Abstract
Atomic force microscopy (AFM) is a method that provides the nanometer-resolution three-dimensional imaging of living cells in their native state in their natural physiological environment. In addition, AFM’s sensitivity to measure interaction forces in the piconewton range enables researchers to probe surface properties, such as elasticity, viscoelasticity, hydrophobicity and adhesion. Despite the growing number of applications of AFM as a method to study biological systems, AFM is not yet an established technique for studying microalgae. Following a brief introduction to the basic principles and operation modes of AFM, this review highlights the major contributions of AFM in the field of microalgae research. A pioneering AFM study on microalgae was performed on diatoms, revealing the fine structural details of diatom frustule, without the need for sample modification. While, to date, diatoms are the most studied class of microalgae using AFM, it has also been used to study microalgae belonging to other classes. Besides using AFM for the morphological characterization of microalgae at the single cell level, AFM has also been used to study the surface properties of microalgal cells, with cell elasticity being most frequently studied one. Here, we also present our preliminary results on the viscoelastic properties of microalgae cell (Dunaliella tertiolecta), as the first microrheological study of microalgae. Overall, the studies presented show that AFM, with its multiparametric characterization, alone or in combination with other complementary techniques, can address many outstanding questions in the field of microalgae.
1. Introduction
Since its invention in 1986, atomic force microscopy (AFM) has grown to be one of the most important method for characterizing biological samples at the nanometer scale [1]. AFM is a versatile tool for the high-resolution three-dimensional imaging, nanomechanical characterization and measurement of inter- and intramolecular forces in living and non-living structures [2]. The AFM probe, which has a nm-sized tip, measures the interatomic forces between the sample surface and the tip apex. Sample preparation for AFM measurements is simple, and there is no need for the freezing, metal coating or staining of the sample. As a result, there is little-to-no damage to the sample, and the functions of biological systems can be preserved. AFM works in both air and liquids; so, physiological buffers and growth media can be used to study living cells. The high resolution of AFM allows the imaging of atoms on hard surfaces and molecules on soft biological samples. The ability of AFM to be used to measure nanomechanical properties has further extended its potential in various research areas, from materials science to biology. The resulting multiparametric images present high-resolution topography, with a local nanomechanical map of the same sample area. Since the invention of AFM, a wide range of biological samples have been studied, from biological macromolecules, including proteins, lipids, DNA and polysaccharides, to cells (see [3,4] and references therein).
Although the number of applications of AFM in biological research has increased exponentially in recent years [3,4,5,6], there are still a limited number of examples of its use in microalgal research. Traditional SEM and TEM have been used for the high-resolution imaging of many microalgal cells [7,8,9], but on dried and coated samples. In contrast, AFM can be used to image cells in their natural aqueous environment, thereby keeping them alive [10,11]. A pioneering AFM experiment on microalgae was performed on dried samples of six diatom species [12]. This study confirmed the ability of AFM to be used to study diatoms without the need to modify the samples. The imaging of live microalgal cells in their native environment soon followed. Almquist and co-workers [13] and Crawford and co-workers [14] utilized AFM to study the nanostructure of the live diatoms, Navicula pelliculosa and Pinnularia viridis, and they were among the first to image live microalgal cells. Although it is mainly used for diatom studies (see [15] and references therein), AFM has also been used in studies of other microalgal classes, including Dinophyceae [16,17], Chlorophyceae [18,19,20,21,22], Chlorodendrophyceae [21,22], Trebouxiophyceae [23,24] and Porphyridiophyceae [25]. In addition to imaging, AFM has also been used to study the micro- and nanomechanical properties of microalgal cells and has already been used to characterize the elastic properties of various microalgal species [13,15,20,21,22,26,27,28,29,30,31,32,33]. The first AFM experiments conducted to measure the elasticity of microalgal cells were performed by Almquist and co-workers [13] on the diatom, Navicula pelliculosa. They showed that the overall hardness and elasticity of the diatom shell resembled that of known silicas and confirmed that AFM is an important complementary method in the study of silica biomineralization. Further, using functionalized tips, AFM can be used to study the distribution of chemical groups on living cells using AFM mode known as chemical force microscopy (CFM) [34]. Pillet and co-workers [20] used CFM with a hydrophobic methyl-terminated tip, which provided hydrophobicity maps on a single living Dunaliella tertiolecta cell. The cells themselves attached to the end of the cantilever and could be used as AFM probes to study cell adhesion. Arce and co-workers [35] used AFM with a cell as a probe to study the adhesion of Navicula sp. cells to hydrophobic and hydrophilic surfaces. Other applications of AFM in the field of microalgae research include studies of the interaction of nanoparticles with microalgae [32,36], the study of colony formation [37], cell disruption studies [38], etc.
