Antibacterial Activity of Bioactive Compounds Extracted from the Egyptian Untapped Green Alga Rhizoclonium hieroglyphicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Origin and Culture Conditions
2.2. Bacterial Strains
2.3. Preparation of the Algal Extracts
2.4. Antibacterial Assay of the Algal Extracts
2.5. Determination of the Minimal Inhibitory Concentrations (MICs) of the Algal Extracts
2.6. Identification of the Bioactive Compounds of the R. hieroglyphicum Ethanolic Extract
2.7. Statistical Analysis
3. Results
3.1. Antibacterial Activity of the Different R. hieroglyphicum Extracts
3.2. Antibacterial Action of the Positive Standard Antibiotics
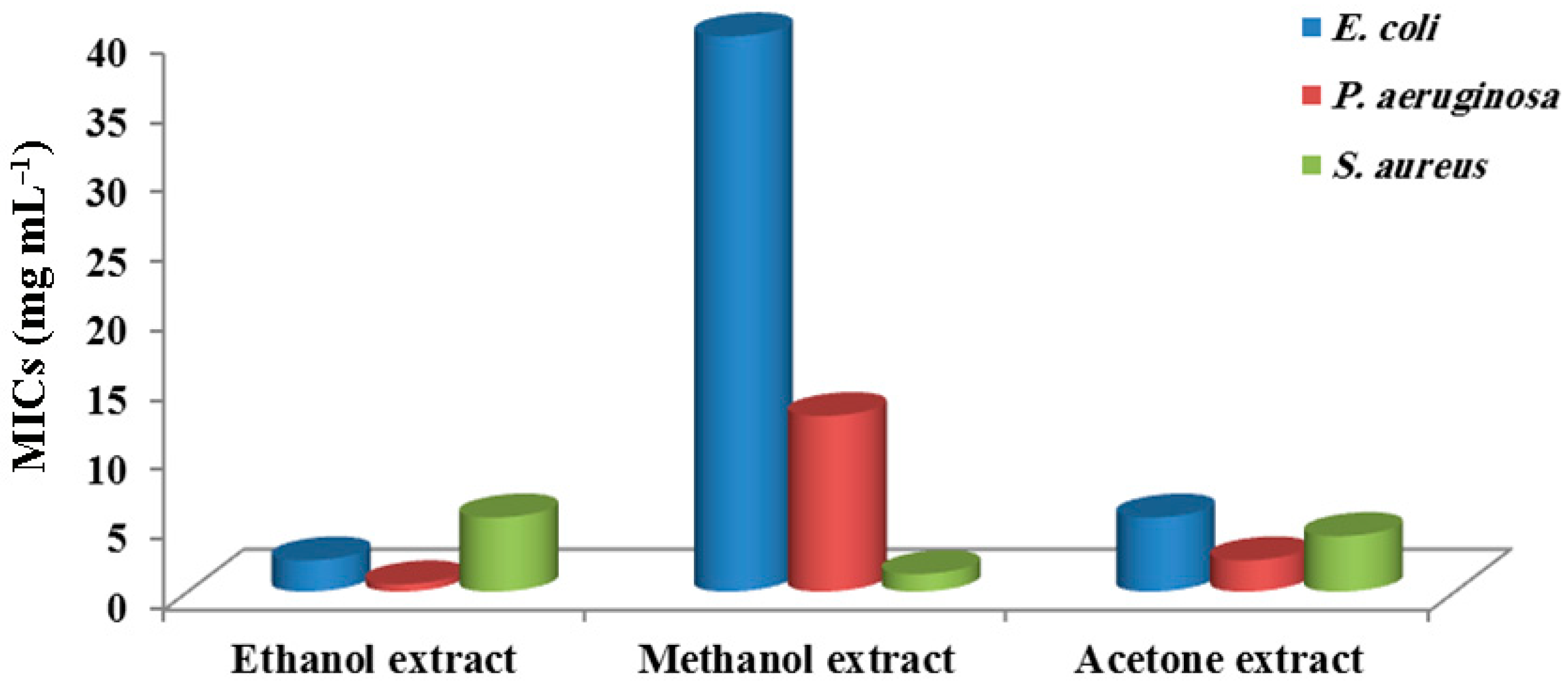
3.3. Minimum Inhibitory Concentrations (MICs) of R. hieroglyphicum Extracts
3.4. Phytochemical Constituents of the Rhizoclonium hieroglyphicum Ethanolic Extract
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leliaert, F.; Boedeker, C. Cladophorales. In Green Seaweeds of Britain and Ireland; Brodie, J., Maggs, C.A., John, D., Eds.; Natural History Museum Publications: London, UK, 2007; pp. 131–183. [Google Scholar]
- Zhao, Z.J.; Zhu, H.; Liu, G.X.; Hu, Z.Y. Rhizoclonium ramosum sp. nov. (Cladophorales, Chlorophyta), a new freshwater algal species from China. Fottea 2016, 16, 12–21. [Google Scholar] [CrossRef]
- Boedeker, C.; Leliaert, F.; Timoshkin, O.A.; Vishnyakov, V.S.; Diaz-Martinez, S.; Zuccarello, G.C. The endemic Cladophorales (Ulvophyceae) of ancient Lake Baikal represent a monophyletic group of very closely related by morphologically diverse species. J. Phycol. 2018, 54, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Saber, A.A.; Ichihara, K.; Cantonati, M. Molecular phylogeny and detailed morphological analysis of two freshwater Rhizoclonium strains from contrasting spring types in Egypt and Italy. Plant Biosyst. 2017, 151, 800–812. [Google Scholar] [CrossRef]
- Ruperez, P.; Ahrazem, O.; Leal, J.A. Potential antioxidant capacity of sulphated polysaccharides from the edible marine brown seaweeds Fucus vesiculosus. J. Agric. Food Chem. 2002, 50, 840–845. [Google Scholar] [CrossRef]
- Mungmai, L.; Jiranusornkul, S.; Peerapornpisal, Y.; Sirithunyalug, B.; Leelapornpisid, P. Extraction, characterization and biological activities of extracts from freshwater macroalga [Rhizoclonium hieroglyphicum (C. Agardh) Kützing] cultivated in northern Thailand. Chiang Mai J. Sci. 2014, 41, 14–26. [Google Scholar]
