Analyzing the Akinete Protein of the Harmful Freshwater Cyanobacterium, Dolichospermum circinale
Abstract
1. Introduction
2. Materials and Methods
2.1. D. circinale Isolation and Culture
2.2. Akinete Preparation
2.3. Protein Extraction from Akinete and Vegetative Cell
2.4. Sample Preparation for Mass Spectrometry
2.5. Analysis of Amino Acid Sequence
2.6. Statistical Analyses
3. Results and Discussion
3.1. Protein Profile of D. circinale
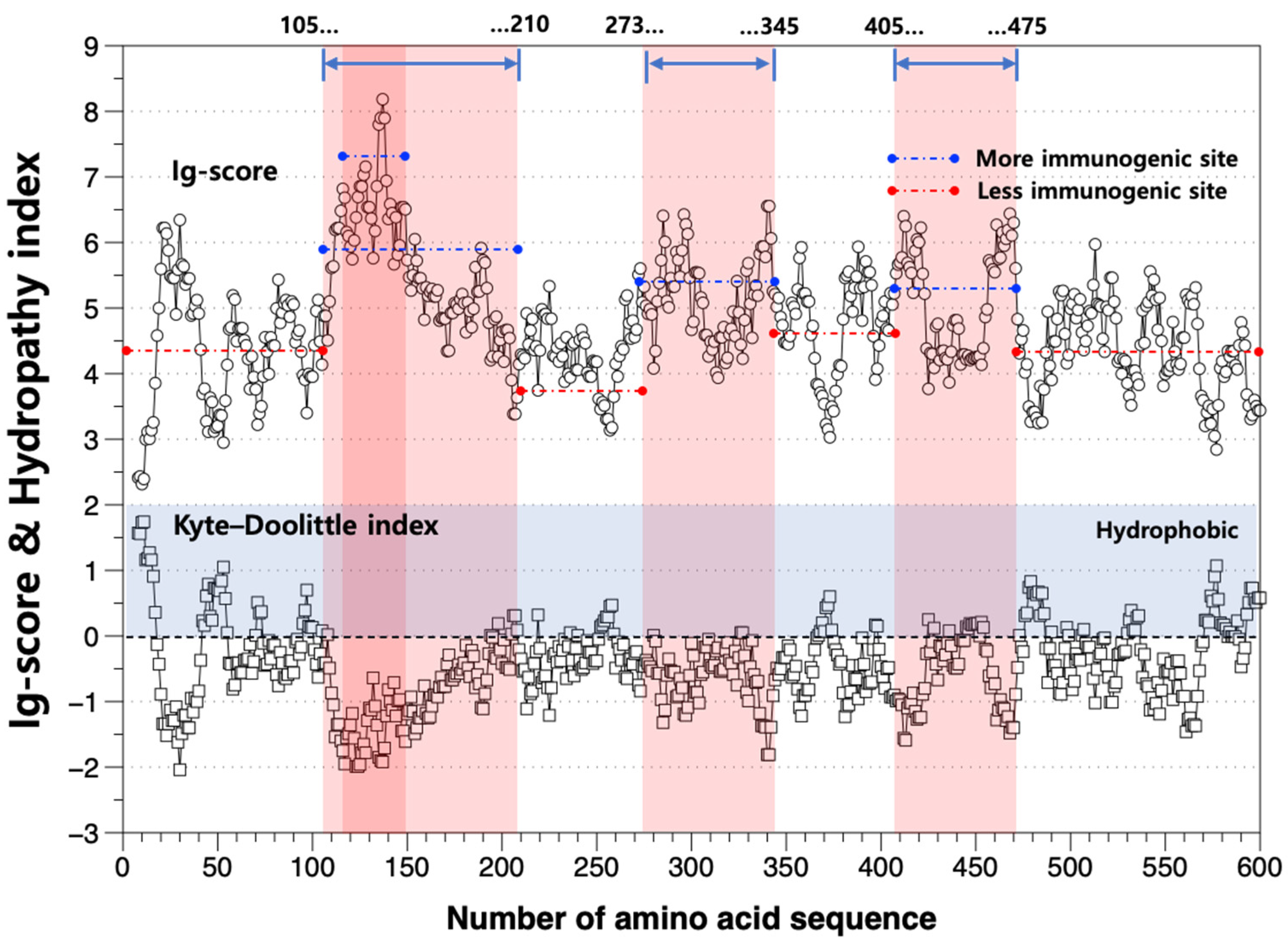
3.2. Amino Acid Sequence of D. circinale Akinete-Specific Protein
3.3. Perspective on Developing the Akinete Screening Technique
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suikkanen, S.; Kaartokallio, H.; Hällfors, S.; Huttunen, M.; Laamanen, M. Life cycle strategies of bloom-forming, filamentous cyanobacteria in the Baltic Sea. Deep Sea Res. Part II Top 2010, 57, 199–209. [Google Scholar] [CrossRef]
- Anderson, D.M.; Wall, D. Potential importance of benthic cysts of Gonyaulax tamarensis and G. excavata in initiating toxic dinoflagellate blooms 1, 2, 3. J. Phycol. 1978, 14, 224–234. [Google Scholar] [CrossRef]
- Andersen, D.M.; Keafer, B.A. An endogenous annual clock in the toxic marine dinoflagellate Gonyaulax tamarensis. Nature 1987, 325, 616. [Google Scholar] [CrossRef]
- Baker, P.D.; Bellifemine, D. Environmental influences on akinete germination of Anabaena circinalis and implications for management of cyanobacterial blooms. Hydrobiologia 2000, 427, 65–73. [Google Scholar] [CrossRef]
- Huber, A.L. Factors affecting the germination of akinetes of Nodularia spumigena (Cyanobacteriaceae). Appl. Environ. Microbiol. 1985, 49, 73–78. [Google Scholar] [CrossRef]
- Ståhl-Delbanco, A.; Hansson, L.-A. Effects of bioturbation on recruitment of algal cells from the “seed bank” of lake sediments. Limnol. Oceanogr. 2002, 47, 1836–1843. [Google Scholar] [CrossRef]
- van Dok, W.; Hart, B.T. Akinete germination in Anabaena circinalis (cyanophta). J. Phycol. 1997, 33, 12–17. [Google Scholar]
- Anderson, D.M.; Glibert, P.M.; Burkholder, J.M. Harmful algal blooms and eutrophication: Nutrient sources, composition, and consequences. Estuaries 2002, 25, 704–726. [Google Scholar] [CrossRef]
- Norris, G.; Mcandrews, J.H. Dinoflagellate cysts from post-glacial lake muds, Minnesota (USA). Rev. Paleobot. Palynol. 1970, 10, 131–156. [Google Scholar] [CrossRef]
- Thiel, T.; Wolk, C.P. Metabolic activities of isolated akinetes of the cyanobacterium Nostoc spongiaeforme. J. Bacteriol. Res. 1983, 156, 369–374. [Google Scholar] [CrossRef]
- Moore, D.; O’Donohue, M.; Garnett, C.; Critchley, C.; Shaw, G. Factors affecting akinete differentiation in Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria). Freshw. Biol. 2005, 50, 345–352. [Google Scholar] [CrossRef]
- Sukenik, A.; Kaplan-Levi, R.N.; Viner-Mozzini, Y.; Lupu, A.; Sela, D. Induction, Isolation and Counting of Akinetes in Aphanizomenon ovalisporum. Bio-protoc 2016, 6, e1808. [Google Scholar] [CrossRef]
