Abstract
The larvae of the genus Chironomus are a common object for hydrobiological studies, as well as a model object for cytogenetics. Morphologically, the species are very similar. One of these species or species complex is Chironomus “annularius”, which has a Holarctic distribution. It has chromosomal banding sequences characteristic of Nearctic and Palearctic populations. Using an integrated method that included morphology, cytogenetics, and molecular genetics, we analyzed populations from Russia, Mongolia, and Armenia. We found through cytogenetics and larval morphology that the populations have high similarity. Molecular genetic studies have shown significant differences between the populations. The genetic distances between the populations, in some cases, exceed the interspecific threshold of 3%, and are 6.5%. In the South Caucasian population (Lake Sevan), a chromosomal banding sequence, h’annD3, that was previously observed only in North America, was found for the first time. The larvae from Lake Sevan have large genetic distances from others, and are morphologically similar to the species Chironomus markosjani Shilova 1983, described from this lake without comparison with Ch. annularius nor an exact description of the karyotype. The sequences of the COI genes from Montenegro (Lake Skadar) and West Siberia (Novosibirsk) found in GenBank may belong to a new undescribed species, or a species not represented in the database. Thus, the analyzed data on Chironomus “annularius” support the presence of the complex of homosequential species under this name.
1. Introduction
Many studies have been devoted to the investigation of the species Chironomus annularius Meigen 1818; however, as a result of a comprehensive analysis, it was shown that this name can unite several species []. The most complete descriptions of the imago morphology and karyotype appropriate to Chironomus annularius Meigen 1818 were presented in several studies [,,]. Due to several discrepancies between the published descriptions, it was recommended to indicate the source used for identification by applying a name format analogous to, for example, Chironomus «annularius» sensu Strenzke (1959) []. The species has a Holarctic distribution [,,,], with the type locality situated in West Germany [,]. The reliable “species” records known from Russia are the following: Yaroslavl, Saratov, Chelyabinsk, Novosibirsk, Irkutsk Oblast’s, Ural, Western Siberia, Altai, Tyva, and Sakha (Yakutia) Republics; Kyrgyzstan (Issyk-Kul Lake); Kazakhstan; Mongolia; North America: USA and Canada [,,].
The karyotype of Chironomus annularius was described from the German populations []. The chromosomal arms A, E, and F were mapped by Keyl [] and Kiknadze et al. [], and arms C and D by Kiknadze et al. [,]. Chromosomal polymorphism in C. «annularius» from the Palearctic was studied later using a non-standard system of chromosome mapping [,,]. The karyotype and chromosomal polymorphism of C. “annularius” has also been studied from Nearctic populations [,,,]. In C. “annularius” from Lake Khubsugul in North Mongolia, the Nearctic banding sequence h’annG3 was found in a homozygous state []. Recently, the karyotype and chromosomal polymorphism of C. “annularius” were studied in detail in South Caucasian populations, and the banding pool was enriched with new sequences []. Researchers of the cytogenetic structure of populations of C. “annularius” noted a high level of chromosomal polymorphism and the presence of endemic sequences in arms A, B, and D, which can affect the phenotypic variability of individuals from different populations [,,].
We noticed C. “annularius” in the list of species in studied published recently that were dedicated to the investigation of chironomids in Europe and China via barcoding [,,,]. Many nucleotide sequences of this species have also been found in genetic information databases (GenBank, BOLD); unfortunately, most of them are very short and are not suitable for subsequent analysis (access on 18 May 2023). In addition to modern techniques, classical ones are also used to improve the accuracy of species identification (for example, morphological); however, the authors of the articles rightly note the high complexity of this approach []. The Chironomus species have giant polytene chromosomes with which we usually can accurately identify their species [,,,]. We already know that for the best results in identifying the species of chironomids, it is necessary to use a comprehensive approach that includes morphological, cytogenetic, and molecular genetic analyses [,,].
In this study, we are trying to understand the structure of a complex species of Chironomus “annularius” using specimens from different populations and databases using a comprehensive approach that involves morphological, cytogenetic, and molecular genetics.
2. Materials and Methods
We used larvae of Chironomus “annularius” of the fourth instar, which were collected in Russia, Mongolia, and Armenia. All of the larvae were fixed in 96% ethanol.
One larva was found in a pond in Pivovariha village (52.265833, 104.452778) near Irkutsk city, Irkutsk region, Russia, in October 2021 (henceforth, the shorter name “Irkutsk” will be used for convenience). The depth at the sample place was 0.3–0.5 m, and the bottom sediments were silted sand on stones (rocks). Floodplain–valley willow sedge-grass landscapes were found. Swamping of the pond was observed. The composition of the aquatic core of the flora includes Rumex aquaticus L. (Polygonaceae), Alisma plantago-aquatica L. (Alismataceae), Sparganium emersum Rehmann (Sparganiaceae), Schoenoplectus lacustris (L.), Palla sp. (Cyperaceae), and Beckmannia syzigachne (Poaceae).
Two larvae were found in an unnamed pond in Dubki village (57.525207, 39.728620), near Yaroslavl City, Yaroslavl region, Russia, in September 2019 (shorter name “Yaroslavl”). The depth at the sample place was 0.5–1.5 m, and the bottom sediments were black silted sand. Willow sedge-grass thickets, natural processes of pond overgrowth, and swamping of the land were observed. The composition of the aquatic core of the flora includes the following: Phragmites australis (Cav.) Trin. ex Steud. (Poaceae), Ceratophyllum demersum L. (Ceratophyllaceae), Lemna trisulca L. (Lemnaceae), Hydrocharis morsus-ranae L. (Hydrocharitaceae), Elodea canadensis Michx. (Hydrocharitaceae), and Salix sp. (Salicaceae).
Two larvae were found in Dörgön Reservoir, Khovd Province, Dörgön District, Mongolia, in August 2018. One of them was in the backwater zone (48.311917, 92.788139) (shorter name “MongoliaBW”). The depth at the sample place was 2 m, the bottom sediments were silted sand. leg. A. Prokin. Another one was found in the dam area (48.324639, 92.807111) (shorter name “MongoliaDam”). The depth of the sample location was 2 m, and the bottom sediments were silted gravel. leg. A. Prokin. Dörgön Reservoir is situated in the Great Lakes Basin desert steppe ecoregion. The shores are overgrowing by reed beds (Phragmites australis (Cav.) Trin. ex Steud. (Poaceae), and in the backwater zone by diverse aquatic flora that were not estimated.
Four larvae were found in Sevan Lake, Gegharkunik Province, Armenia. Two of them were in the east part of the “Noratus” section (40.385583, 45.234778) (shorter name “SevanN”). The depth at the sample place was 10 m, and the bottom sediments were black silt. leg. A. Prokin. Two larvae were found in the east part, the “Tsapatakh (Babadzhan)” section (40.439111, 45.416472) (shorter name “SevanB”). The depth of the sample location was 20 m, and the bottom sediments were black silt. leg. A. Prokin. Lake Sevan is situated in the Eastern Anatolian montane steppe ecoregion. In the depths of 10–20 m, only two species of aquatic plants have been registered: Chara globularis Thuiller and Chara papillosa Kützing (both Characeae).
All of the larvae were fixed in 96% ethanol. We determined the age of the larvae using a standard method []. All of the head capsules of the studied larvae were placed on a slide in a Fora-Berlese solution; the morphological terminology offered by Sæther [] was used. Karyotype analysis was performed on all collected larvae using the aceto-orcein method []. For the microscopy analysis, a light microscope (Micromed-6 C. LOMO, St-. Petersburg, Russia) was used with an objective of ×100 and a digital camera (ToupCam 5.1., ToupTek Photonics, Hangzhou, China). For chromosome banding sequences identification, cytomaps were used [,,,].
