristics, we found above in the analysis of our surveillance data nontrivial heterogeneity in predictive ability (coefficient values and standard deviation estimates) between different WWTPs. Unmeasured variables such as population mobility, access to clinical testing centers, or population willingness to utilize clinical testing could account for some of the variation. Wastewater storage conditions are another possible cause. To shed light on what causes such unexplained large differences between WWTPs, we conducted a stability study, as described in the Materials and Methods section. The summary statistic means (SDs) for each temperature (4 °C, −20 °C, and −80 °C) [averaging across all times] of 39,400 (8560) for baseline, 18,600 (8600) for 4 °C, 2970 (5420) for −20 °C, and 3360 (1550) for −80 °C).
We found that storage temperature had a large and significant impact on the measured RNA. The length of time of storage had a less noticeable and not significant impact. We subtracted each baseline measurement from each storage measurement, yielding a negative number in all cases. We transformed the response by taking the negative and then the logarithm. We then fit a linear mixed regression model with no intercept term, a (linear) time trend term, a factor variable for the three storage conditions, and a random intercept effect for each sampling unit, with no interactions included; that is, we fit
for
, TempIndicator
j an indicator/dummy variable for the three temperature storage conditions,
i.i.d.
random effects,
,
i.i.d.
, and
the fixed effects of interest
. The coefficient estimates for each of the three storage conditions were significant; the coefficients for −20 °C and −80 °C were not significantly different than each other, but are different than the coefficient for 4 °C. The negative exponentiated coefficient estimates (
p-values) are −16,200 (2
) for 4 °C, −32,300 (
) for −20 °C, and −32,300 (
) for −80 °C. The time trend term was not significant. The means and standard deviations (over the four samples) at each of the 12 different conditions are presented in
Table 8. The Day 0 average (sd) is 39,400 (8560). It is easy to see at a glance the effect of storing below 4 °C.
These data provide important guidelines for sample submission and storage whereby 4 °C stored samples showed less degradation than a −20 °C or −80 °C freeze and thaw. Under these principles we advocate long term storage at −80 °C and shipping temperatures under refrigeration (4 °C).
4. Discussion
We performed wastewater surveillance for SARS-CoV-2 across the state of Minnesota and undertook efforts to apply statistical analyses to inform how to best interpret and apply these data by studying the predictive relationship of wastewater virus levels to clinical case counts that can be used for public health decision making. For instance, it is clear that if clinical case counts demonstrate a large surge, then the ability of wastewater to demonstrate the same surge is important and beneficial; so in this sense a strong statistical relationship between the wastewater measurements and case counts would be meaningful. Our estimates of error variance and regression coefficients revealed substantial variability across WWTPs in our program. These findings suggest that the relationship between COVID-19 incidence in clinical patients and SARS-CoV-2 RNA in wastewater may be treatment plant-specific, and future work will need to continue investigating how to appropriately normalize data from different plants to allow for cross-plant comparisons. We acknowledge that in the current study, usage of areas with discrepant boundaries for defining case counts and wastewater levels could create data accuracy issues. Future work will assess whether using more refined location data for clinical case counts (rather than binning by zip codes) could aid in improving model performance.
The variation we observed is not accounted for by the size of the catchment area or the size of the corresponding population. This suggests that, at present, COVID-19 WWBE may need to be viewed at the individual plant level for its most impactful application as a regional metric for disease burden. Additionally, studies such as ours involving community level analyses represent challenges in that the available individual level data are rather limited, and the conclusions may not apply in an extremely data rich environment. However, in infectious disease modeling, we believe that true data rich environments (in which necessarily model parameters and immunological landscapes all stay constant over time) are exceedingly rare.
Prompted by variability observed in community-based studies, we set out to determine if we could identify actionable items that would boost accuracy (i.e., diminish variance). Our surveillance study period over the year of 2022 was characterized by mixed SARS-CoV-2 lineages and immunity from either/both exposure or primary and booster vaccines. Future models including vaccination data should adapt updated categories of vaccination classification, for example, a time-since-vaccination variable, to account for the waning of immunity over time. Further considerations are seasonal, regional, climatological, and sample temperature effects [
40]. SARS-CoV-2 has year-round disease activity and as the pandemic has progressed, the levels and severity (i.e., hospitalization and deaths) has shown a trend toward higher incidence in the autumn and winter in both the Northern and Southern hemispheres [
41,
42]. We have not correlated large precipitation events with peaks or troughs in wastewater levels of SARS-CoV-2; however, other studies have observed that rainfall can have a diluting effect that can be calibrated using other chemical indicators [
43]. This raises an important consideration for normalizing wastewater data that has been undertaken using both treatment center flow rates and fecal indicators [
26,
43,
44,
45,
46,
47]. Consistent with some studies in the wastewater literature [
28,
37,
38], the current study finds that the predictive performance does not improve after involving PMMoV in the normalization. Interestingly, adding lagged virus concentrations to the model generally worsened the predictive performance. Using the vaccination rate in the model has been shown to improve the predictions. However, the results are not uniformly consistent across different forecast horizons. Due to the relatively low number of data points for each WWTP (
Table 1), nonlinear or machine learning methods are not effective, and in fact our very parsimonious linear regression models performed best.
The temperature during the transit of wastewater to treatment sites can vary based on the location and infrastructure of the municipality and represents a variable that is difficult to control. Toward identifying temperature variables we could control, we asked whether the storage condition of the sample from which we quantify viral load would impact analytics. To answer this question, we acquired four large volume samples obtained across four different time periods and partitioned them into aliquots that were stored under room temperature, refrigerated, or frozen conditions for defined lengths of time (
Table 8). We observed that freeze-thawing led to greater RNA degradation than maintaining the sample at 4 °C. This has important ramifications and provides guidance for the analytical community whereby samples can be stored onsite under refrigerated conditions and shipped on ice packs. These conditions are easily achievable for any geographic region as opposed to frozen storage that requires dedicated equipment for cold chain storage.
Our multiplex gene analysis has enabled some view of circulating variants due to the polymorphic nature of the spike gene/protein where gain and loss of the amino acids at positions 69 and 70 (H69V70) can lead to presence or absence of a S-gene signal by qRT PCR with the TaqPath™assay [
48,
49]. Efforts are underway in our lab to expand and more definitively define the wastewater population in regards to SARS-CoV-2 variants using genomic surveillance [
37,
50,
51]. Such approaches can enable correlative relationships between a circulating variant or a mixed occurrence of variants and clinical specimen testing [
52] and will enhance our statistical predictive accuracy.
Our program has operated continuously since 2020, and the data presented in this manuscript represent a broad geographic area. Our findings provide new guidance for statistical modeling and prediction along with storage procedures for wastewater samples and are additive to the fields of epidemiology and pathogen surveillance.