- Kwon, D.; Kim, K.; Jo, H.; Lee, S.D.; Yun, S.M.; Park, C. Environmental factors affecting akinete germination and resting cell awakening of two cyanobacteria. Appl. Microsc. 2023, 53, 1–13. [Google Scholar] [CrossRef]
- Legrand, B.; Miras, Y.; Beauger, A.; Dussauze, M.; Latour, D. Akinetes and ancient DNA reveal toxic cyanobacterial recurrences and their potential for resurrection in a 6700-year-old core from a eutrophic lake. Sci. Total Environ. 2019, 687, 1369–1380. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Moon, C.-H.; Cho, H.-J. Comparison of panning and sodium polytungstate methods for separating dinoflagellate cysts. J. Fish Aquat. Sci. 2008, 41, 228–231. [Google Scholar] [CrossRef]
- Kang, Y.J. Distribution of Dinoflagellate Cyst and Germination Using SPT Solution in Eutrophic Region of Southern Coastal Waters of Korea; Pukyong National University: Busan, Republic of Korea, 2007. [Google Scholar]
- D’agostino, P.M.; Woodhouse, J.N.; Makower, A.K.; Yeung, A.C.; Ongley, S.E.; Micallef, M.L.; Moffitt, M.C.; Neilan, B.A. Advances in genomics, transcriptomics and proteomics of toxin-producing cyanobacteria. Environ. Microbiol. Rep. 2016, 8, 3–13. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Zhang, Y.; Peng, W. Microbial community structure and functional properties in permanently and seasonally flooded areas in Poyang Lake. Sci. Rep. 2020, 10, 1–11. [Google Scholar]
- Rasmussen, J.P.; Monis, P.T.; Saint, C.P. Early Detection of Cyanobacterial Toxins Using Genetic Methods; American Water Works Association Research Foundation: Denver, CO, USA, 2008. [Google Scholar]
- Tsao, H.-W.; Michinaka, A.; Yen, H.-K.; Giglio, S.; Hobson, P.; Monis, P.; Lin, T.-F. Monitoring of geosmin producing Anabaena circinalis using quantitative PCR. Water Res. 2014, 49, 416–425. [Google Scholar] [CrossRef]
- Ueno, Y.; Nagata, S.; Tsutsumi, T.; Hasegawa, A.; Watanabe, M.F.; Park, H.-D.; Chen, G.-C.; Chen, G.; Yu, S.-Z. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogs 1996, 17, 1317–1321. [Google Scholar] [CrossRef]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative real-time PCR for determination of microcystin synthetase E copy numbers for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef]
- Ramm, J.; Lupu, A.; Hadas, O.; Ballot, A.; Rücker, J.; Wiedner, C.; Sukenik, A. A CARD-FISH protocol for the identification and enumeration of cyanobacterial akinetes in lake sediments. FEMS Microbiol. Ecol. 2012, 82, 23–36. [Google Scholar] [CrossRef]
- Wootton, J.C.; Federhen, S. Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 1993, 17, 149–163. [Google Scholar] [CrossRef]
- Kodama, Y.; Shumway, M.; Leinonen, R. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2011, 40, D54–D56. [Google Scholar] [CrossRef]