The studied material was deposited in the collection of Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences (IBIW), Borok, Russia.
Seven larvae (Irkutsk—1 larva, Yaroslavl—1, MongoliaBW—1, MongoliaDam—1, SevanN—2 larvae, SevanB—1) were studied karyologically and morphologically, and were used for DNA extraction with the «M-sorb-OOM» (Sintol, Moscow) kit with magnetic particles according to the manufacturer’s protocol. For amplification, the COI (cytochrome oxidase subunit I) primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3’) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were used (Eurogen, Moscow) (Folmer et al. 1994). An amplification reaction was carried out in 25 μL of reaction mixture (1x buffer, 1.5 μM MgCl2, 0.5 mM of each primer, 0.2 μM dNTP of each nucleotide, 17.55 μL of deionized water, 1 μL of template DNA, and 1 unit of Taq-polymerase (Evrogen, Moscow, Russia)). PCR analysis was performed at 94 °C (3 min), followed by 30 cycles at 94 °C (15 s), 50 °C (45 s), 72 °C (60 s), and a final at 72 °C (8 min). For the PCR products, the visualization involved a 1% agarose gel and purification by ethanol and ammonium acetate (3 M). Both of the strands were sequenced on an Applied Biosystems 3500 DNA sequencer (Thermo Scientific, Waltham, MA, USA), following the manufacturer’s instructions.
For alignment of the COI nucleotide sequences, we used MUSCLE in the MEGA6 software []. To calculate pairwise genetic distances (Kimura 2-parameter) with codon position preferences: 1st, 2nd, 3rd and noncoding sites the MEGA6 software was used. []. The program MrBayes v.3.2.6 was used for the Bayesian analysis [,], with previously suggested settings [,] of 1,000,000 iterations and 1000 iterations of burn-in, nst = 6 (GTR + I + G). The phylogenetic trees resulting from the Bayesian inference analyses were visualized and edited using FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 25 August 2023). For the estimation of the number of haplotypes, we used DNA SP v.6 software []; to create a network of haplotypes, we used PopArt 1.7 software with the median-joining algorithm [].
In addition, twenty-five COI gene sequences of species of the genus Chironomus from “GenBank” and “Barcode of Life Data Systems” (BOLD) were analyzed. The accession numbers of the used sequences in GenBank and BOLD are the following: Chironomus “annularius” (MZ626747, MZ657059, BSCHI604-17, AF192189, BSCHI129-17, XJDQD1011-18, OP927527, OP927683, XJDQD1026-18, JDQD1025-18, MT534990, MT535187, MT535143), C. bernensis (Wülker and Klötzli, 1973) (AF192188), C. commutatus (Keyl, 1960) (AF192187), C. cingulatus (Meigen, 1830) (AF192191), C. dilutus (Shobanov, Kiknadze, and Butler, 1999) (KF278335), C. melanotus (Keyl, 1961) (OL546775), C. muratensis Ryser and Scholl, 1983 (AF192194, KC250748), C. plumosus (Linnaeus, 1758) (KF278217), C. pallidivittatus (Malloch, 1915) (AF110164), C. piger (Strenzke, 1959) (AF192202), and C. riparius (Meigen, 1804) (KR756187). In the phylogenetic analysis, the COI gene sequence of Drosophila melanogaster (Meigen, 1830) (HQ551913), was used as an outgroup.
In this study, we used an integrated method including larval morphology, cytogenetics, and molecular genetics, with the attribution of karyotype and COI gene sequences from Germany as a reference for Chironomus annularius Meigen 1818, which was closest to the type locality.
3. Results
3.1. Morphological Characters of C. “annularius”
The morphological characteristics of the larvae (highly chitinized structures of the head capsule and the structure of tubules on the last segments of the body) from all of the studied places satisfied the description of C. annularius [,,]. In the mentum structure of the larvae from Lake Sevan, the fourth lateral tooth is slightly shorter than the neighboring ones; on the eighth body segment, there are short ventral tubules with rounded ends.
The investigation of paratypes of C. markosjani Shilova, 1983 from the collection of IBIW also showed its conspecifics with C. annularius, with some differences in line drawings in the original description [] resulting from insufficient quality. Images of the details of C. “annularius” larvae are presented in Appendix A. The mentum of one larva was damaged during specimen preparation (Appendix A. Specimen: Armenia SevanB (OR342387)).
3.2. Karyotype of C. “annularius” from All Studied Populations
The chromosome set of the species is 2n = 8. The chromosomal arm combination AB, CD, EF, and G corresponds to the Chironomus “thummi” cytocomplex. The chromosomes are AB and CD—metacentric, EF—submetacentric, and G—telocentric (Figure 1). The arm G homologues are not paired in the Palearctic, and are paired in the Nearctic populations. Four nucleoli (N) are clearly visible: one nucleolus in arm C, two in arm E, and one in arm G. In addition to the permanent nucleoli, there is a fluctuating nucleus at arm A (region 2d–3a), which can be found in most larvae of previously studied populations in a homo- or heterozygous state [].
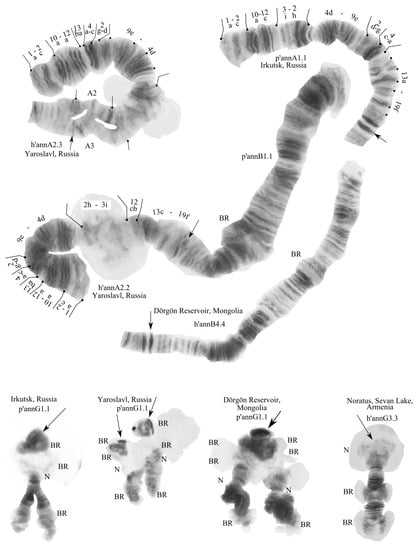
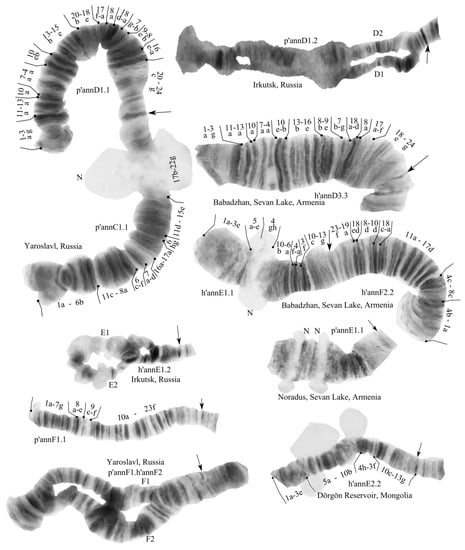
Figure 1.
Chromosomes of Chironomus “annularius” from Russia, Mongolia, and Armenia. Arrows indicate centromeric bands; p’annA1.1, h’annA2.3, etc., genotypic combinations of banding sequences in chromosome arms; N, nucleolus; and prefix p’—Palearctic sequences, h’—Holarctic sequences; BR, Balbiani rings. The inscriptions on the chromosome pictures, “Irkutsk, Russia”, etc., are the designations of the collection sites.
We found 16 banding sequences (Table 1) that form five banding sequence (zygotic) combinations.

Table 1.
Chromosomal banding sequences of C. “annularius” from the studied populations.
One zygotic combination was found in Irkutsk: annA1.1.B1.2.C1.1.D1.2.E1.2.F2.2.G1.1.