- Mohamed, S.S.; Saber, A.A. Antifungal potential of the bioactive constituents in extracts of the mostly untapped brown seaweed Hormophysa cuneiformis from the Egyptian coastal waters. Egypt. J. Bot. 2019, 59, 695–708. [Google Scholar] [CrossRef]
- Ibrahim, R.Y.M.; Hammad, H.B.I.; Gaafar, A.A.; Saber, A.A. The possible role of the seaweed Sargassum vulgare as a promising functional food ingredient minimizing aspartame-associated toxicity in rats. Int. J. Environ. Health Res. 2022, 32, 752–771. [Google Scholar] [CrossRef]
- El-Sheekh, M.; Fathy, A.A.; Saber, H.; Saber, A.A. Medicinal and pharmaceutical applications of seaweeds. Egypt. J. Bot. 2023, 63, 1–29. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; Daboor, S.M.; Swelim, M.A.; Mohamed, S. Production and characterization of antimicrobial active substance from Arthrospira platensis. Iran. J. Microbiol. 2014, 6, 112–119. [Google Scholar]
- El-Sheekh, M.; Hassan, L.H.; Morsi, H.H. Evaluation of antimicrobial activities of blue-green algae mediated silver and gold nanoparticles. Rend. Lincei. 2021, 32, 747–757. [Google Scholar] [CrossRef]
- Ghazala, B.; Shameel, M.; Choudhary, M.I.; Shahzad, S.; Leghari, S.M. Phycochemistry and bioactivity of certain freshwater green algae of Sindh. Pak. J. Bot. 2003, 35, 695–704. [Google Scholar]
- Elshouny, W.A.; El-Sheekh, M.M.; Sabae, S.Z.; Khalil, M.A.; Badr, H.M. Antimicrobial activity of Spirulina platensis against aquatic bacterial isolates. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 1203–1208. [Google Scholar] [CrossRef]
- Abd El-Hack, M.E.; Abdelnour, S.A.; Alagawany, M.; Abdo, M.; Sakr, M.A.; Khafaga, A.F.; Mahgoub, S.A.; Elnesr, S.S.; Gebriel, M.G. Microalgae in modern cancer therapy: Current knowledge. Biomed. Pharmacother. 2019, 11, 42–50. [Google Scholar] [CrossRef]
- Tsvetanova, F.; Yankov, D. Bioactive compounds from red microalgae with therapeutic and nutritional value. Microorganisms 2022, 10, 2290. [Google Scholar] [CrossRef] [PubMed]
- Ismail, G.; El-Sheekh, M.M.; Gheda, S.; Samy, R. Antimicrobial, antioxidant and antiviral activities of biosynthesized silver nanoparticles by phycobiliproteins crude extract of the cyanobacteria Spirulina platensis and Nostoc linckia. Bionanoscience 2021, 11, 355–370. [Google Scholar] [CrossRef]
- Ismail, A.; Ktari, L.; Romdhane, Y.; Aoun, B.; Sadok, S.; Boudabous, A.; El Bour, M. Antimicrobial fatty acids from the green alga Ulva rigida (Chlorophyta). BioMed Res. Int. 2018, 2018, 3069595. [Google Scholar] [CrossRef]
