Abstract
The heavy metal cadmium poses severe threats to both ecosystems and human health. Utilizing genetic engineering to enhance the microbial capability for efficient cadmium accumulation has emerged as a pivotal research direction. This study constructed a genetically engineered bacterium capable of expressing multivalent phytochelatins with a self-assembly ability and explored its efficacy in cadmium adsorption. Molecular biology techniques were adopted to fuse the recombinant human ferritin (rHF) gene and the synthetic phytochelatin (EC) gene, known for its robust adsorption capacity for heavy metals. The expression vector was constructed. Escherichia coli (E. coli) served as the host cell to express multivalent nanochelator rHF-ECs tailored for high-efficiency heavy metal adsorption. The results reveal the successful soluble expression of the recombinant fusion protein in E. coli cells, forming self-assembled multivalent nanoparticles with a size of about 13 nm, and the target protein rHF-EC20 (monomer) could adsorb approximately 9.2 μmol of Cd2+ in vitro. Moreover, this recombinant strain demonstrated cadmium adsorption across a temperature range of 16–45 °C and a pH range of 5–9, with the optimal performance observed at pH 7.0 and 37 °C. Compared with the control strain, the recombinant strain BL21 (FLE), expressing nano-chelating peptides, achieves an adsorption rate of 80% for Cd2+ at 60 min, resulting in an approximately 18% increase in the Cd2+ enrichment efficiency. The maximum adsorption capability of cadmium reached 12.62 mg per gram of dry cell weight. This work indicated that the synthesis of multivalent chelating peptides in E. coli cells could efficiently enhance the bioaccumulation of the heavy metal cadmium, which renders novel avenues and methodologies for addressing cadmium pollution, offering promising prospects for environmental remediation.
1. Introduction
Cadmium, a natural heavy metal characterized by strong mobility, possesses a high toxicity to human health. The discharge of waste and sewage in industries such as electronics, chemicals, and photovoltaics as well as cadmium-laden dust and smoke deposition in the atmosphere have led to a substantial accumulation of cadmium in the environment. Moreover, as this metal traverses water bodies and food chains and accumulates, it further adversely affects the human respiratory, cardiovascular, gastrointestinal, reproductive, renal, and nervous systems, posing a grave threat to ecological balance and human health [1,2]. Therefore, efficiently reducing the content of cadmium in the living environment or eliminating its bioavailability has emerged as an imperative issue demanding human intervention.
In recent years, substantial research efforts have focused on the environmental remediation of cadmium pollution. Traditional physical and chemical treatment methods include ion exchange, dialysis, reverse osmosis, ozonation, nanofiltration, and chemical precipitation [3,4]. Meanwhile, there is growing interest in utilizing natural or modified biomass materials as well as carbonizing biomass into materials with adsorption properties for the adsorption and fixation of Cd2+ [5,6]. In recent years, hydrogel-based adsorbents have been designed and applied for the removal of heavy metals from water [7,8]. Microorganisms, pivotal in ecosystems, exhibit diverse mechanisms for cadmium adsorption and storage, including cell membrane/wall surface adsorption, intracellular aggregation, mineralization precipitation, and intracellular conversion [9,10]. Compared with conventional treatment methods, microorganisms offer advantages like a superior adsorption efficiency, absence of secondary pollution, and environmental compatibility [11,12,13]. Naturally occurring bacteria, fungi, seaweed, and algae have demonstrated the capability to adsorb and accumulate heavy metals [14,15,16,17]. Due to their small volume, ease of cultivation, and rapid reproduction, they have been extensively used for the remediation of heavy metal pollution in the environment. However, naturally isolated microorganisms usually possess drawbacks in their adsorption capacity and stress resistance. With the increasingly stringent requirements of national pollutant emission standards, challenges persist in the adaptability and efficiency of employing these microorganisms for heavy metal treatment. In response to this issue, recent research has focused on leveraging genetic engineering to enhance the efficiency of microbial cells in treating heavy metals [18,19,20]. This involves introducing dominant genes associated with the enrichment and degradation of heavy metals in microbial cells into recipient strains with a robust reproductive capacity and ease of manipulation. Candidate genes involved in the adsorption and transport of cadmium ions include cadmium transporters, metallothionein capable of binding to heavy metal ions, and synthetic phytochelatins (ECs). A previous study has verified the cadmium transport activities of mercury superfamily proteins (MerC, MerE, MerF, and MerT) [21]. Furthermore, the overexpression of these transporters has been shown to elevate the Cd uptake [22]. Metallothioneins (MTs) are intracellular proteins with a low molecular weight and high cysteine content that exhibit a robust adsorption capacity for heavy metals [23]. Organisms that overexpress metallothionein and biomaterials based on metallothionein have been extensively utilized in treating heavy metals [24,25,26]. Ying Cai et al. successfully expressed both the cadmium transporter protein and pea metallothionein in E. coli cells, resulting in a 50% increase in cadmium ion accumulation in stem cells per gram compared to wild strains [27]. ECs are heavy metal binding peptides that mimic phytochelatins. With a structure similar to that of the cell detoxifying agent glutathione, ECs contain repeating dipeptide units composed of glutamic acid and cysteine {(Glu-Cys) n Gly (n: repetition times)}. These two amino acids can efficiently chelate heavy metals through carboxyl and thiol groups [28]. Bae et al. fused and expressed synthetic chelating peptide ECs, which can adsorb heavy metals, with ice nucleation protein INPNC and successfully displayed ECs on the surface of E. coli and Moraxella sp. cell membranes for the treatment of Hg2+ in aquatic environments. The results show that the Moraxella sp. engineered bacterium has a better ability to adsorb Hg2+ than the E. coli engineered bacterium, which is ten times higher [29]. Yu et al. showed metallothionein on the surface of the strain Alishewanella sp. WH16-1-MT, which doubled the adsorption capacity of the strain for Cd2+. The engineered strain was further used to remediate a cadmium-contaminated paddy soil, effectively improving the resistance of rice to cadmium and reducing the accumulation of cadmium in rice [30]. Moreover, Zhu et al. constructed a genetically engineered strain that could efficiently enrich heavy metals by displaying a fusion protein (SynHM) carrying a 6*His-tag, a cysteine-rich short peptide, and metallothionein on the surface of E. coli cells. The adsorption efficiency of this strain for Cd2+ can reach 50 mg/L [31].