Two different zygotic combinations were found in Yaroslavl: annA2.2.B1.1.C1.1.D1.1.E1.1.F.1.2.G1.1 and annA2.3.B1.1.C1.1.D1.1.E1.1.F1.1.G1.1. One zygotic combination was found in both Mongolian populations: annA1.1.B4.4.C1.1.D2.2.E2.2.F1.1.G1.1. One zygotic combination was found in all larvae from the Armenian population: annA2.2.B4.4.C1.1.D3.3.E1.1.F2.2.G3.3.
All of the banding sequences were described earlier [,,,]. Some of them were previously found only in the Nearctic populations of C. “annularius”, and have some differences from the Palearctic karyotypes.
Arm A—the three banding sequences found were the following: p’annA1 1a-2c 10a-12c 3i-2h 4d-9e 2d-g 4c-a 13a-19f C, h’annA2 1a-2c 10a-12a 13ba 4a-c 2g-d 9e-4d 2h-3i 12cb 13c-19f C, and p’annA3 1a-2c 10a-12c 3i-2h 4d-9e 2d-g 4c-a 13a-16c 19d-16d 19ef C. Sequence p’annA3 was found only in the heterozygous form with h’annA2. Table 1.
Arm B—three banding sequences were found: p’annB1, h’annB2 and p’annB4; all of them are not mapped.
Arm C—in all studied populations was found only one banding sequence: p’annC1 1a-6b 11c-8a 6c-f 7a-d 16a-17a 6gh 11d-15e 17b-22g C.
Arm D—three banding sequences were found: p’annD1 1a-3g 11a-13a 10a 7a-4a 10e-b 13b-15e 20b-17a 8a 18d-a 7g-b 9e-8b 16e-a 20c-24g C, p’annD2 1a-3g 11a-16e 8b-10e 4a-7g 18a-d 8a 17a-f 18e-24g C, and h’annD3 1a-3g 11a-13a 10a 7a-4a 10e-b 13b-16e 8b-9e 7b-g 18a-d 8a 17a-f 18e-24g C. D3 is a fixed Nearctic banding sequence.
Arm E—two banding sequences were found: h’annE1 1a-3e 5a-e 4gh 10b-6a 4f-3f 10c-13g C and h’annE2 1a-3e 5a-10b 4h-3f 10c-13g C. The morphological features of E arm in larvae from the Sevan populations are more similar to the Nearctic populations by the number of nucleoli (Figure 1).
Arm F—two banding sequences were found: p’annF1 1a-7g 8a-e 9c-f 10a-23f C and h’annF2 1a-4b 8c-4c 17d-11a 18a-c 10d-8d 18de 19a-23f C. The mapping of arm F was conducted according to [].
Arm G—two banding sequences were found: p’annG1 and h’annG3.
3.3. COI Gene Sequences and Phylogenetic Analysis of C. “annularius” from All Studied Populations
All of the obtained COI gene sequences of C. “annularius” from all studied populations were deposited in GenBank with the following accession numbers:
Irkutsk—GenBank accession number OR342379, length—620 bp.
Yaroslavl—GenBank accession number OR342373, length—378 bp.
MongoliaBW—GenBank accession number OR342375, length—674 bp.
MongoliaDam—GenBank accession number OR342376, length—613 bp.
SevanN—GenBank accession number OR342381, length—613 bp.
SevanN—GenBank accession number OR342383, length—613 bp.
SevanB—GenBank accession number OR342387. length—613 bp.
Percentage of nucleotides A: 26; T: 36; G: 18; C: 20.
3.3.1. Genetic Distances between Chironomus “annularius” Obtained with K2P
In the GenBank database, only one sample of C. annularius (AF192189) from the vicinity of Novosibirsk city was identified via karyology []. The results were unexpected. The minimal distance of 3.8% was between COI gene sequences from the following samples: Novosibirsk (AF192189), China (XJDQD1026-18, XJDQD1025-18), Sweden (BSCHI129-17), and Germany (OP927683, OP927527) (Table 2). The distance between sequences from Novosibirsk (AF192189), Irkutsk (OR342379), Yaroslavl (OR342373), and MongoliaBW (OR342375) was 4.4%; MongoliaDam (OR342376)—4.7%, and Sevan (OR342381, OR342383, OR342387)—7%. The minimal distance was between the sequence from Yaroslavl (OR342373) and the same numbers as in the case of Novosibirsk, China (XJDQD1026-18, XJDQD1025-18), Sweden (BSCHI129-17), and Germany (OP927683, OP927527), which was 0.5%. This was repeated for other samples: Irkutsk—0.5%, Irkutsk (OR342379)—0.5%, and MongoliaDam (OR342376)—1.6%. The minimal distance between the sequence from MongoliaBW (OR342375), Finland (MZ626747, MZ657059), and Sweden (BSCHI604-17) was 0.8%. The minimal distance between all sequences from Sevan Lake (OR342381, OR342383, OR342387) and China (ON975033) was 1.9%. In addition, we noticed that the sequences of C. annularius (MT534990, MT535143, and MT535187) from Skadar Lake (Balkan Peninsula, Montenegro) have very large distances between all of the analyzed samples, from 5.2 to 7%.

Table 2.
The pairwise genetic distances (K2P) between COI gene sequences of C. “annularius” from different populations.
3.3.2. Analysis of the Phylogenetic Tree
The unexpected results were repeated on the phylogenetic tree. We distinguished four clusters, the formation of which cannot be explained by geographical location (Figure 2). The first cluster includes sequences from Mongolia (OR342375, OR342376), Finland (MZ626747, MZ657059), and Sweden (BSCHI604-17), and, in addition, Chironomus muratensis (KC250748) from Sweden. The second cluster includes sequences from Irkutsk (OR342379), Yaroslavl (OR342373), China (XJDQD1011-18, XJDQD1026-18, XJDQD1025-18), and Germany (OP927683, OP927527). The third cluster includes sequences from Sevan Lake (OR342381, OR342383, OR342387) and China (ON975033), and, in addition, Chironomus bernensis (JN016851) from Saratov reg., Russia. The fourth cluster includes sequences from Skadar Lake (Balkan Peninsula) (MT534990, MT535143, and MT535187), C. bernensis (AF192188), and C. commutatus (AF192187) from the vicinities of Novosibirsk city. Separate from the second cluster is C. annularius (AF192189), from Novosibirsk city.

Figure 2.
Bayesian tree of the analyzed samples of Chironomus (Camptochironomus) spp. inferred from COI gene sequences. Species names, GenBank accession numbers, and group names are shown to the right of the branches. Support p-values are given if they exceed 0.3. “Black dots” —sequences obtained in this study.
3.3.3. Analysis of the Haplotype Network
The populations of C. annularius have a complicated structure (Figure 3). A total of 13 COI haplotypes were defined, which were divided into four main haplogroups. Three of these haplogroups are significantly distanced from each other in correspondence to geographical localities, excluding one Chinese haplotype (Hap_1). These are the groups from Sevan Lake, Novosibirsk, and Skadar Lake. The Sevan Lake haplogroup is separated from the Novosibirsk haplotype by 37 mutation steps, and from the Skadar Lake group by 42 mutation steps. The Skadar Lake haplogroup is separated from the Novosibirsk haplotype by 32 mutation steps. The minimal amount of mutation steps to the nearest haplotype is from 17 to 24. The central haplogroup is diverse, and includes the populations from Mongolia, Yaroslavl, Irkutsk, Finland, Sweden, Germany, and China.