- Santoyo, S.; Rodríguez-Meizoso, I.; Cifuentes, A.; Jaime, L.; García-Blairsy Reina, G.; Señorans, F.J.; Ibáñez, E. Green processes based on the extraction with pressurized fluids to obtain potent antimicrobials from Haematococcus pluvialis microalgae. LWT-Food Sci. Technol. 2009, 42, 1213–1218. [Google Scholar] [CrossRef]
- Guedes, A.C.; Barbosa, C.R.; Amaro, H.M.; Pereira, C.I.; Malcata, F.X. Microalgal and cyanobacterial cell extracts for use as natural antibacterial additives against food pathogens. Int. J. Food Sci. Technol. 2011, 46, 862–870. [Google Scholar] [CrossRef]
- Desbois, A.P.; Mearns-Spragg, A.; Smith, V.J. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria including multi-resistant Staphylococcus aureus (MRSA). Mar. Biotechnol. 2009, 11, 45–52. [Google Scholar] [CrossRef]
- Jeevanantham, G.; Vinoth, M.; Hussain, J.M.; Muruganantham, P.; Ahamed, A.K. Biochemical characterization of five marine cyanobacteria species for their biotechnological applications. J. Pharmacogn. Phytochem. 2019, 8, 35–40. [Google Scholar]
- Gutierrez, R.M.P.; Baez, E.G. Diterpenoids from the freshwater green algae Rhizoclonium hieroglyphicum with antibacterial activity. J. Asian Nat. Prod. Res. 2011, 13, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Ilavarasi, A.; Mubarakali, D.; Praveenkumar, R.; Baldev, E.; Thajuddin, N. Optimization of various growth media to freshwater microalgae for biomass production. Biotechnology 2011, 10, 540–545. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; El Sabagh, S.; Abou-El-Souod, G.W.; Elbeltagy, A. The effect of different growth conditions on the biomass and chemical constituents of Chlorella vulgaris. Egypt. J. Exp. Biol. Bot. 2018, 14, 121–131. [Google Scholar]
- Abubakar, A.R.; Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pridham, T.G.; Lindenfelser, L.A.; Shotwell, O.L.; Stodola, F.H.; Penedict, R.G.; Foley, C.; Jackson, P.W.; Zaumeyer, W.J.; Perston, W.H.; Miychell, J.W. Antibiotics against plant diseases. I. Laboratory and greenhouse survey. Phytopathology 1956, 46, 568–575. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Marrez, D.A.; Sultan, Y.Y.; Embaby, M.A. Biological activity of the cyanobacterium Oscillatoria brevis extracts as a source of nutraceutical and biopreservative agents. Int. J. Pharmacol. 2017, 13, 1010–1019. [Google Scholar] [CrossRef]
- Hosseini, N.; Akhavan, A.; Nowruzi, B. Detection and relation of polyketide synthase (PKSs) genes with antimicrobial activity in terrestrial cyanobacteria of Lavasan. Iran. J. Med. Microbiol. 2019, 12, 419–431. [Google Scholar] [CrossRef]