In nature, a plethora of proteins can precisely self-assemble and form various nanostructures, such as nanowires and protein nanocages. Among these, due to its unique structure and stable properties, the human ferritin heavy chain (rHF) has been widely used as a structural unit in the design and synthesis of nanodevices, showcasing its multifaceted function. Human-derived ferritin rHF primarily facilitates the storage and dynamic regulation of iron elements within the human body, exhibiting highly conserved biochemical and structural characteristics [32]. The rHF subunit contains 180 amino acids, consisting of a 5-α-helix composition, spontaneously assembled into a 24-polymer nanocage structure in cells, with an outer diameter of 12 nm and an inner diameter of 8 nm. The N-terminus of each subunit is exposed on the outer side of the cage. The overall spatial structure presents a 4-3-2 symmetry [33]. It has been verified that ferritin has an extremely high thermal stability, with a Tm value of up to 85 °C. It can achieve soluble expression and self-assembly in hosts, making it an ideal self-assembled nanoelement [34]. The successful engineering of ferritin proteins as potential nanoreactors for synthesizing/incorporating nanoparticles and as nanocarriers for tumor detection and therapy has been documented [35,36].
E. coli stands as an ideal microorganism for wastewater treatment due to its rapid growth and clear genetic background, making it a current research hotspot to enrich heavy metals through genetic engineering modification. Upon the enrichment of heavy metals, achieving subsequent non-hazardous treatment is straightforward through solidification and stabilization, including the encapsulation of pollutants into inert substrate materials (such as cement and lime), organic binders like asphalt and other thermoplastic materials, and thermally hardened organic polymers such as urea, phenolic plastics, and epoxides. These approaches aim to convert pollutants into less soluble, less migratory, or less toxic forms, thereby abating the risk of harm to ecosystems [37,38,39]. The design concept and system are shown in Figure 1.
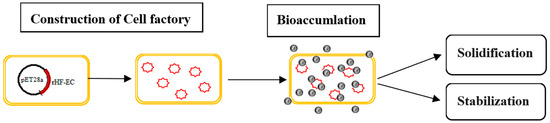
Figure 1.
Design concept of cadmium treatment with Escherichia coli cell factory. Yellow boxes represent E. coli cells, the red circles represent nanochelator rHF-ECs, and black balls represent cadmium ions.
This study selected 20 repetitive units of artificial plant chelating peptide EC20 and the coding gene of human transferrin rHF for gene fusion through the peptide linker coding sequence. By overexpressing the fusion protein in E. coli cells, a 24-polymer multivalent, nano-chelating peptide self-assembled in the cells for heavy metal treatment. This recombinant strain was used to investigate the adsorption capacity and characteristics of Cd2+ in simulated wastewater, providing new research ideas and highly effective biomaterials for the bioremediation of cadmium-contaminated environments.
2. Materials and Methods
2.1. Material
2.1.1. Strain and Plasmid
The host bacteria E. coli DH5α and E. coli BL21 (DE3) and the plasmids pET28a and pET28a-rHF were preserved in the laboratory of this study. The plasmid pUC57-L-EC20 carried a linker peptide sequence (GGGSGGGS) and the fusion gene EC20. Endonucleases BamH I and Xho I were introduced at both ends of the gene to recognize sites. The codon optimization and synthesis were fulfilled by Genscript Biotech Corporation (Nanjing, China).
2.1.2. Main Reagent
The plasmid extraction kit and DNA purification kit were purchased from Beijing Solarbio Biotechnology Co., Ltd. (Beijing, China); the protein gel electrophoresis kit was from Crystal Biology; PrimeSTAR Max DNA Polymerase, restriction endonuclease, and T4 DNA ligase came from Takara Bio (Beijing, China); and the cadmium detection kit was purchased from Henan Suijing Environmental Protection Technology Co., Ltd. (Luoyang, China). All other reagents were analytical grade.
2.1.3. Culture Medium
The LB medium for E. coli culture comprised the following: 0.5% yeast extract, 1% Tryptone, 1% NaCl, and a solid plate with 2% agar. The protein expression self-induction culture medium was prepared according to the formula of Gawron et al. [40], including Tryptone, 10 g/L; yeast extract, 5 g/L; Na2HPO4·12H2O, 9 g/L; KH2PO4, 3.4 g/L; NH4Cl, 2.68 g/L; Na2SO4, 0.71 g/L; MgSO4, 0.24 g/L; glycerol, 6 mL/L; glucose, 0.5 g/L; and α-D-Lactose, 2 g/L. When using the above culture media, kanamycin sulfate with a final mass concentration of 0.1 mg/L was added.
2.2. Method
2.2.1. Construction of Fusion Protein Gene Expression Vector
Based on the nucleotide sequence of rHF (Genbank: M97164.1), specific primers were designed: FP (5′-GATCCATATGACGACCGCGTCCACC-3′, underlined as Nde Ⅰ restriction site) and RP (5′-TACCGGATCCGGGCGCTCCCATCTTGCG-3′, underlined as BamH Ⅰ restriction site). The PCR amplification was performed using the plasmid pET28a-rHF as a template. The PCR product was detected using 1% agarose gel electrophoresis, and the target gene rHF was recovered with a DNA purification kit. The fragment was digested with Nde I and BamH I and recovered for later use. Simultaneously, the plasmid pUC57-L-EC20 was digested by BamH I and Xho I. After the reaction, the L-EC20 gene fragment was recovered for later use. The digested products of the rHF gene and the L-EC20 gene obtained in the above steps were ligated to the pET-28a (+) vector double digested with Nde I and Xho I using T4 DNA ligase. The ligation products were transformed into E. coli DH5α competent cells, and monoclones were selected for colony PCR validation. Clones with preliminary validation were sent to Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) for sequencing. The correctly sequenced recombinant plasmid vector was named pET28a-rHF-EC20.