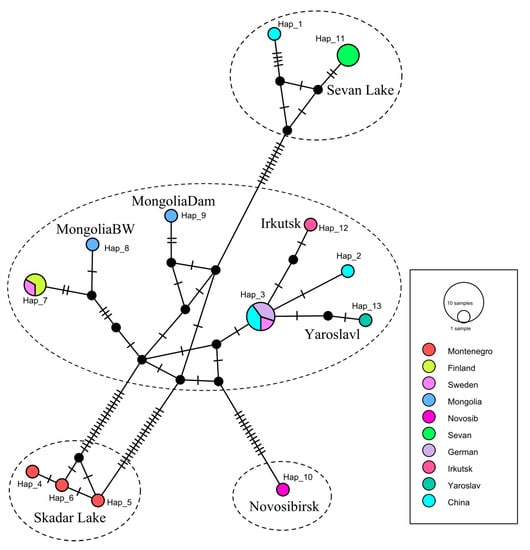
Figure 3.
Median-joining network showing phylogenetic relationships within C. annularius sensu lato. Each bar represents a single mutational change. The diameter of the circles is proportional to the number of individuals in each haplotype sampled (see open circles with numbers). Black dots represent hypothetical intermediate haplotypes. Discontinuous lines indicate populations with large genetic distances and mutation steps. Novosibirsk, Irkutsk, etc.—names of populations.
4. Discussion
The results showed that the studied taxon is the species complex. Usually, species and larval forms are confined to the type of water bodies or specific environmental conditions [,,]. In reviewing the literature, Spies and Sæther [] found that larvae of C. annularius sensu lato are found in water bodies of various types, including small freshwater bodies, the profundal zone of lakes, and saline waters such as the Baltic Sea littoral, and are widely spread geographically; it could be that different species were identified as C. “annularius”. It is known that Chironomus has high variability at the chromosomal and biochemical levels of organization, allowing it to inhabit various types of water bodies [,,,,]. The cytogenetic distances between two distant reservoirs (for example lakes with similar conditions) can be minimal, and vice versa, the distances between two neighboring reservoirs (for example a lake and a river) can be very large, with karyoforms even arising [,,].
Our samples from Russia, Armenia, and Mongolia, were studied using a comprehensive approach, including morphological, cytogenetic, and molecular genetic analyses. In addition, we used COI gene sequences of Ch. “annularius” from the GenBank and BOLD databases, realizing that there may be errors in species identification, as mentioned earlier [,].
Initially, we performed a morphological analysis, and found out that all of the larvae generally correspond to the description of Kiknadze et al. [,]. However, one minor detail in the mentum structure of the larvae from Lake Sevan attracted our attention. The fourth lateral tooth is slightly shorter than the neighboring ones; this may be a demonstration of ecological plasticity. In another way, it could indicate the presence of a different species of Chironomus markosjani Shilova 1983 that only inhabits this lake [,,]. The authors note that a characteristic feature of this species is the “bathophilus type” larval form: on the eighth segment, there are short ventral tubules with blunt ends []. In our samples from Lake Sevan, the ends were rounded. We found a slide with this species paratype in the collection of A.I. Shilova (IBIW RAS), but due to its degradation, we could not perform a comparative morphological analysis. The genus Chironomus includes many sibling species that are poorly separated on the basis of larval morphology. For example, many species of chironomids were found in Lake Sevan at the beginning of the last century (the 1930s); the following species of Chironomus were found: C. plumosus (at a depth of no more than 10 m), C. tentans (in the sublittoral at a depth of 21–30 m), and C. thummi Kief. (riparius) (in profundal at a depth of 31–40 m) []. However, then in 1969, adult midges obtained from collected larvae were analyzed, and it turned out that individuals with the “plumosus type” (ventral tubules of a similar length as the corresponding segment, lateral tubules of segment 7 are also present) of larva were identified incorrectly; they were actually C. tentans, and individuals with the “semireductus type” (short ventral tubuli) of larva were actually C. plumosus []. The larvae of chironomids, which were previously identified as C. thummi (riparius), turned out to be exactly C. markosjani, with the “bathophilus type” of larva []. After analyzing adult midges of C. annularius from different geographical zones, Belyanina [] noted the high variability in their color, came to the conclusion that there are no reliable morphological criteria, and then suggested using cytogenetics. At first, C. plumosus was considered a single polytypical species after use of the cytogenetic approach began; more than ten new species were described, and the ways of their evolution were established [].
Based on the results of cytogenetic analysis, we found that all banding sequences of all studied larvae belong to the chromosome banding pool of C. “annularius” [,,,]. We found through banding sequences and zygotic combinations that A1.1, A2.2, B1.1, B1.2., C1.1, D1.1, D1.2, D2.2, E1.1, E1.2, E2.2, F2.2, and G1.1 previously were observed in almost all of the studied Palearctic populations except the Nearctic [,]. The banding sequence combinations h’annA2 p’annA3 from Yaroslavl and p’annB4.4 from Armenia and Mongolia were previously found only in the South Caucasian (Georgian) population []. The rare banding sequence p’annF1.1 from Yaroslavl and Mongolia was previously found only in one South Caucasian (Georgian) and the Kazakhstan populations [,]. The banding sequence h’annG3.3 was previously considered Nearctic; after finding it in C. “annularius” larvae from Lake Khubsugul (Mongolia), it began to be considered Holarctic []. The most interesting finding was the Nearctic sequence n’annD3.3; previously, it was found exclusively in North America (USA and Canada) []. We found it in Sevan Lake, together with p’annG3.3 and h’annE1.1, which is morphologically very similar to the Nearctic population (Figure 1). It should be noted that sequence n’annD3.3 was already noted earlier by Belyanina [] in the Mangystau region, Kazakhstan. Unfortunately, the author used his own mapping system, and did not provide images of this chromosomal arm. In the diagram of the position of the chromosomal inversion presented by the author, the break point “16” corresponds to region “15a-e.20b-18e” []; this corresponds to the break point n’annD3. The other break point does not match n’annD3; it was designated by the author as “11”, but should be located on the border of the regions “12–13”. Therefore, we cannot accept the finding of Belyanina D3 [] corresponding to n’annD3 []. In another study, Pankratova [] shows the karyotype of C. markosjani (Chironomus forma larvalis bathophilus) from Lake Sevan (Noratus, deep—25 m.); morphologically, it is very similar to the one we found, but its quality is very low, and we could not make a precise comparison (Appendix B). We have only a description of its karyotype. The author referred to the chromosomal set 2n = 8, and noted that all of the chromosomes are closely conjugated; nucleolus organizers are present in all chromosomes, and the largest of them is on chromosome IV []. In C. “annularius” sensu Strenzke (1959), the homologues of chromosome IV are only partially paired in the Palearctic [], and the centromeric region contains a clearly visible heterochromatized centromere band [].
The analysis of the results of the genetic p-distances, Bayesian tree, and network of haplotypes was also interesting. It was shown that the distances between the COI gene sequences of C. commutatus and C. bernensis were comparable to the intraspecific distances (5.1%) obtained for the samples of C. “annularius”. Despite the fact that these species belong to different cytocomplexes, the data obtained by us are consistent with earlier investigations on this theme [,,,]. Another COI gene sequence of C. bernensis (JN016851) from Saratov reg. appeared to be close to sequences of C. “annularius” (markosjani) from Sevan Lake (OR342381, OR342383, OR342387). Perhaps the length of 535 bp is insufficient for an accurate identification of the sequence of C. bernensis (JN016851), or it could be a result of interspecific hybridization and horizontal transfer of mitochondrial genes, with fixation in one of the parental species in the population [,,]. We have already shown that hybridization of species from different cytocomplexes is possible []. Sample C. muratensis (KC250748) from Sweden may also be misidentified.