- Park, Y.M.; Squizzato, S.; Buso, N.; Gur, T.; Lopez, R. The EBI search engine: EBI search as a service—Making biological data accessible for all. Nucleic Acids Res. 2017, 45, W545–W549. [Google Scholar] [CrossRef]
- Li, X.; Dreher, T.W.; Li, R. An overview of diversity, occurrence, genetics and toxin production of bloom-forming Dolichospermum (Anabaena) species. Harmful Algae 2016, 54, 54–68. [Google Scholar] [CrossRef]
- Stein, J.R. Handbook of Phycological Methods: Culture Methods and Growth Measurements; CUP archive; Cambridge University Press: Cambridge, UK, 1979; Volume 1. [Google Scholar]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171. [Google Scholar] [CrossRef]
- Park, C.-H.; Park, M.-H.; Kim, K.H.; Park, J.-H.; Kwon, D.-R.; Kim, N.Y.; Lim, B.-J.; Hwang, S.-J. Akinete germination chamber: An experimental device for cyanobacterial akinete germination and plankton emergence. Harmful Algae 2018, 72, 74–81. [Google Scholar] [CrossRef]
- Egeler, P.; Römbke, J.; Meller, M.; Knacker, T.; Nagel, R. Bioaccumulation test with Tubificid Sludgeworms in artificial media–development of a standardisable method. Hydrobiologia 1999, 406, 271–280. [Google Scholar] [CrossRef]
- Li, R.; Watanabe, M.; Watanabe, M.M. Taxonomic studies of planktic species of Anabaena based on morphological characteristics in cultured strains. Hydrobiologia 2000, 438, 117–138. [Google Scholar] [CrossRef]
- Park, C.H. Study on the Akinete Life Cycle and Vegetative Cell Dynamics in a Harmful Cyanobacterium, Dolichospermum circinale (Nostocales); Konkuk University: Seoul, Republic of Korea, 2018. [Google Scholar]
- Guillard, R.R. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Springer: Berlin/Heidelberg, Germany, 1975; pp. 29–60. [Google Scholar]
- Kruger, N.J. The Bradford method for protein quantitation. In The Protein Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2009; pp. 17–24. [Google Scholar]
- Shevchenko, A.; Tomas, H.; Havli, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856. [Google Scholar] [CrossRef]
- Maier, T.; Kostrzewa, M. Fast and reliable MALDI-TOF MS-based microorganism identification. Chim. Oggi 2007, 25, 68. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015, 44, D733–D745. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 2014, 43, D571–D577. [Google Scholar] [CrossRef]
- Pisitkun, T.; Hoffert, J.D.; Saeed, F.; Knepper, M.A. NHLBI-AbDesigner: An online tool for design of peptide-directed antibodies. Am. J. Physiol. Cell Physiol. 2011, 302, C154–C164. [Google Scholar] [CrossRef]
- Cardemil, L.; Wolk, C.P. The polysaccharides from heterocyst and spore envelopes of a blue-green alga. Methylation analysis and structure of the backbones. Biol. Chem. 1976, 251, 2967–2975. [Google Scholar] [CrossRef]
- Cardemil, L.; Wolk, C.P. Polysaccharides from the envelopes of heterocysts and spores of the blue-green algae Anabaena variabilis and Cylindrospermum licheniforme. J. Phycol. 1981, 17, 234–240. [Google Scholar] [CrossRef]