2.2.2. Expression and Purification of Recombinant Fusion Protein
The correctly sequenced recombinant plasmid pET28a-rHF-EC20 was transformed into E. coli BL21 (DE3) competent cells to obtain the recombinant strain BL21 (FLE). Meanwhile, pET28a was transformed into the same strain to serve as the control strain BL21 (28a). Single colonies were picked from LB agar plates and inoculated into a 5 mL LB liquid medium containing 100 mg/mL kanamycin. The cultures were incubated overnight at 37 °C and 180 rpm to prepare seed solutions. These seed solutions were then inoculated at 5% (volume fraction) into a 300 mL auto-induction medium containing the same antibiotic at 37 °C for 16 h. Then, the bacterial cells were collected by centrifugation at 4 °C and 12,000 rpm for 5 min. Part of the bacterial cells was resuspended in a pre-cooled cell disruption buffer, followed by sonication under ice bath conditions for 30 min (working duration: 3 s; interval time: 3 s; power: 300 W) until the bacterial suspension became clear. The suspension was centrifuged again at 12,000 rpm for 30 min to collect the supernatant and precipitate for SDS-PAGE analysis. The supernatant passed through a 0.45 μm filter membrane, and the recombinant protein in the filtered supernatant was purified using a Ni-NTA column through affinity chromatography. The purified protein was concentrated and desalted to determine its concentration before being stored at −80 °C for future use.
2.2.3. Morphological Characterization of Recombinant Fusion Protein
The purified target protein was diluted to lower the salt ion concentration. Then, 10 μL of the protein solution was pipetted onto a carbon membrane and dried at room temperature. Next, it was negatively stained with 1% phosphotungstic acid solution (pH 7.0) for three minutes, followed by rinsing with deionized water 2–3 times. After drying at room temperature, the morphology of the protein was observed using the transmission electron microscope FEI Tecnai G2 spirit Biotwin (Thermo Fisher Scientific lnc., Waltham, MA, USA).
2.2.4. Adsorption of Recombinant Protein rHF-EC20 to Cadmium
A total of 2 mg of purified target protein rHF-EC20 was mixed with 100 mg of Ni-NTA agarose resin loaded with Ni2+ and reacted for 30 min to ensure full binding to the resin. The resin was washed with ddH2O to remove unbound target protein. The eluate was collected after centrifugation, and the protein content was detected to calculate the amount of protein fixed to the resin. The immobilized target protein was mixed with 100 mg/L cadmium chloride and shaken at room temperature for 30 min. The supernatant was collected after centrifugation and washed once with a small amount of ddH2O. The cadmium content was determined using a cadmium quantification detection kit. Finally, the adsorption capacity of the target protein to cadmium was calculated.
2.2.5. Adsorption of Recombinant Strain to Cadmium
Influence of Induction Time on Cadmium Adsorption by Recombinant Strain
Single colonies of the recombinant strain BL21 (FLE) and control strain BL21 (28a) were picked and inoculated into 5 mL LB liquid medium containing kanamycin and cultured overnight to prepare seed solutions. These seed solutions were then inoculated at 5% (volume fraction) into 200 mL of auto-induction medium supplemented with cadmium chloride with a final concentration of 50 mg/L. The cultures were incubated at 37 °C for 24 h, with continuous sampling every four hours to determine the OD and the adsorption rate of the strains to cadmium.
Optimal Adsorption pH and Temperature
- (1)
- Optimal adsorption temperature: The recombinant strain BL21 (FLE), auto-induced for expression, was collected after centrifugation and resuspended in Tris-HCl (pH 7.0). The bacterium with a density of 3 g/L was added to cadmium chloride with a final concentration of 50 mg/L. The strains were separately placed on a shaker at 16, 25, 37, and 45 °C and incubated for two hours at 150 rpm. Samples were collected into nitric acid-soaked centrifuge tubes and centrifuged at 12,000 rpm for two minutes. The bacterial cells were dried, digested with nitric acid, and then quantitatively analyzed for their cadmium content using cadmium detection kits.
- (2)
- Optimal adsorption pH: The recombinant strain BL21 (FLE) were collected after centrifugation and resuspended in Tris-HCl buffer at pH values of 5.0, 6.0, 7.0, 8.0, and 9.0 at the optimal temperature. The bacterial density was 3 g/L. After adding cadmium chloride with a final concentration of 50 mg/L, the strains were separately incubated at 150 r/min for two hours. The bacterial cells were collected, and a quantitative analysis was performed on the adsorbed cadmium following the above procedure. The adsorption rate A was calculated using the formula A = Ce/C0 × 100%, where C0 represents the initial concentration of cadmium chloride (in mg), and Ce denotes the cadmium ions adsorbed by the recombinant strains (in mg).
Time Adsorption Curve of Recombinant Strain
Under optimal adsorption conditions, the self-induced expression of recombinant strain BL21 (FLE) and control strain BL21 (28a) were separately placed in 50 mg/L cadmium chloride solutions and continuously cultured for 120 min on a shaker at 150 rpm. Samples were taken at 5, 10, 20, 40, 60, and 120 min time points for detection, and the adsorption rates at these time points were calculated.
Equilibrium Adsorption Capacity of Recombinant Strain to Cadmium
Under optimal adsorption conditions, the recombinant strain BL21 (FLE), auto-induced for expression, and control strain BL21 (28a) were separately placed in cadmium chloride solutions with final concentrations of 20, 40, 60, 80, 100, and 120 mg/L and continuously cultured for 120 min on a shaker at 150 r/min. Samples were taken to determine the adsorption quantity of cadmium. The typical Langmuir model was used to calculate the equilibrium enrichment amount (qm) of strains for Cd2+ using the formula q = qm Ce/(K + Ce), where Ce represents the equilibrium concentration of Cd2+ in the solution (mg/L), q is the enrichment amount of the strain for Cd2+ (mg/g), qm stands for the maximum enrichment capacity (mg/g), and K denotes the dissociation constant (mg/L). The values of qm and K can be obtained graphically with the rearranged form of the above formula: Ce/q = Ce/qm + K/qm.