The average distances within the analyzed COI gene sequences of C. “annularius” were 3.7%; this is significantly higher than the accepted 3% threshold [,,]. We found minimal genetic distances within clusters (Figure 2) of sequences from Sevan Lake (OR342381, OR342383, OR342387)—0%, including China (ON975033)—0.9%. Within the cluster of Skadar Lake (MT535143, MT535187, MT534990), the distance was 0%. The average genetic distance within a large cluster with sequences from Irkutsk, Yaroslavl, Mongolia, Finland, Sweden, Germany, and China (XJDQD1011-18, XJDQD1025-18, XJDQD1026-18, MZ626747, MZ657059, BSCHI129-17, BSCHI604-17, OP927527, and OP927683) was 1.3%, and with an additional sequence from Novosibirsk, it was 1.7%. The distances between clusters were as follows: Novosibirsk (AF192189), Sevan Lake (OR342381, OR342383, OR342387), and China (ON975033)—6.9%; Novosibirsk (AF192189) and Skadar Lake (MT535143, MT535187, MT534990)—6.1%; Skadar Lake (MT535143, MT535187, MT534990), Sevan Lake (OR342381, OR342383, OR342387), and China (ON975033)—6.7%. As we know, the interspecific distances for COI gene sequences in Chironomus, in most cases, varied from 9 to 20%, and from 1 to 4% in rare cases []. For other Diptera, the threshold value of the intraspecific distance is 4–5% for genetically separate species of Tanytarsus (van der Wulp, 1874) (Chironominae) [], and 5–8% for Polypedilum (Kieffer, 1912) (Chironominae) []. In one of the last studies on Skadar Lake, for the species Hydrometra stagnorum Linnaeus, 1758 (Heteroptera, Hydrometridae) it was shown that a high 6.1% intraspecific k2-p-distance could signify the presence of cryptic species; this requires more detailed investigation [].
The large genetic distances we have found may be, for example, the result of an incorrect identification of species, or from the presence of cryptic species. It is known that the GenBank and BOLD databases of genetic information contain about 65% of sequences without species-level assignments, the so-called “dark taxa” of all Chironomidae recorded from Germany []. For the sequence from Novosibirsk (AF192189), we found the remark, “It is possible the specimen sequenced in Guryev et al. (2001) may have been a misidentified C. muratensis as it is closest to that species in GenBank accessions” []. It has already been shown that Anopheles culicifacies (Giles, 1901) consists of four haplotypes, one of them being a primary vector of malaria in Sri Lanka, species A; species B is a non- or poor vector []. Species Culex palpalis (Taylor, 1912) was originally identified from Lake Victoria (Australia) as Culex annulirostris (Skuse, 1889), and accurately identified only through COI gene sequencing [].
During a study of Chironomus populations from Germany, in C. bernensis was found a cryptic species. The authors first studied the larvae cytogenetically, and then performed a molecular genetic analysis of the COI gene sequences of C. bernensis; it turned out that some of the sequences were combined with C. bernensis, and other sequences with C. heterodentatus (Konstantinov 1956) and C. acutiventris (Wülker and Ryser, 1983) []. In the same study, the authors presented the sequences of C. annularius, which were combined with sequences of C. annularius from Novosibirsk []; unfortunately, no cytogenetic analysis was performed for it [].
The analysis of the haplotype network of C. “annularius” (Figure 3) also showed a large number of mutational steps between these clusters, as well as a discrepancy between the phylogenetic data and the geographical location of the studied sequences. For Culex mosquito (Meigen, 1818) (Culicidae), it was shown that the complex pattern of the haplotype network of COI gene sequences in most of the species indicates that there is less association between certain haplotypes and geographic locations [].
We have already noted earlier that several species may be hidden under the name “C. annularius” []. Therefore, based on the data presented, we can assume that there are cryptic species among our specimens that can be identified only using a molecular genetic approach.
We assumed that specimens from the largest cluster with COI gene sequences from Irkutsk, Yaroslavl, China, Mongolia, Finland, Sweden, and Germany, where this species was first described from, correspond to species Chironomus annularius (Meigen, 1818). On the haplotype network (Figure 3), the Novosibirsk, Sevan Lake, and Skadar Lake clusters are separated from the central cluster, and may belong to potential homosequential cryptic species, with the available name Ch. markosjani Shilova, 1983 for the Sevan species. The known example of such Chironomus species that are difficult to identify even cytogenetically, are Chironomus agilis2 (Kiknadze, Siirin, and Filippova, 1991) and Chironomus agilis, which differ only in the centromeric band []. Previously, 62 chromosome banding sequences were known for C. tentans (Fabricius, 1805); only 6 of them were Holarctically distributed, 39 were endemic to the Palearctic, and 17 were endemic to the Nearctic; then, from the Nearctic population, a distinct species of C. dilutus (Shobanov, Kiknadze, and Butler, 1999) was described via morphology []. In the GenBank, the genetic distances between the COI gene sequences of C. tentans and C. dilutus is about 6%. There is a dispute about two species of C. nuditarsis (Keyl, 1961) and C. curabilis (Beljanina, Sigareva, and Loginova, 1990), which also have a difference only in the size of the centromeric disk [].
The specimen from Novosibirsk was identified only by the larval karyotype, is labeled as Chironomus “annularius” sensu Strenzke 1959, and requires additional refinement of the species identity according to the adult form.
The nucleotide sequences of C. “annularius” from Skadar Lake, according to the BOLD specimen voucher, were obtained from the adult midge. The authors of the study did not provide the nomenclature they used to determine the species of Chironomus, but they especially noticed the difficulty of determining species by morphological characteristics [], which in this species can be highly variable []. Earlier, it was noted that there may be many cryptic species of chironomids in the studied area, some of which could only be identified up to the genus level, and endemic species are also characteristic of this region [].
There are several facts in favor of the cryptic species C. markosjani (Shilova, 1983) living in Lake Sevan. It has minor morphological differences (Appendix A), the presence of chromosomal sequences characteristic of Nearctic populations, and large genetic p-distances with other specimens that are more than twice the threshold values (Table 2). On the Bayesian tree (Figure 2) and haplotype network (Figure 3), together with a sample from China, they form a separate branch with a high level of significance. At the same time, the sample from China (ON975033) may represent a geographically isolated population of this cryptic species, or an invasion from Armenia.
5. Conclusions
The detailed results of the morphological, cytogenetic, and molecular genetic analyses allowed us to obtain fresh insight on Chironomus annularius sensu lato, and allowed us to assume the existence of potential homosequential cryptic species. All of the larvae studied by us were identified with chromosomal banding sequences characteristic for C. “annularius”. A sequence that was previously found exclusively in the Nearctic region was found for the first time in larvae from Lake Sevan, and should be named Holarctic h’annD3, as in the case of h’annG3 []. The phylogenetic tree has the largest cluster, which includes COI gene sequences from Irkutsk, Yaroslavl, China, Mongolia, Finland, Sweden, and Germany, which most probably matches a widespread species Chironomus annularius (Meigen, 1818), because of high similarity with the reference German population that is closest to the type locality of the species. The species corresponding to the sequences from Novosibirsk and Skadar Lake require clarification and a first description, respectively, due to the large genetic distances exceeding the threshold values. The sequences from Lake Sevan also have large genetic distances from other studied specimens, and may belong to the previously described Chironomus markosjani in the mass inhabiting the deep-water part of this lake. In the future, for a more detailed and comprehensive examination of this complex species, it is necessary to use the adult and larval stages of development, and in addition to the COI gene sequence, nuclear genes must be used. When publishing data, it is necessary to specify exactly which characteristics were used in the identification of the species.