Despite numerous studies having been conducted to date using a variety of analytical techniques and approaches, many aspects of the unique field of microalgae have remained unexplored. AFM, with its multiparametric and multifunctional characterization, i.e., for imaging native biostructures with exceptional nano- and sub-nanometric-scale resolution and mapping surface properties alone or in combination with other complementary techniques, can address many open questions in the world of microalgae. Following a brief introduction to the basic principles and operation modes of AFM, this review highlights the major contributions and future perspectives that this emerging research field offers in the field of microalgae research.
2. AFM—Basic Principles and Modes of Operation
Atomic force microscopy is a type of scanning probe microscopy (SPM) and is one of the most commonly used ones. For the invention of the scanning tunneling microscope, the first type of SPM technique to be invented, Gerd Binnig and Heinrich Rohrer won the Nobel Prize in Physics and showed that it could be used to image single atoms [39]. This was followed by the invention of a variety of other scanning probe techniques, including AFM. AFM’s basic principle is based on the scanning of a sharp tip over a sample’s surface [1]. AFM not only provides three-dimensional topographic images of surfaces with nanometer to angstrom resolution, but it can also be used to study the forces between single molecules and the physical and mechanical properties of samples. The concept on which all scanning probe microscopes are based is to scan a probe above a sample surface, while monitoring the interaction between the probe and the surface. An AFM uses probe consisting of a nm-sized tip attached to the end of a flexible cantilever, and a laser light focused on the back of the cantilever is used to monitor the deflection of the cantilever through a four-quadrant photodiode detector (Figure 1). An XYZ scanner, usually made of piezoelectric material, raster scans the sample versus the tip in a line-by-line manner, while trying to keep the same distance between the tip and the sample [40,41]. The resulting three-dimensional topographic image is quantitative along all axes. Besides measuring topography, AFM can also be used to assess the sample’s properties, such as elasticity, viscoelasticity, adhesion and hydrophobicity. To perform these measurements, force–distance curves are acquired while the tip is pushed towards the sample, and then retracted back. The force–distance curves collected during this measurement can provide spatially resolved maps of the surface properties of the sample [42].
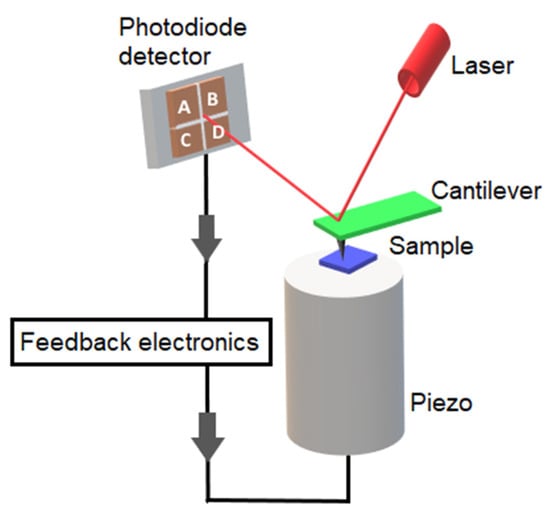
Figure 1.
Schematic presentation of atomic force microscope.