2.3. Statistical Analyses
In this study, all experiments were performed in three replicates. All measurements were repeated three times, and the data were represented as the mean ± standard deviation (SD). SPSS V19.0 (IBM, New York, NY, USA) software was used to conduct analysis variance, and p < 0.05 stands for statistical significance.
3. Result and Analysis
3.1. Construction of Fusion Gene Expression Vector and Analysis of Fusion Protein Expression
The rHF and EC20 genes were enzymatically cleaved and ligated into the prokaryotic expression vector pET28a to obtain the recombinant plasmid pET28a-rHF-EC20. After sequencing verification, the plasmid was transformed into E. coli BL21 (DE3) to obtain the recombinant strain E. coli BL21 (FLE). The fusion gene rHF-EC20 has a full length of 748 bp, encoding 248 amino acids. With the N-terminal His-tag, the fusion protein has a total length of 266 amino acids, with a theoretical molecular weight of approximately 30.8 ku and an isoelectric point of approximately 4.9. After continuous cultivation of the recombinant strain E. coli BL21 (FLE) in the auto-induction medium at 37 °C for 16 h, the bacterial cells were collected using centrifugation. The cells were crushed using sonication and centrifuged to harvest the supernatant and precipitate. The recombinant protein was purified using a Ni-NTA column. The samples were separately taken for SDS-PAGE analysis. The results are shown in Figure 2. The target protein mainly exists in the supernatant, indicating the soluble expression of the recombinant protein in E. coli. The protein molecular weight is around 30 ku, consistent with the predicted theoretical value.
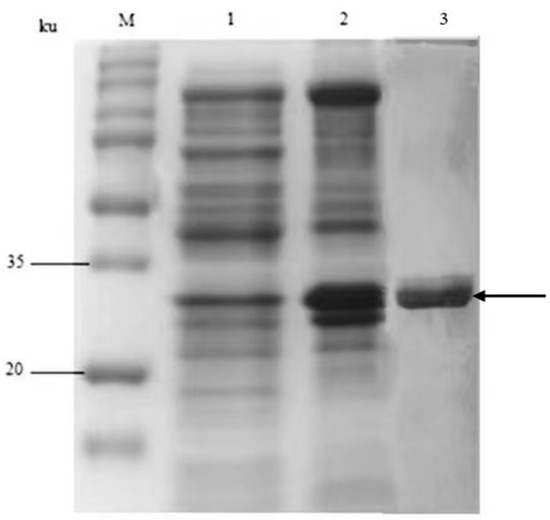
Figure 2.
SDS-PAGE analysis of the recombinant fusion protein rHF-EC20. M: Protein standard molecular weight; 1: precipitate; 2: supernatant; and 3: purified protein; the band indicated by the black arrow represents the target recombinant protein rHF-EC20.
3.2. Morphological Analysis of Fusion Protein
The purified fusion protein was negatively stained with phosphotungstic acid, and its morphology was observed using transmission electron microscopy (TEM). As shown in Figure 3, uniformly sized nanoparticles were observed under the electron microscope, implying that the fusion protein rHF-EC20 auto-assembled into nanospheres in E. coli, with a size of approximately 13 nm, consistent with the expectation.
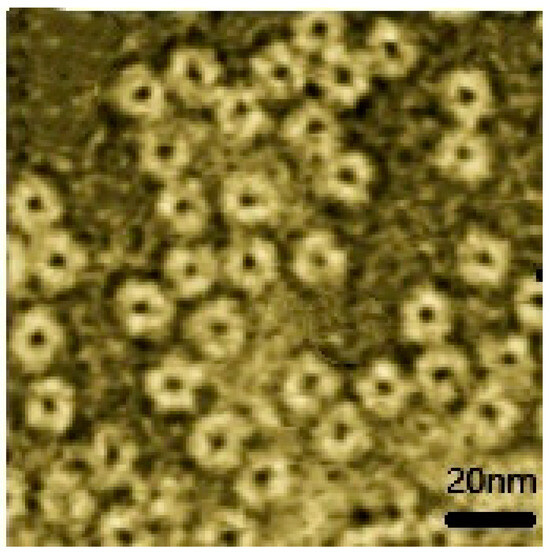
Figure 3.
TEM analysis of the recombinant fusion protein rHF-EC20.
3.3. Adsorption Capacity of Recombinant Fusion Protein rHF-EC20 to Cadmium
The purified target protein was immobilized onto Ni-NTA agarose through a His-tag to treat cadmium chloride. It was calculated that 1 μmol of the target protein rHF-EC20 (monomer) could adsorb approximately 9.2 μmol of Cd2+. Xu et al. conducted adsorption experiments on Cd2+ using the EC20 fusion protein in vitro. The results show that theoretically, one molecule of EC20 fusion protein can bind to ten molecules of Cd2+ [41]. The findings of this study agree with this, demonstrating the ability of the recombinant protein to bind Cd2+.
3.4. Adsorption Characteristics of Engineered Bacterium E. coli BL21 (FLE) to Cadmium
3.4.1. Impact of Induction Time on Cadmium Adsorption by Engineered Strain
The recombinant engineered strain E. coli BL21 (FLE) and control strain E. coli BL21 (28a) were inoculated into auto-induction media containing cadmium and continuously cultured for 24 h. The adsorption characteristics of strains for cadmium were determined throughout the entire cultivation period. The results shown in Figure 4 reveal that the growth curve of the target strain is generally consistent with that of the control strain BL21. This verifies that under the same antibiotic and heavy metal stresses, the introduction of the target gene into BL21 does not significantly affect cell growth. During the initial 12 h of cultivation, there was no evident difference in the cadmium adsorption efficiency between the target and control strains. However, between 12 to 16 h, as the cell density increased, the target strain exhibited a higher cadmium adsorption efficiency than the control strain. After continuous cultivation for 16 h, E. coli BL21 (FLE) achieved an adsorption efficiency of 78.13% for Cd2+ in the solution, and the control strain was only 62.32%. After continuous cultivation for 20 h, the target strain E. coli BL21 (FLE) reached its maximum cadmium adsorption capacity. However, after 24 h, the adsorption efficiency of cadmium by the engineered strains decreased. This may be attributed to the excretion of heavy metal ions from the cells through specialized “pumps” or transporters, such as P-type ATPase and CDF (Cation Diffusion Facilitator) family proteins [42,43,44]. When the concentration of heavy metals in the cells of microorganisms reaches their tolerance capacity, the optimal detoxification method is to expel heavy metals from the cells.