Author Contributions
V.B., A.P. and E.M. developed the concept of this study; A.P., V.B, T.M., E.M. and S.H. collected the samples; E.M. designed and carried out the molecular analyses; V.B., A.P., T.M. and E.M. performed project administration and carried out the karyological and statistical analysis; V.B. and A.P. wrote the manuscript, with input from E.M. and S.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was performed in the framework of the following state assignment: theme Nos. 121050500046-8, 121051100099-5, 121051100109-1, 0279-2021-0005 (121032300224-8).
Data Availability Statement
Data supporting reported results can be obtained upon request from the corresponding author (V.B.).
Acknowledgments
The material in Mongolia was collected during an expedition of the hydrobiological team of the ‘Joint Russian–Mongolian complex biological expedition of the Russian and Mongolian Academies of Sciences’, and in Armenia as a part of research program of the ‘Joint Russian–Armenian biological expedition of the Russian and Armenian Academies of Sciences’. We are grateful to the Mongolian and Armenian colleagues and the crew of the vessel “Hydrolog” for their assistance in carrying out the research. The authors are grateful to B.A. Levin, M.I. Malin (IBIW RAS), M.Kh. Karmokov (IEMT RAS), V.G. Kuznetsova, N.A. Shapoval (ZIN RAS), and Yu.V. Bespalaya (FECIAR RAS) for their help and consultation during all stages of the investigation and manuscript preparation.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
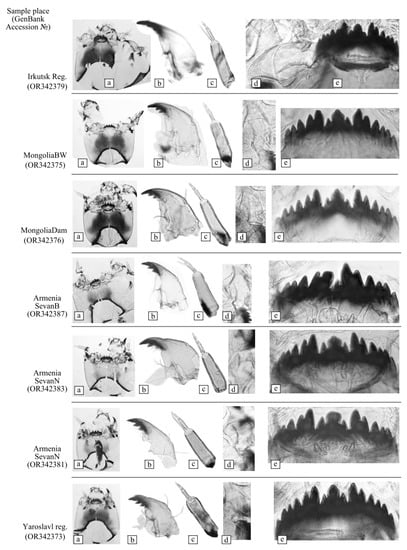
Figure A1.
Larval morphology of Chironomus annularius sensu lato from studied populations: (a) head capsule, (b) mandible, (c) antenna, (d) ventromental plate, (e) mentum.
Appendix B
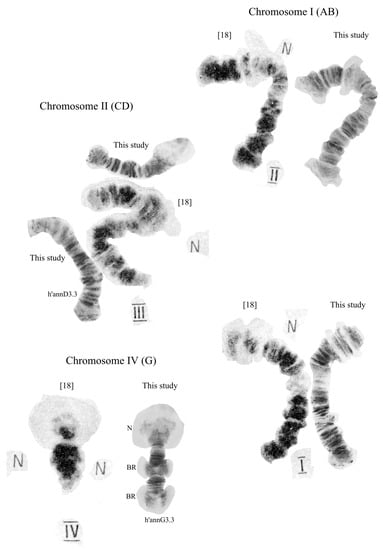
Figure A2.
Comparison of karyotypes of Chironomus annularius (this study) and Chironomus forma larvalis bathophilus (markosjani) [] from Sevan Lake.
References
- Spies, M.; Sæther, O.A. Notes and Recommendations on Taxonomy and Nomenclature of Chironomidae (Diptera). Zootaxa 2004, 752, 1. [Google Scholar] [CrossRef]
- Strenzke, K. Revision Der Gattung Chironomus Meig. i. Die Imagines von 15 Norddeutschen Arten Und Unterarten. Arch. Hydrobiol. 1959, 56, 1–42. [Google Scholar]
- Keyl, H.-G.; Keyl, I. Die Cytologische Diagnostik Der Chironomiden. I. Bestimmungstabelle Fur Die Gattung Chironomus Auf Grund Der Speicheldrusen-Chromosomen. Arch. Hydrobiol. 1959, 56, 43–57. [Google Scholar]
- Keyl, H.G. Chromosomenevolution Bei Chironomus. II. Chromosomenumbauten Und Phylogenetische Beziehungen Der Arten. Chromosoma 1962, 13, 464–514. [Google Scholar] [CrossRef]
- Shobanov, N.A. The Genus Chironomus Meigen (Diptera, Chironomidae) (Systematics, Biology, Evolution). Ph.D. Thesis, Zoological Institute of Russian Academy of Sciences, Saint-Petersburg, Russia, 2000. [Google Scholar]
- Karmokov, M.K. Karyotype Characteristics and Chromosomal Polymorphism of Chironomus “annularius” Sensu Strenzke (1959) (Diptera, Chironomidae) from the Caucasus Region. Comp. Cytogenet. 2018, 12, 267–284. [Google Scholar] [CrossRef]
- Kiknadze, I.I.; Istomina, A.G.; Golygina, V.V.; Gunderina, L.I. Karyotypes of Palearctic and Holarctic Species of the Genus Chironomus; Kiknadze, I.I., Ed.; Russian Academy of Sciences, Siberian Branch, Federal Research Center Institute of Cytology and Genetics, Academic Publishing House “GEO”: Novosibirsk, Russia, 2016; ISBN 9785990885325. [Google Scholar]
- Kiknadze, I.I.; Shilova, A.I.; Shobanov, N.A.; Zelentsov, N.I.; Grebenyuk, L.P.; Istomina, A.G.; Pratasov, V.A. Karyoypes and Larval Morphology in Tribe Chironomini. Atlas.; Nauka Publish: Novosibirsk, Russia, 1991; pp. 1–115. [Google Scholar]
- Kiknadze, I.I.; Istomina, A.G.; Gunderina, L.I.; Aimanova, K.G.; Savvinov, D.D. Banding Sequence Pools of Chironomid of Yakutian Permafrost. Tribe Chironomini; Nauka: Novosibirsk, Russia, 1996; pp. 1–166. [Google Scholar]
- Kiknadze, I.I.; Istomina, A.G.; Golygina, V.V. The Karyotype and Chromosome Polymorphism of the Holarctic Species Chironomus «annularius» Sensu Strenzke, 1959 (Diptera, Chironomidae). Euroasian Entomol. J. 2012, 11, 95–114. [Google Scholar]
- Belyanina, S.I. A Comparative Karyotypical Characteristics of Chironomus annularius (Diptera, Chironomidae) from Different Geographic Zones. Zool. Zhurnal 1981, 60, 1030–1039. [Google Scholar]
- Michailova, P.V. The Polytene Chromosomes and Their Significance to the Family Chironomidae, Diptera. Acta Zool. Fenn. 1989, 186, 1–107. [Google Scholar]
- Petrova, N.A.; Michailova, P.V. The Population-Karyological Studies of Some Chironomidae Species (Diptera, Chironomidae). Tsitologiia 1986, 28, 727–734. [Google Scholar]
- Butler, M.G.; Kiknadze, I.I.; Cooper, J.K.; Siirin, M.T. Cytologically Identified Chironomus Species from Lakes in North Dakota and Minnesota, USA. In Chironomids: From Genes to Ecosystems; Cranston, P.S., Ed.; CSIRO Publications: Canberra, Australia, 1995; pp. 31–38. [Google Scholar]
- Kiknadze, I.I.; Gunderina, L.I.; Butler, M.G.; Wuelker, W.F.; Martin, J. Chromosomes and Continents. In Biosphere Origin and Evolution; Springer: Boston, MA, USA, 2008; pp. 349–369. [Google Scholar] [CrossRef]
- Kiknadze, I.I.; Butler, M.G.; Gunderina, L.I.; Istomina, A.G.; Gusev, V.D.; Nemytikova, L.A. Chromosomal Evolution of Nearctic and Palearctic Chironomus Species (Diptera: Chironomidae). In Proceedings of the XV International Symposium on Chironomidae, Saint Paul, MA, USA, 12–15 August 2003; pp. 203–221. [Google Scholar]
- Petrova, N.A.; Zhirov, S.V.; Yerbayeva, E.A. Description of Three Species of Chironomids (Diptera, Chironomidae) from Lake Khubsugul, Mongolia (Morphological and Karyological Aspects). EUROASIAN Entomol. J. 2014, 13, 445–450. [Google Scholar]