There are three major imaging modes in AFM: contact, intermittent contact and non-contact. In contact mode, the AFM tip is in continuous contact with the sample surface, while the probe raster scans the surface, and the interaction forces between the tip and the sample is repulsive. There are two modes of imaging in contact mode: constant force and constant height mode. In constant force mode, the tip height is continually adjusted using a piezoelectric scanner to maintain a specified deflection (force), while in constant height mode, the height of the scanner is constant, and deflection is monitored. During contact-mode imaging, the applied force and the frictional force can damage soft biological samples, and thus, the force must be carefully controlled. In intermittent contact mode (also called AC mode or tapping mode), the cantilever is oscillated by a few nm, and the probe lightly touches the sample. The forces between the tip and the sample induce changes in the resonant behavior of the cantilever, and obtained images are based on frequency, phase shifts and amplitude changes. The intermittent contact mode reduces the contact and lateral forces between the tip and the sample, thus preserving soft biological samples [40,41]. The third, least frequently used AFM mode of operation is non-contact (NC) mode, where the oscillating cantilever never touches the sample, and the forces between tip and the sample are attractive. In non-contact mode, a cantilever is oscillated near its resonant frequency (usually from 100 to 400 kHz), and the detection scheme, such as in the intermittent contact mode, is based on changes in the resonant frequency, phase or amplitude of the cantilever during scanning.
The high force sensitivity of AFM (down to 10−12 N) is one of its main benefits [40,43,44,45]. This allows AFM to be used in so-called force spectroscopy mode, in which force–distance curves are generated based on the deflection of the cantilever as it moves towards and away from the sample. The force–distance curve can be further converted into a force–indentation curve, the analysis of which can provide information about the surface’s properties, such as elasticity, viscoelasticity, adhesion and hydrophobicity. This type of force mapping can be conducted at multiple locations on the (x, y) plane to obtain spatially resolved maps of properties and interactions [3].
AFM force spectroscopy is most often used to quantify and map the elastic properties of samples (Young’s modulus). Elasticity is measured by indenting a tip into the sample and recording a force–distance (FD) curve. Different contact mechanical models can be used to derive Young’s modulus, and the oldest model developed by Hertz utilizes a spherical indenter applied to a perfectly flat, isotropic and homogeneous sample and is widely used [46]. To obtain the Young’s modulus of the sample, the FD curve must be transformed into a force–indentation curve, which depicts the force required to indent a sample at a certain depth [47]. The Young’s modulus of the sample can then be calculated as follows:
where E is Young’s modulus of the sample; ν is the Poisson’s ratio of the sample; F is the force applied by the cantilever; r is the radius of the cantilever tip; δ is the indentation depth. Several alternative models for measuring elasticity have been further developed, such as the Sneddon, Derjaguin–Muller–Toporov (DMT) and Johnson–Kendall–Roberts models (JKR) [48,49]. The selection of the right model for analysis is not trivial, since most biological samples do not fully satisfy all assumptions for existing contact mechanics models.
The most widely used way to analyze cell mechanics is to obtain an apparent elastic modulus, considering that the cell is purely elastic. However, in mechanical characterization experiments on cells, it has been noticed that the mechanical properties of cells cannot be fully described as purely elastic, as confirmed by hysteresis between the approach and retract parts of the AFM force–indentation curve [50]. Because cells display both elastic and viscous behaviors, they are best defined as viscoelastic [51]. As a result, evaluating this viscoelastic behavior is important for understanding the complexity of cells [52,53]. For viscoelastic characterization in the frequency domain, in the AFM setup (also known as AFM microrheology), the cantilever is oscillated with a small fixed amplitude at several frequencies, either during the indentation period or during the scanning process. The complex elastic modulus (E*(ω)) is calculated as follows:
where E′(ω) is the elastic (storage) modulus; E″(ω) is the viscous (loss) modulus; ω is the frequency of cantilever oscillation; δ is the indentation; ν is the Poisson’s ratio of the sample; θ is half-open angle of the AFM probe; . The storage and loss moduli together form the complex elastic modulus of the material, E*(ω). For materials that are purely elastic, the force is in phase with the input deformation, and the loss modulus (E″(ω)) is 0, while for a purely viscous material, the induced stress is out of phase with the input deformation, and the storage modulus E′(ω) is 0. As a result, the value of the loss tangent (loss tangent = E″(ω)/E′(ω)) can be utilized as an indicator of a solid-like or liquid-like behavior [50,54].