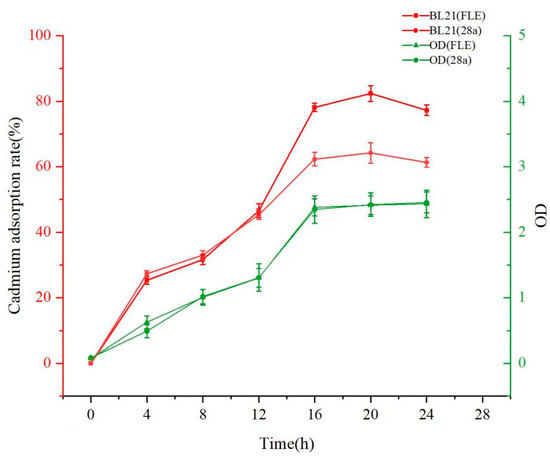
Figure 4.
Curves of the growth of engineered strains and their adsorption to cadmium.
3.4.2. Optimal Adsorption Temperature of Engineered Bacterium BL21 (FLE)
The determination results of the optimal adsorption temperature of the recombinant engineered strain E. coli BL21 (FLE) for Cd2+ are shown in Figure 5. The optimal adsorption temperature for Cd2+ by the recombinant strain is 37 °C. After continuous adsorption for two hours, the adsorption rate reached 82.4%. Conversely, low temperatures are unfavorable for E. coli adsorption of Cd2+. After continuous adsorption at 16 °C for two hours, the adsorption rate was merely 26.3%. By comparison, adsorption experiments at 45 °C result in a decline in the adsorption efficiency. The optimal growth temperature for E. coli is 37 °C, Prior research delineates heavy metal adsorption by microorganisms into stages encompassing heavy metal ion adsorption onto cell membrane surfaces, transmembrane transport, and intracellular detoxification [45]. A range of transport proteins and enzymes orchestrates the intricate process of heavy metal transport. Existing research indicates that temperature substantially influences gene expression and protein synthesis within cells. Microbial activities surge within an appropriate temperature range, enhancing both microbial metabolism and enzyme activity, thereby expediting the bioremediation process of heavy metals [46,47]. Conversely, low temperatures hamper the rapid synthesis of functional proteins and can impair the permeability of cell membranes. Additionally, higher environmental temperatures may disrupt the activities of metal transporters and related enzymes, potentially impeding the transport and adsorption of Cd2+ [48].
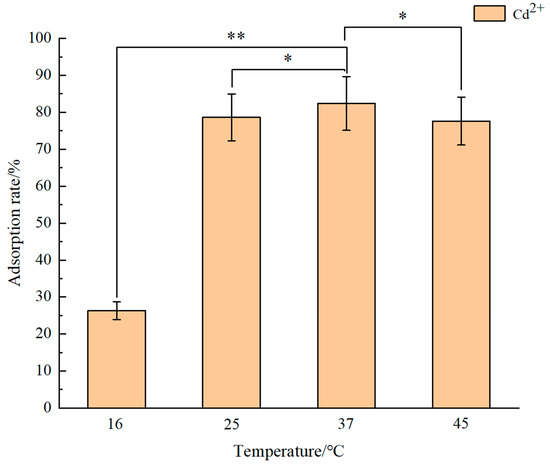
Figure 5.
The optimal adsorption temperature of the engineered bacterium BL21 (FLE). Different asterisks mean significant difference between treatments (* p < 0.05, ** p < 0.01).
3.4.3. Optimal Adsorption pH of Engineered Bacterium E. coli BL21 (FLE)
The recombinant strain was subjected to different pH buffer solutions to determine its adsorption efficiency for Cd2+. The results show that the recombinant strain E. coli BL21 (FLE) exhibits a high adsorption efficiency for Cd2+ in the pH range of 5–7. Specifically, treatment in a pH 7.0 solution for two hours results in the highest adsorption efficiency for Cd2+, reaching 82.4%. In contrast, under pH 9.0 environmental conditions for two hours, the adsorption efficiency is only 69.2% (Figure 6). The pH exerts considerable influence on the redox and solubility of heavy metals. Under neutral and acidic conditions (pH < 7), negatively charged functional groups on the microbial cell surface, such as hydroxyl, phosphate, carboxyl, and amide groups, can readily bind with Cd2+. In contrast, when the pH levels exceed 8, Cd(OH)2 precipitates out of wastewater. Cadmium concentrations decrease rapidly, and the cellular uptake of Cd2+ also drops [7].
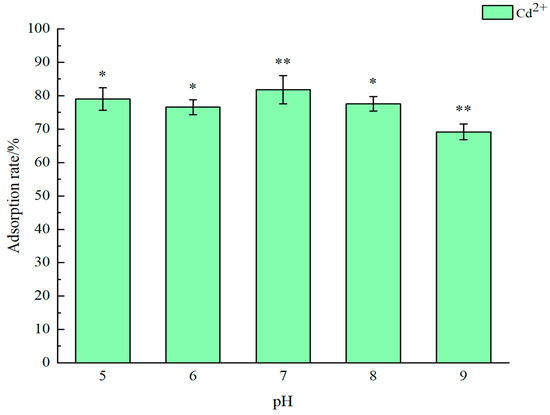
Figure 6.
The optimum adsorption pH of engineered bacterium BL21 (FLE). Different asterisks mean significant difference between treatments (* p < 0.05, ** p < 0.01).