- Al-Abdulameer, S.H. The wide spectrum activity of Chlorella and Spirulina extracts on the viability of pathogenic and environmental bacteria in Baghdad, Iraq. Egypt. J. Aquat. Biol. Fish. 2022, 26, 1–11. [Google Scholar] [CrossRef]
- Rani, B.V.; Kumari, B.V.; Nair, S.G. Studies on antibacterial activity of two filamentous algae against a few human pathogens. J. Theor. Biol. 2018, 13, 39–42. [Google Scholar]
- Morsi, H.H.; El-Sabbagh, S.M.; Mehesen, A.A.; Mohamed, A.D. Bioactivity of natural compounds extracted from Scenedesmus obliquus toward some pathogenic bacteria. IJAFE 2022, 6, 1–11. [Google Scholar]
- Yotova, T.T.; Georgieva, A.; Lliev, I.; Ivanova, A.; Pilarski, P.; Toshkova, R. Antitumor and antimicrobial activity of fatty acids from green microalga Coelastrella sp. BGV. S. Afr. J. Bot. 2022, 151, 394–402. [Google Scholar] [CrossRef]
- Sudalayandi, K.; Kumar, A.; Sessler, R.; Sayre, R.T.; Falcoa, V.; Ihemere, U.; Ndunguru, J.; Narayanan, N. Determination of fatty acids and proteins from the fresh water alga Chlamydomonas reinhardtii cc2137 and its antagonism against aquatic bacteria. Pak. J. Bot. 2012, 44, 2139–2144. [Google Scholar]
- Pohl, C.H.; Kock, J.L.F.; Thibane, V.S. Antifungal free fatty acids: A review. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; pp. 61–71. [Google Scholar]
- Zheng, C.J.; Yoo, J.-S.; Lee, T.-G.; Choc, H.-Y.; Kim, Y.-H.; Kim, W.-G. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 2005, 579, 5157–5162. [Google Scholar] [CrossRef] [PubMed]
- Kabara, J.J.; Swieczkowski, D.M.; Conley, A.J.; Truant, J.P. Fatty acids and derivatives as antimicrobial agents. Antimicrob. Agents Chemother. 1972, 2, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Q.; O’Connor, C.J.; Roberton, A.M. Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2003, 36, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Seidel, V.; Taylor, P.W. In vitro activity of extracts and constituents of Pelagonium against rapidly growing mycobacteria. Int. J. Antimicrob. Agents 2004, 23, 613–619. [Google Scholar] [CrossRef]
- Cepas, V.; Gutiérrez-Del-Río, I.; López, Y.; Redondo-Blanco, S.; Gabasa, Y.; Iglesias, M.J.; Soengas, R.; Fernández-Lorenzo, A.; López-Ibáñez, S.; Villar, C.J.; et al. Microalgae and cyanobacteria strains as producers of lipids with antibacterial and antibiofilm activity. Mar. Drugs 2021, 19, 675. [Google Scholar] [CrossRef]
- Afzal, S.; Yadav, A.K.; Poonia, A.K.; Choure, K.; Yadav, A.N.; Pandey, A. Antimicrobial therapeutics isolated from algal source: Retrospect and prospect. Biologia 2023, 78, 291–305. [Google Scholar] [CrossRef]

| Algal Extracts | Conc. (mg mL–1) | E. coli | P. aeruginosa | S. aureus |