- Graham, L.E.; Wilcox, L.W. Algae; Prentice Hall: Upper Saddler River, NJ, USA, 2000; Volume 25, p. 29. [Google Scholar]
- Maheshwari, R.; Bharadwaj, G.; Bhat, M.K. Thermophilic fungi: Their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000, 64, 461–488. [Google Scholar] [CrossRef]
- Raugei, G.; Ramponi, G.; Chiarugi, P. Low molecular weight protein tyrosine phosphatases: Small, but smart. Cell Mol. Life Sci. 2002, 59, 941–949. [Google Scholar] [CrossRef]
- da Silva Aires, R.; Steindorff, A.S.; Ramada, M.H.S.; de Siqueira, S.J.L.; Ulhoa, C.J. Biochemical characterization of a 27 kDa 1, 3-β-D-glucanase from Trichoderma asperellum induced by cell wall of Rhizoctonia solani. Carbohydr. Polym. 2012, 87, 1219–1223. [Google Scholar] [CrossRef]
- Zhou, R.; Wolk, C.P. Identification of an akinete marker gene in Anabaena variabilis. J. Bacteriol. Res. 2002, 184, 2529–2532. [Google Scholar] [CrossRef]
- Qiu, Y.; Gu, L.; Brözel, V.; Whitten, D.; Hildreth, M.; Zhou, R. Unique proteomes implicate functional specialization across heterocysts, akinetes, and vegetative cells in Anabaena cylindrica. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bryant, D.A. The Molecular Biology of Cyanobacteria; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006; Volume 1. [Google Scholar]
- Flores, F.G. The Cyanobacteria: Molecular Biology, Genomics, and Evolution; Horizon Scientific Press: Poole, UK, 2008. [Google Scholar]
- Perez, R.; Forchhammer, K.; Salerno, G.; Maldener, I. Clear differences in metabolic and morphological adaptations of akinetes of two Nostocales living in different habitats. Microbiology 2016, 162, 214–223. [Google Scholar] [CrossRef]
- Singh, H.; Sunita, K. A biochemical study of spore germination in the blue-green alga Anabaena doliolum. J. Exp. Bot. 1974, 25, 837–845. [Google Scholar] [CrossRef]
- Zheng, L.; Donovan, W.P.; Fitz-James, P.C.; Losick, R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988, 2, 1047–1054. [Google Scholar] [CrossRef]
- Abhyankar, W.; de Koning, L.J.; Brul, S.; de Koster, C.G. Spore proteomics: The past, present and the future. FEMS Microbiol. Lett. 2014, 358, 137–144. [Google Scholar] [CrossRef][Green Version]
- Setlow, B.; Hand, A.; Setlow, P. Synthesis of a Bacillus subtilis small, acid-soluble spore protein in Escherichia coli causes cell DNA to assume some characteristics of spore DNA. J. Bacteriol. 1991, 173, 1642–1653. [Google Scholar] [CrossRef]
- Zhang, G.; An, Y.; Zabed, H.M.; Guo, Q.; Yang, M.; Yuan, J.; Li, W.; Sun, W.; Qi, X. Bacillus subtilis Spore Surface Display Technology: A Review of Its Development and Applications. J. Microbiol. Biotechnol. 2019, 29, 179–190. [Google Scholar]
- Abel-Santos, E. Endospores, sporulation and germination. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 163–178. [Google Scholar]
- Ow, S.Y.; Cardona, T.; Taton, A.; Magnuson, A.; Lindblad, P.; Stensjö, K.; Wright, P.C. Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J. Proteome Res. 2008, 7, 1615–1628. [Google Scholar] [CrossRef]