3.4.4. Evaluation of Adsorption Efficiency of Recombinant Strain to Cd2+
Under the optimal adsorption conditions, the adsorption efficiencies of the recombinant genetically engineered strain BL21 (FLE) and control strain BL21 (28a) were determined at different time points (Figure 7a). Throughout the entire time process, the recombinant strain BL21 (FLE) consistently demonstrates a higher adsorption efficiency for Cd2+ compared to the control strain. The recombinant strain BL21 (FLE), expressing nano-chelating peptides, achieves an adsorption rate of 80% for Cd2+ at 60 min, whereas the control strain BL21 (28a) only reaches 62%, resulting in an approximately 18% increase in the Cd2+ enrichment efficiency. These results indicate that the overexpression of multivalent phytochelatins in E. coli cells effectively enhances the adsorption efficiency and tolerance of the strain to Cd2+, thereby improving the adsorption efficiency of heavy metals. As shown in Figure 7b, the results of the kinetic study regarding BL21 (FLE) reveal that during the initial adsorption stage (0–30 min), E. coli exhibits a swift adsorption rate for Cd2+. At this stage, heavy metal ions adhere to cell membrane surfaces primarily through processes like ion exchange and complexation with chemical groups in biomolecules on the membrane surfaces, featured by rapid and large-scale increases. Subsequently, the adsorption rate gradually decelerates, reaching equilibrium at 60 min. Some heavy metals bound to the membrane surfaces are transported into the cells via metal transporters. Within the cells, these metals interact with heavy metal chelating proteins and form precipitates within the cytoplasm, thereby mitigating the impact of soluble heavy metal ions on cells. This phase represents the rate-limiting step throughout the entire adsorption process [45].
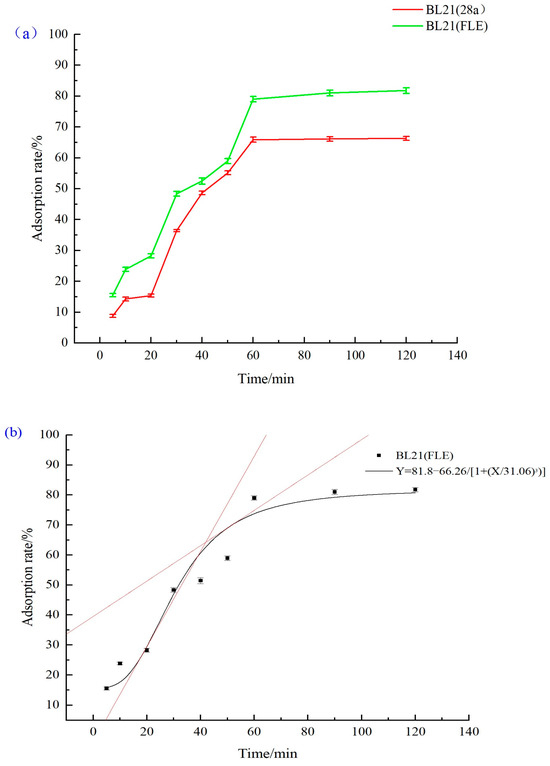
Figure 7.
(a) The temporal profile curves of cadmium adsorption by the strains; (b) the kinetic profile curve of cadmium adsorption by the strain BL21 (FLE).
3.4.5. Equilibrium Adsorption Capacity of Recombinant Strain to Cd2+
This experiment analyzed the enrichment behavior of the recombinant strain BL21 (FLE) and the control strain BL21 (28a) at different Cd2+ concentrations. As shown in Figure 8, the maximum enrichment of Cd2+ by the recombinant strain BL21 (FLE) and the control strain BL21 (28a) can be calculated based on the Langmuir model formula. The equilibrium enrichment of the control strain BL21 (28a) is 9.27 mg/g, and that of the recombinant strain BL21 (FLE) is 12.62 mg/g. Meanwhile, the equilibrium curve shows that in solutions with high concentrations of Cd2+, the recombinant genetically engineered bacterium presents a higher resistance and adsorption capacity to cadmium. However, due to the absence of the secretion mechanism, proteins expressed in E. coli cells mainly reside in the cytoplasm. Therefore, the genetically engineered bacterium in this study aimed to promote heavy metal adsorption by elevating the expression levels of intracellular heavy metal chelating peptides. However, the results indicate that compared with the control strain, the engineered strain merely augments the cadmium adsorption efficiency by approximately 18%. This may stem from the ineffective transport of heavy metal ions into cells.
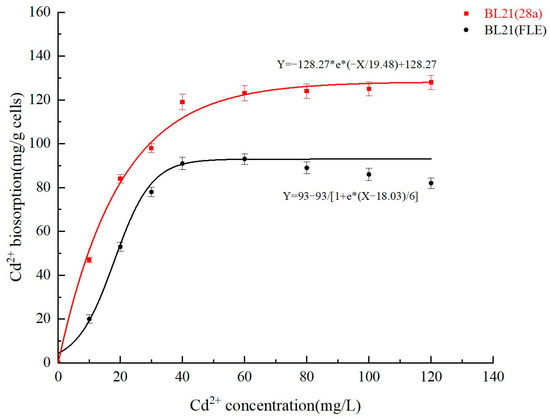
Figure 8.
The equilibrium curve of cadmium adsorption by strains.
4. Conclusions
This work demonstrates the feasibility of employing genetically engineered E. coli cell factories to reinforce the efficient bioaccumulation of heavy metal cadmium via the overexpression of a fusion protein, rHF-EC. Moreover, the recombinant proteins can self-assemble into multivalent nanochelators with a size of about 13 nm within cells, effectively adsorbing heavy metal cadmium both intracellularly and extracellularly. The target protein rHF-EC20 (monomer) could adsorb approximately 9.2 μmol of Cd2+ in vitro. The adsorption capacity varies with temperature and pH. The engineered strain BL21 (FLE) exhibits a superior performance in cadmium biosorption at pH 7.0 and 37 °C. Meanwhile, a notable 18% increase in cadmium accumulation implies that the strain that produces rHF-ECs maintains significant advantages over the control host cells.
Considering the findings of the above research, promoting heavy metal adsorption by elevating the expression levels of intracellular heavy metal chelating peptides emerges as a promising strategy. Nonetheless, this approach faces limitations due to the ineffective transport of heavy metal ions into cells. Future research can focus on tethering chelating peptides to the surface of cell membranes and co-expressing cadmium transporters alongside this nanochelator within cells to further augment the adsorption and transport of heavy metals [49]. Such advancements hold the potential to expand the utility of genetically engineered E. coli in environmental pollution control [50].