|---|---|---|---|---|
| ethanolic extract | 40 | 15.2 ± 0.18 a | 18.2 ± 0.22 a | 13.2 ± 0.17 a |
| 20 | 13.3 ± 0.17 b | 16.3 ± 0.25 b | 11.3 ± 0.23 a | |
| 10 | 11.3 ± 0.26 b | 14.3 ± 0.27 c | 8.28 ± 0.39 b | |
| 5 | 7.21 ± 0.17 c | 10.2 ± 0.09 d | – | |
| methanolic extract | 40 | 7.12 ± 0.05 a | 10.2 ± 0.08 a | 15.1 ± 0.06 a |
| 20 | – | 8.12 ± 0.04 b | 13.2 ± 0.13 a | |
| 10 | – | – | 10.3 ± 0.29 b | |
| 5 | – | – | 9.2 ± 0.12 c | |
| acetone extract | 40 | 13.2 ± 0.19 a | 14.3 ± 0.19 a | 13.3 ± 0.18 a |
| 20 | 11.3 ± 0.20 b | 12.2 ± 0.11 b | 11.3 ± 0.18 a | |
| 10 | 8.32 ± 0.17 b | 10.3 ± 0.26 c | 9.20 ± 0.18 b | |
| 5 | – | 8.15 ± 0.11 d | 7.15 ± 0.11 c |
| Antibiotics | Average Diameters of Inhibition Zones (mm) | ||
|---|---|---|---|
| E. coli | P. aeruginosa | S. aureus | |
| ampicillin (10 μg) | – | – | 7.37 ± 0.31 a |
| amoxicillin (25 μg) | 19.1 ± 0.33 b | – | 21.2 ± 0.26 a |
| cefadroxil (30 μg) | – | – | 10.2 ± 0.18 a |
| doxycycline (30 μg) | 15.2 ± 0.22 b | 9.11± 0.08 c | 19.2 ± 0.22 a |
| cefoxitin (30 μg) | 9.19 ± 0.19 b | – | 11.2 ± 0.19 a |
| ofloxacin (5 μg) | 16.4 ± 0.36 b | 6.12 ± 0.0 c | 13.4 ± 0.29 a |
| vancomycin (30 μg) | – | – | 11.2 ± 0.22 a |
| No. | Compounds Identified | RT (min) | Peak Area (%) | Norm (%) |
|---|---|---|---|---|
| 1 | 2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy-trimethylsilane | 32.214 | 9.272 | 100.00 |
| 2 | Octadecanoic acid, ethyl ester (stearic acid C18:0) | 30.644 | 8.030 | 86.60 |
| 3 | 2-[2-[2-[2-[2-[2-[2-(2methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy-trimethylsilane | 34.375 | 5.720 | 61.69 |
| 4 | tert-butyl-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy] ethoxy]dimethylsilane | 29.088 | 5.318 | 57.36 |
| 5 | Hexadecanoic acid, ethyl ester (palmitic acid C16:0) | 27.698 | 5.161 | 55.66 |
| 6 | {2,2-dimethyl-5-[2-(2-trimethylsilylethoxymethoxy)propyl][1,3]dioxolan-4-yl}methanol | 32.059 | 4.532 | 48.88 |
| 7 | 9,12,15-octadecatrienoic acid, 2-[(trimethylsilyl)oxy]-1-[[(trimethylsilyl)oxy]methyl]ethyl ester, (Z,Z,Z)-(α-linolenic acid C18:3; ω−3) | 34.540 | 4.427 | 47.75 |
| 8 | 1,4,7,10,13,16,19-heptaoxa-2-cycloheneicosanone | 28.988 | 3.827 | 41.28 |
| 9 | (1S,14S)-bicyclo[12.10.0]-3,6,9,12,15,18,21,24-octaoxatetracosane | 34.495 | 2.927 | 31.57 |
| 10 | Oleic acid, ethyl ester (C18:1, ω–9) | 36.456 | 2.752 | 29.68 |
| 11 | 1,2-hexanediol, 2-methyl- | 23.246 | 2.702 | 29.14 |
| 12 | 1,2-benzenedicarboxylic acid, diisooctyl ester | 34.020 | 1.578 | 17.02 |
| 13 | Cyclotetrasiloxane, octamethyl- | 8.405 | 1.473 | 15.89 |