- Brandão-Dias, P.F.; Rosi, E.J.; Shogren, A.J.; Tank, J.L.; Fischer, D.T.; Egan, S.P. Fate of environmental proteins (eProteins) from genetically Engineered crops in Streams is controlled by Water pH and ecosystem metabolism. Environ. Sci.Technol. 2021, 55, 4688–4697. [Google Scholar] [CrossRef]
- Pandey, S.; Rai, R.; Rai, L.C. Proteomics combines morphological, physiological and biochemical attributes to unravel the survival strategy of Anabaena sp. PCC7120 under arsenic stress. J. Proteom. 2012, 75, 921–937. [Google Scholar] [CrossRef]
- Panda, B.; Basu, B.; Rajaram, H.; Kumar Apte, S. Methyl viologen responsive proteome dynamics of Anabaena sp. strain PCC7120. Proteomics 2014, 14, 1895–1904. [Google Scholar] [CrossRef]
- Battchikova, N.; Muth-Pawlak, D.; Aro, E.-M. Proteomics of cyanobacteria: Current horizons. Curr. Opin. Biotechnol. 2018, 54, 65–71. [Google Scholar] [CrossRef]
- Sound, J.K.; Bellamy-Carter, J.; Leney, A.C. The increasing role of structural proteomics in cyanobacteria. Essays Biochem. 2023, 67, 269–282. [Google Scholar]

| No. | Amino Acid Sequence | No. |
|---|---|---|
| 1 | MKWVTFISLL LLFSSAYSRG VFRRDTHKSE IAHRFKDLGE EHFKGLVLIA | 50 |
| 51 | FSQYLQQCPF DEHVKLVNEL TEFAKTCVAD ESHAGCEKSL HTLFGDELCK | 100 |
| 101 | VASLRETYGD MADCCEKQEP ERNECFLSHK DDSPDLPKLK PDPNTLCDEF | 150 |
| 151 | KADEKKFWGK YLYEIARRHP YFYAPELLYY ANKYNGVFQE CCQAEDKGAC | 200 |
| 201 | LLPKIETMRE KVLASSARQR LRCASIQKFG ERALKAWSVA RLSQKFPKAE | 250 |
| 251 | FVEVTKLVTD LTKVHKECCH GDLLECADDR ADLAKYICDN QDTISSKLKE | 300 |
| 301 | CCDKPLLEKS HCIAEVEKDA IPENLPPLTA DFAEDKDVCK NYQEAKDAFL | 350 |
| 351 | GSFLYEYSRR HPEYAVSVLL RLAKEYEATL EECCAKDDPH ACYSTVFDKL | 400 |
| 401 | KHLVDEPQNL IKQNCDQFEK LGEYGFQNAL IVRYTRKVPQ VSTPTLVEVS | 450 |
| 451 | RSLGKVGTRC CTKPESERMP CTEDYLSLIL NRLCVLHEKT PVSEKVTKCC | 500 |
| 501 | TESLVNRRPC FSALTPDETY VPKAFDEKLF TFHADICTLP DTEKQIKKQT | 550 |
| 551 | ALVELLKHKP KATEEQLKTV MENFVAFVDK CCAADDKEAC FAVEGPKLVV | 600 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Park, C.-H.; Hwang, S.-J. Analyzing the Akinete Protein of the Harmful Freshwater Cyanobacterium, Dolichospermum circinale. Water 2023, 15, 2746. https://doi.org/10.3390/w15152746
Kim K, Park C-H, Hwang S-J. Analyzing the Akinete Protein of the Harmful Freshwater Cyanobacterium, Dolichospermum circinale. Water. 2023; 15(15):2746. https://doi.org/10.3390/w15152746
Chicago/Turabian StyleKim, Keonhee, Chae-Hong Park, and Soon-Jin Hwang. 2023. "Analyzing the Akinete Protein of the Harmful Freshwater Cyanobacterium, Dolichospermum circinale" Water 15, no. 15: 2746. https://doi.org/10.3390/w15152746
APA StyleKim, K., Park, C.-H., & Hwang, S.-J. (2023). Analyzing the Akinete Protein of the Harmful Freshwater Cyanobacterium, Dolichospermum circinale. Water, 15(15), 2746. https://doi.org/10.3390/w15152746