- Pankratova, V.Y.; Chubareva, L.A.; Petrova, N.A. On the Taxonomy of Chironomus Species (Chironomidae) from Lake Sevan. Tr. Zool. Inst. Akad. Nauk. SSSR 1980, 95, 50–55. [Google Scholar]
- Chimeno, C.; Rulik, B.; Manfrin, A.; Kalinkat, G.; Hölker, F.; Baranov, V. Facing the Infinity: Tackling Large Samples of Challenging Chironomidae (Diptera) with an Integrative Approach. PeerJ 2023, 11, e15336. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Zhao, Y.M.; Guo, B.X.; Li, C.H.; Sun, B.J.; Lin, X.L. Comparative Analysis of Mitogenomes of Chironomus (Diptera: Chironomidae). Insects 2022, 13, 1164. [Google Scholar] [CrossRef]
- Gadawski, P.; Montagna, M.; Rossaro, B.; Giłka, W.; Pešić, V.; Grabowski, M.; Magoga, G. DNA Barcoding of Chironomidae from the Lake Skadar Region: Reference Library and a Comparative Analysis of the European Fauna. Divers. Distrib. 2022, 28, 2838–2857. [Google Scholar] [CrossRef]
- Gadawski, P.; Rossaro, B.; Giłka, W.; Montagna, M.; Zawal, A.; Grabowski, M. First Insights into the Diversity and Ecology of Non-Biting Midges (Diptera: Chironomidae) of the Unique Ancient Skadar Lake Basin (Montenegro/Albania). J. Great Lakes Res. 2022, 48, 538–550. [Google Scholar] [CrossRef]
- Martin, J. Chromosomes as Tools in Taxonomy and Phylogeny of Chironomidae (Diptera). Entomol. Scand. 1979, 10, 67–74. [Google Scholar]
- Keyl, H.-G. Chromosomenevolution Bei Chironomus I. Strukturabwandlungen an Speicheldrüsen-Chromosomen. Chromosoma 1961, 12, 26–47. [Google Scholar] [CrossRef]
- Zamani, A.; Fric, Z.F.; Gante, H.F.; Hopkins, T.; Orfinger, A.B.; Scherz, M.D.; Bartoňová, A.S.; Pos, D.D. DNA Barcodes on Their Own Are Not Enough to Describe a Species. Syst. Entomol. 2022, 47, 385–389. [Google Scholar] [CrossRef]
- DeSalle, R.; Egan, M.G.; Siddall, M. The Unholy Trinity: Taxonomy, Species Delimitation and DNA Barcoding. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1905–1916. [Google Scholar] [CrossRef]
- Bolshakov, V.V.; Movergoz, E.A. Karyotype and COI Gene Sequences of Chironomus melanotus Keyl, 1961 from the Yaroslavl Region, Russia, and the Difficulties with Its Identification Using GenBank and BOLD Systems. Comp. Cytogenet. 2022, 16, 161–172. [Google Scholar] [CrossRef]
- Ilyinskaya, N.B. The Development of 4th Instar Larvae and Diapause. In Chironomus plumosus L. (Diptera, Chironomidae). Systematics, Morphology, Ecology, Production; Sokolova, N.Y., Ed.; Nauka Publishers: Moscow, Russia, 1983; pp. 167–188. [Google Scholar]
- Sæther, O. Glossary of Chironomid Morphology Terminology (Diptera: Chironomidae). Entomol. Scand. Suppl. 1980, 14, 1–51. [Google Scholar]
- Dyomin, S.Y. Variability of the Degree of Condensation of Polytene Chromosomes in the Cells of Different Organs of Chironomus plumosus Larvae from Nature. Ph.D. Thesis, Institute of Cytology of the USSR Academy of Sciences, Leningrad, Russia, 1989. [Google Scholar]
- Dévai, G.; Wülker, W.F.; Scholl, A. Revision Der Gattung Chironomus Meigen (Diptera). IX. C. Balatonicus Sp. n. Aus Flachsee Balaton. Acta Zool. Acad. Sci. Hung. 1983, 29, 357–374. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; van der Mark, P.; Huelsenbeck, J.P. Bayesian Phylogenetic Analysis Using MRBAYES. In The Phylogenetic Handbook; Cambridge University Press: Cambridge, UK, 2009; pp. 210–266. [Google Scholar]
- Karmokov, M.K. Karyotype Characteristics, Chromosomal Polymorphism and Gene COI Sequences of Chironomus Heteropilicornis Wülker, 1996 (Diptera, Chironomidae) from the South Caucasus. Comp. Cytogenet. 2019, 13, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Bolshakov, V.V.; Prokin, A.A.; Artemenko, S. V Karyotype and COI Gene Sequence of Chironomus heteropilicornis Wülker, 1996 (Diptera, Chironomidae) from the Gydan Peninsula, Russia. Comp. Cytogenet. 2021, 15, 447–458. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Webb, C.J.; Scholl, A. Identification of Larvae of European Species of Chironomus Meigen (Diptera: Chironomidae) by Morphological Characters. Syst. Entomol. 1985, 10, 353–372. [Google Scholar] [CrossRef]
- Shilova, A.I. A New Species of the Genus Chironomus (Diptera, Chironomidae) from Lake Sevan. Zool. Zhurnal 1983, 62, 245–251. [Google Scholar]
- Guryev, V.; Makarevitch, I.; Blinov, A.; Martin, J. Phylogeny of the Genus Chironomus (Diptera) Inferred from DNA Sequences of Mitochondrial Cytochrome b and Cytochrome Oxidase I. Mol. Phylogenet. Evol. 2001, 19, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Gunderina, L.I.; Kiknadze, I.I.; Golygina, V.V. Differentiation of the Cytogenetic Structure of Natural Populations in the Plumosus Group of Sibling Species Chironomus balatonicus, Chironomus entis, Chironomus muratensis, Chironomus nudiventris (Chironomidae: Diptera). Genetica 1999, 35, 606–614. [Google Scholar]
- Shobanov, N.A.; Bolshakov, V.V. Karyofund of Chironomus plumosus (L.) (Diptera, Chironomidae): V. Terminal and Interstitial Populations. Cell Tissue Biol. 2011, 5, 196–201. [Google Scholar] [CrossRef]
- Bolshakov, V.; Movergoz, E.; Stolbov, V. Karyotypes and COI Gene Sequences of Chironomus agilis2, Ch. balatonicus, and Camptochironomus tentans (Diptera, Chironomidae) from Kurchatskoe Lake, Tyumen Region, Russia. Diversity 2022, 14, 1044. [Google Scholar] [CrossRef]
- Bolshakov, V.V.; Andreeva, A.M. Peculiarities of Structural Organization of Hemoglobin of Chironomus plumosus L. (Diptera: Chironomidae). J. Evol. Biochem. Physiol. 2012, 48, 265–271. [Google Scholar] [CrossRef]
- Osmulski, P.A.; Leyko, W. Structure, Function and Physiological Role of Chironomus Haemoglobin. Comp. Biochem. Physiol. Part B Comp. Biochem. 1986, 85, 701–722. [Google Scholar] [CrossRef]