| 14 | Tetradecanoic acid, ethyl ester (myristic acid C14:0) | 22.881 | 0.823 | 8.88 |
| 15 | 4-ethylbenzoic acid, 2-butyl ester | 7.014 | 0.778 | 8.39 |
| 16 | 2-dimethylsilyloxytridecane | 11.071 | 0.775 | 8.36 |
| 17 | [1,1′-bicyclopropyl]-2-octanoic acid, 2′-hexyl-, methyl ester | 32.549 | 0.721 | 7.78 |
| 18 | Cyclopentasiloxane, decamethyl | 11.706 | 0.629 | 6.78 |
| 19 | 3,7,11,15-tetramethyl-2-hexadecen-1-ol (phytol) | 29.548 | 0.616 | 6.65 |
| 20 | 1-propanol, 3,3′-oxybis- | 10.260 | 0.530 | 5.71 |
| 21 | 3-deoxyglucose | 8.765 | 0.435 | 4.69 |
| 22 | Cyclohexasiloxane, dodecamethyl- | 14.847 | 0.428 | 4.61 |
| 23 | 3-Isopropoxy-1,1,1,7,7,7-hexamethyl-3,5,5-tris(trimethylsiloxy)tetrasiloxane | 17.573 | 0.325 | 3.51 |
| 24 | Phenol, 2,2′-methylenebis[6-(1,1-dimethylethyl)-4-methyl- | 32.945 | 0.321 | 3.46 |
| 25 | 1,3,5-pentanetriol, 3-methyl | 10.185 | 0.301 | 3.24 |
| 26 | Bicyclo[2.1.1]hexan-2-ol, 2-ethenyl- | 6.419 | 0.276 | 2.98 |
| 27 | Octaethylene glycol monododecyl ether | 15.212 | 0.245 | 2.64 |
| 28 | Octasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyl- | 35.541 | 0.242 | 2.61 |
| 29 | Cyclononasiloxane, octadecamethyl | 30.759 | 0.228 | 2.46 |
| 30 | Ethyl oleate | 30.294 | 0.225 | 2.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morsi, H.H.; El-Sabbagh, S.M.; Mehesen, A.A.; Mohamed, A.D.; Al-Harbi, M.; Elkelish, A.; El-Sheekh, M.M.; Saber, A.A. Antibacterial Activity of Bioactive Compounds Extracted from the Egyptian Untapped Green Alga Rhizoclonium hieroglyphicum. Water 2023, 15, 2030. https://doi.org/10.3390/w15112030
Morsi HH, El-Sabbagh SM, Mehesen AA, Mohamed AD, Al-Harbi M, Elkelish A, El-Sheekh MM, Saber AA. Antibacterial Activity of Bioactive Compounds Extracted from the Egyptian Untapped Green Alga Rhizoclonium hieroglyphicum. Water. 2023; 15(11):2030. https://doi.org/10.3390/w15112030
Chicago/Turabian StyleMorsi, Hanaa H., Sabha M. El-Sabbagh, Ahlam A. Mehesen, Ahmed D. Mohamed, Maha Al-Harbi, Amr Elkelish, Mostafa M. El-Sheekh, and Abdullah A. Saber. 2023. "Antibacterial Activity of Bioactive Compounds Extracted from the Egyptian Untapped Green Alga Rhizoclonium hieroglyphicum" Water 15, no. 11: 2030. https://doi.org/10.3390/w15112030
APA StyleMorsi, H. H., El-Sabbagh, S. M., Mehesen, A. A., Mohamed, A. D., Al-Harbi, M., Elkelish, A., El-Sheekh, M. M., & Saber, A. A. (2023). Antibacterial Activity of Bioactive Compounds Extracted from the Egyptian Untapped Green Alga Rhizoclonium hieroglyphicum. Water, 15(11), 2030. https://doi.org/10.3390/w15112030