Author Contributions
G.L. and L.Y. conceived the study. L.T., D.W., Y.L., X.H. and M.W. performed the experiments, L.Y. wrote the draft of the manuscript. X.S. and G.L. critically reviewed the full manuscript content. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Gansu Province Key R&D Plan under Grant No. 21YF5FA129; Gansu Province Key R&D Plan under Grant No. 22YF7FG188; Gansu Province Natural Science Foundation (No. 22JR11RG222) and Hexi University Research Initiation Project (No. KYQD2020018).
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pratush, A.; Kumar, A.; Hu, Z. Adverse effect of heavy metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: A review. Int. Microbiol. 2018, 21, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Sun, J.; Sun, M.; Shi, X.; He, Z.; Gong, Z.; Yao, M.; Sun, Y.; Xu, X.; Sui, H. Research progress on cadmium toxicity and prevention measures. J. Toxicol. 2022, 36, 517–520. [Google Scholar]
- Barakat, M.A. New trends in removing heavy metals from industrial wastewater. Arab. J. Chem. 2011, 4, 361–377. [Google Scholar] [CrossRef]
- Al-Rashdi, B.A.M.; Johnson, D.J.; Hilal, N. Removal of heavy metal ions by nano-filtration. Desalination 2013, 315, 2–17. [Google Scholar] [CrossRef]
- Priyanka, R.; Sharma, A.C.; Sunil, K.S.; Geng, L.; Nasim, A.; Darren, M.; Benjamin, S.H. Nanocellulose from Spinifex as an Effective Adsorbent to Remove Cadmium (II) from Water. ACS Sustain. Chem. Eng. 2018, 6, 3279–3290. [Google Scholar]
- Guo, Z.; Zhang, X.; Kang, Y.; Zhang, J. Biomass-Derived Carbon Sorbents for Cd (II) Removal: Activation and Adsorption Mechanism. ACS Sustain. Chem. Eng. 2017, 5, 4103–4109. [Google Scholar] [CrossRef]
- Khan, S.A.; Siddiqui, M.F.; Khan, T.A. Ultrasonic-assisted synthesis of polyacrylamide/bentonite hydrogel nanocomposite for the sequestration of lead and cadmium from aqueous phase: Equilibrium, kinetics and thermodynamic studies. Ultrason. Sonochem. 2020, 60, 104761. [Google Scholar] [CrossRef] [PubMed]
- Muya, F.N.; Sunday, C.E.; Baker, P.; Iwuoha, E. Environmental remediation of heavy metal ions from aqueous solution through hydrogel adsorption: A critical review. Water Sci. Technol. 2016, 73, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xiao, C.; Chi, R. Remediation of soil cadmium pollution by biomineralization using microbial-induced precipitation: A review. World J. Microbiol. Biotechnol. 2021, 37, 208–213. [Google Scholar] [CrossRef]
- Saumya, A.; Ankur, S.; Vipin, K. Recent advancements in cadmium-microbe interactive relations and their application for environmental remediation: A mechanistic overview. Environ. Sci. Pollut. Res. Int. 2023, 30, 17009–17038. [Google Scholar]
- Alabssawy, A.N.; Hashem, A.H. Bioremediation of hazardous heavy metals by marine microorganisms: A recent review. Arch. Microbiol. 2024, 206, 103. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Samanta, S.; Pandit, S.; Naaz, T.; Banerjee, S.; Rawat, J.M.; Chaubey, K.K.; Saha, R.P. An Overview of Bacteria-Mediated Heavy Metal Bioremediation Strategies. Appl. Biochem. Biotechnol. 2024, 196, 1712–1751. [Google Scholar] [CrossRef] [PubMed]
- Joseph, L.; Jun, B.; Flora, J.; Park, C.; Yoon, Y. Removal of heavy metals from water sources in the developing world using low-cost materials: A review. Chemosphere 2019, 229, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zeng, G.; Niu, Q.; Liu, Y.; Zhou, L.; Jiang, L.; Tan, X.; Xu, P.; Zhang, C.; Cheng, M. Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: A mini review. Bioresour. Technol. 2017, 224, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Znad, H.; Awual, M.R.; Martini, S. The Utilization of Algae and Seaweed Biomass for Bioremediation of Heavy Metal-Contaminated Wastewater. Molecules 2022, 27, 1275. [Google Scholar] [CrossRef] [PubMed]
- Salama, E.S.; Roh, H.S.; Dev, S.; Khan, M.A.; Abou-Shanab, R.A.I.; Chang, S.W.; Jeon, B.H. Algae as a green technology for heavy metals removal from various wastewater. World J. Microbiol. Biotechnol. 2019, 35, 75. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, N.; Chelliapan, S.; Kamyab, H.; Thirugnana, S.; Othman, N.; Nasri, N.S. Treatment of Wastewater Using Seaweed: A Review. Int. J. Environ. Res. Public Health 2018, 15, 2851. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Tao, H. Cell surface engineering of microorganisms towards adsorption of heavy metals. Crit. Rev. Microbiol. 2015, 41, 140–149. [Google Scholar] [CrossRef]
- Hansda, A.; Kumar, V.; Anshumali, A. comparative review towards potential of microbial cells for heavy metal removal with emphasis on biosorption and bioaccumulation. World J. Microbiol. Biotechnol. 2016, 32, 170. [Google Scholar] [CrossRef]
- Saravanan, A.; Kumar, P.S.; Ramesh, B.; Srinivasan, S. Removal of toxic heavy metals using genetically engineered microbes: Molecular tools, risk assessment and management strategies. Chemosphere 2022, 298, 134341. [Google Scholar] [CrossRef]
- Thévenod, F.; Fels, J.; Lee, W.K.; Zarbock, R. Channels.transporters and receptors for cadmium and cadmium complexes in eukaryotic cells: Myths and facts. Biometals 2019, 32, 469–489. [Google Scholar] [CrossRef]
- Chang, J.D.; Huang, S.; Konishi, N. Overexpression of the manganese/cadmium transporter OsNRAMP5 reduces cadmium accumulation in rice grain. J. Exp. Bot. 2020, 71, 5705–5715. [Google Scholar] [CrossRef] [PubMed]