- Bolshakov, V. V Variability of Karyofunds of Chironomus plumosus (Diptera, Chironomidae) from the Rybinsk Reservoir during the Period of Climate Warming. Inl. Water Biol. 2021, 14, 256–262. [Google Scholar] [CrossRef]
- Shilova, A.I.; Zelentsov, N.I. To the Chironomid Fauna of Lake Sevan (Diptera, Chironomidae). Biol. Inl. Waters. Inf. Bull. 1988, 79, 48–52. [Google Scholar]
- Makarevich, I.F.; Berezikov, E.V.; Guryev, V.P.; Blinov, A.G. Molecular Phylogeny of the Chironomus Genus Deduced from Nucleotide Sequences of Two Nuclear Genes, Ssp160 and the Globin 2b Gene. Mol. Biol. 2000, 34, 606–612. [Google Scholar] [CrossRef]
- Demin, A.G.; Polukonova, N. Divergence Time Estimation of Chironomids Chironomus Genus (Diptera) Using the «molecular Clock» Hypothesis. Entomol. Parasitol. Res. Volga Reg. 2008, 7, 8–13. [Google Scholar]
- Shobanov, N.A.; Zotov, S.D. Cytological Aspects of the Phylogeny of the Genus Chironomus Meigen (Diptera, Chironomidae). Entomol. Rev. 2001, 80, 180–193. [Google Scholar]
- Papusheva, E.; Gruhl, M.C.; Berezikov, E.; Groudieva, T.; Scherbik, S.V.; Martin, J.; Blinov, A.; Bergtrom, G. The Evolution of SINEs and LINEs in the Genus Chironomus (Diptera). J. Mol. Evol. 2004, 58, 269–279. [Google Scholar] [CrossRef]
- Guryev, V.P.; Blinov, A.G. Phylogenetic Relationships among Holarctic Populations of Chironomus entis and Chironomus plumosus in View of Possible Horisontal Transfer of Mitochondrial Genes. Russ. J. Genet. 2002, 38, 239–243. [Google Scholar] [CrossRef]
- Polukonova, N.V.; Demin, A.G.; Miuge, N.S.; Shaĭkevich, E.V.; Djomin, A.G.; Mugue, N.S.; Shaikevich, E.V.; Demin, A.G.; Miuge, N.S.; Shaĭkevich, E. V Comparison of Chironomus usenicus and Chironomus curabilis with Species of the Group plumosus (Diptera) Inferred from the Mitochondrial DNA Gene COI and by the Polytene Chromosomes Banding Pattern. Russ. J. Genet. 2009, 45, 1029–1035. [Google Scholar] [CrossRef]
- Bolshakov, V.; Prokin, A.; Pavlov, D.; Akkizov, A.; Movergoz, E. Karyotypes and COI Gene Sequences of Chironomus sp. Le1 (Kiknadze and Salova, 1996), Ch. laetus (Belyanina and Filinkova, 1996) and Their Hybrid from the Yamal Peninsula, Arctic Zone of Russia. Insects 2022, 13, 1112. [Google Scholar] [CrossRef]
- Proulx, I.; Martin, J.; Carew, M.; Hare, L. Using Various Lines of Evidence to Identify Chironomus Species (Diptera: Chironomidae) in Eastern Canadian Lakes. Zootaxa 2013, 3741, 401–458. [Google Scholar] [CrossRef] [PubMed]
- Ekrem, T.; Willassen, E.; Stur, E. A Comprehensive DNA Sequence Library Is Essential for Identification with DNA Barcodes. Mol. Phylogenet. Evol. 2007, 43, 530–542. [Google Scholar] [CrossRef]
- Kondo, N.I.; Ueno, R.; Ohbayashi, K.; Golygina, V.V.; Takamura, K. DNA Barcoding Supports Reclassification of Japanese Chironomus Species (Diptera: Chironomidae). Entomol. Sci. 2016, 19, 337–350. [Google Scholar] [CrossRef]
- Lin, X.; Stur, E.; Ekrem, T. Exploring Genetic Divergence in a Species-Rich Insect Genus Using 2790 DNA Barcodes. PLoS ONE 2015, 10, e0138993. [Google Scholar] [CrossRef]
- Song, C.; Lin, X.-L.; Wang, Q.; Wang, X.-H. DNA Barcodes Successfully Delimit Morphospecies in a Superdiverse Insect Genus. Zool. Scr. 2018, 47, 311–324. [Google Scholar] [CrossRef]
- Rewicz, T.; Tończyk, G.; Trębicki, Ł.; Gadawski, P.; Mamos, T.; Pešić, V.; Zawal, A.; Grabowski, M. DNA Barcode-Based Survey Documents Underestimated Diversity and Intricate Phylogeographic Patterns of Aquatic Heteroptera in an Endangered Balkan Biodiversity Hotspot: Ancient Lake Skadar Basin. Biodivers. Conserv. 2023. [Google Scholar] [CrossRef]
- Morinière, J.; Balke, M.; Doczkal, D.; Geiger, M.F.; Hardulak, L.A.; Haszprunar, G.; Hausmann, A.; Hendrich, L.; Regalado, L.; Rulik, B.; et al. A DNA Barcode Library for 5,200 German Flies and Midges (Insecta: Diptera) and Its Implications for Metabarcoding-based Biomonitoring. Mol. Ecol. Resour. 2019, 19, 900–928. [Google Scholar] [CrossRef]
- Martin, J. Personal Web Page. 2023. Available online: http://www.chironomidae.net/Martin/NACytfiles/Sp3d.htm (accessed on 20 August 2023).
- Weeraratne, T.C.; Surendran, S.N.; Reimer, L.J.; Wondji, C.S.; Perera, M.D.B.; Walton, C.; Parakrama Karunaratne, S.H.P. Molecular Characterization of Anopheline (Diptera: Culicidae) Mosquitoes from Eight Geographical Locations of Sri Lanka. Malar. J. 2017, 16, 234. [Google Scholar] [CrossRef] [PubMed]
- Batovska, J.; Blacket, M.J.; Brown, K.; Lynch, S.E. Molecular Identification of Mosquitoes (Diptera: Culicidae) in Southeastern Australia. Ecol. Evol. 2016, 6, 3001–3011. [Google Scholar] [CrossRef] [PubMed]
- Pfenninger, M.; Nowak, C.; Kley, C.; Steinke, D.; Streit, B. Utility of DNA Taxonomy and Barcoding for the Inference of Larval Community Structure in Morphologically Cryptic Chironomus (Diptera) Species. Mol. Ecol. 2007, 16, 1957–1968. [Google Scholar] [CrossRef]
- Panda, D.; Barik, T.K. Molecular Characterization and Genetic Divergence of Seven Culex Mosquito (Diptera: Culicidae) Species Using Mt COI Gene from Odisha State, India. J. Basic Appl. Zool. 2022, 83, 41. [Google Scholar] [CrossRef]
- Shobanov, N.A.; Kiknadze, I.I.; Butler, M.G. Palearctic and Nearctic Chironomus (Camptochironomus) tentans (Fabricius) Are Different Species (Diptera, Chironomidae). Entomol. Scand. 1999, 30, 311–322. [Google Scholar]
- Kiknadze, I.I.; Michailova, P.V.; Istomina, A.G.; Golygina, V.V.; Panis, L.I.; Krastanov, B. The Chromosomal Polymorphism and Divergence of Populations in Chironomus nuditarsis Str. (Diptera, Chironomidae). Tsitologiia 2006, 48, 595–609. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).