- Coyle, P.; Philcox, J.C.; Carey, L.C.; Rofe, A.M. Metallothionein: The multipurpose protein. Cell. Mol. Life Sci. 2002, 59, 627–647. [Google Scholar] [CrossRef]
- Esser-Kahn, A.P.; Iavarone, A.T.; Francis, M.B. Metallothionein-cross-linked hydrogels for the selective removal of heavy metals from water. J. Am. Chem. Soc. 2008, 130, 15820–15822. [Google Scholar] [CrossRef] [PubMed]
- Okasha, H.; Abdel-Motleb, A.; Abdel-Wareth, M.T.A. Metallothionein expression in Aspergillus exposed to environmentally relevant concentrations of heavy metals at different pH levels. Environ. Sci. Pollut. Res. Int. 2021, 28, 49936–49948. [Google Scholar] [CrossRef]
- Nordberg, M.; Nordberg, G.F. Metallothionein and Cadmium Toxicology-Historical Review and Commentary. Biomolecules 2022, 12, 360. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhao, X.; Deng, X. Bioaccumulation of heavy metal cadmium in wastewater by genetically engineered bacteria. Technol. Water Treat. 2006, 32, 26–29. [Google Scholar]
- Bae, W.; Chen, W.; Mulchandania, A.; Mehra, R.K. Enhanced bioaccu-mulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 2000, 70, 518–524. [Google Scholar] [CrossRef]
- Bae, W.; Mulchandania, A.; Chen, W. Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J. Org. Chem. 2002, 88, 223–227. [Google Scholar] [CrossRef]
- Yu, Y.; Shi, K.; Li, X.; Luo, X.; Wang, M.; Li, L.; Wang, G.; Li, M. Reducing cadmium in rice using metallothionein surface-engineered bacteria WH16-1-MT. Environ. Res. 2022, 203, 111801. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, B.; Yu, Q. Genetic engineering-facilitated co-assembly of synthetic bacterial cells and magnetic nanoparticles for efficient heavy metal removal. ACS Appl. Mater. Interfaces 2020, 12, 22948–22957. [Google Scholar] [CrossRef] [PubMed]
- Theil, E.C. Ferritin: The protein nanocage and iron biomineral in health and in disease. Inorg. Chem. 2013, 52, 12223–12233. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Chakrabarti, P. Self-Assembly of Ferritin: Structure, Biological Function and Potential Applications in Nanotechnology. Adv. Exp. Med. Biol. 2019, 1174, 313–329. [Google Scholar] [PubMed]
- Wu, J.; Li, Y.; Wu, H.; Zhang, H.; Sha, X.; Ma, J.; Yang, R. The application of ferritin in transporting and binding diverse metal ions. Food Chem. 2024, 439, 138132. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wu, Z.; Yuan, X. Overexpression of human-derived soluble transferrin receptor sTfR antigen and preparation and application of polyclonal antibodies. J. Biol. 2023, 40, 111–115. [Google Scholar]
- Zhao, X.; Zhou, Y.; Zhang, Y.; Zhang, Y. Ferritin: Significance in viral infections. Rev. Med. Virol. 2024, 34, e2531. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Tyrer, M.; Hills, C.D.; Yang, X.M.; Carey, P. Immobilisation of heavy metal in cement-based solidification/stabilisation: A review. Waste Manag. 2009, 29, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Malviya, R.; Chaudhary, R. Factors affecting hazardous waste solidification/stabilization: A review. J. Hazard. Mater. 2006, 137, 267–276. [Google Scholar] [CrossRef]
- Malviya, R.; Chaudhary, R. Leaching behavior and immobilization of heavy metals in solidified/stabilized products. J. Hazard. Mater. 2006, 137, 207–217. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Xu, Z.; Bae, W.; Mulchandani, A.; Mehra, R.K.; Chen, W. Heavy metal removal by novel CBD-EC20 sorbents immobilized on cellulose. Biomacromolecules 2002, 3, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Drees, S.L.; Beyer, D.F.; Lenders-Lomscher, C.; Lübben, M. Distinct functions of serial metal-binding domains in the Escherichia coli P1B-ATPase CopA. Mol. Microbiol. 2015, 97, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Kolaj-Robin, O.; Russell, D.; Hayes, K.A.; Pembroke, J.T.; Soulimane, T. Cation Diffusion Facilitator family: Structure and function. FEBS Lett. 2015, 589, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Montanini, B.; Blaudez, D.; Jeandroz, S.; Sanders, D.; Chalot, M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: Improved signature and prediction of substrate specificity. BMC Genom. 2007, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Avanbakht, V.; Alavi, S.A.; Zilouei, H. Mechanisms of heavy metal removal using microorganisms as biosorbent. Water Sci. Technol. 2014, 69, 1775–1787. [Google Scholar] [CrossRef] [PubMed]
- Kuriki, Y. Response to temperature shifts of expression of the amp gene on pBR322 in Escherichia coli K-12. J. Bacteriol. 1987, 169, 2294–2297. [Google Scholar] [CrossRef] [PubMed]
- Vortuba, J.; Pazlarova, J.; Dvorakova, M.; Vachova, L.; Strnadova, M.; Kucerova, H.; Vinter, V.; Zourabian, R.; Chaloupka, J. External factors involved in the regulation of synthesis of an extracellular proteinase in Bacillus megaterium: Effect of temperature. Appl. Microbiol. Biotechnol. 1991, 35, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J. Effects of temperature on cell membranes. Symp. Soc. Exp. Biol. 1988, 42, 237–258. [Google Scholar]
- Leong, Y.K.; Chang, J.S. Bioremediation of heavy metals using microalgae: Recent advances and mechanisms. Bioresour. Technol. 2020, 303, 122886. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C. Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 2009, 27, 195–226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).