2.1. Methods of Sample Preparation for Microalgae
AFM sample preparation is rather simple, and there is no need for freezing or coating samples with metals or staining. The samples must, however, be strongly attached to a substrate to be able to withstand lateral forces during scanning. Biomolecules in a solution are deposited on extremely flat surfaces (typically mica or graphite) and are held to the surface using weak forces (electrostatic and/or van der Waals forces). However, the immobilization of whole cells is not so easy to achieve due to the large size of cells and their weak binding to surfaces. Various immobilization strategies have been established for studies on living microalgal cells. Glass coverslips coated with poly-L-lysine [55,56] or with polyethylenimine (PEI) [14,19,20,21,22] have been used for the imaging of live microalgal cells. Growing cells directly on a mica or glass substrate is another way to immobilize them [13,57]. Pletikapić and co-workers [29] used a direct drop deposition method, which was optimized for marine samples [57,58] for imaging diatoms in both air and seawater. An interesting approach was presented by Gebeshuber and co-workers [59] using freshwater snails that feed on algae. In this approach, different diatom species were grown on glass slides in the presence of snails, and only diatoms that produced strongest adhesive remained on the glass slide. Recently, Evans and co-workers [60] used a 3D printed array to mechanically immobilize microalgal cells. Alternatively, chemical fixation can be used to facilitate the attachment of cells to the substrate; however, it can result in substantial sample denaturation. For the AFM analysis of cleaned diatom frustules, prior to AFM measurements, the organic material is removed with sulfuric acid [28,61,62,63] or hydrogen peroxide to prevent silicified frustules from dissolving in strong acids [7]. Cleaned diatoms are then transferred to a substrate (glass slide or mica), which is usually modified with poly-L-lysine.
2.2. Advantages and Limitations of AFM
The main advantage of AFM over traditionally used SEM to study microalgae is that AFM allows the study of living microalgal cells in their natural physiological environment. This is because sample preparation for AFM does not involve the drying, metal coating or staining of the sample, which potentially alters the properties of the sample. In addition, AFM can be conducted in both air and liquids, allowing the imaging of microalgal cells in their growth medium. The resulting topographic AFM images of the sample are three-dimensional, while SEM provides two-dimensional images. Further, AFM can resolve very small differences in height on the cell surface due to its high vertical resolution. Besides imaging, AFM can also provide information about various surface properties, including mechanical properties, such as elasticity and viscoelasticity, and also, hydrophobicity and adhesion, which cannot be assessed via electron microscopy. Despite the great potential of AFM in the study of microalgae, there are also some limitations. The maximum scanning area of an AFM image can be a disadvantage of AFM, since AFM can only be used to scan a maximum area (XY) of about 100 µm × 100 µm and a maximum height in the order of 10–20 µm. SEM, however, can image an area in the order of mm2, with a depth of field in the order of millimeters. Therefore, it is not possible to image the entire cell if the microalgal cells are larger than the maximum scan area and higher than the maximum vertical range of the AFM instrument used. However, different parts of those larger cells can be imaged via AFM. Another limitation of standard AFM is the low speed of AFM measurement, which leads to a low temporal resolution, as standard AFM instruments require several minutes for a typical scan. Therefore, it is difficult to follow fast dynamic processes, while SEM is able to scan in near real time. However, this can be bridged by using a high-speed AFM that can even image surfaces at video rates. Overall, SEM and AFM can complement each other very well and can provide a wealth of information about a sample.
3. Nanomorphology of Microalgal Cells
Various techniques have been employed to characterize the ultrastructure of microalgal cells, such as scanning electron microscopy (SEM) and transmission electron microscopy (TEM) [7,8,9]. However, these techniques usually require dried samples (non-living dead cells). On the contrary, the use of AFM enables the imaging of living cells in their natural aqueous environment [10].
The first AFM experiment on microalgae was performed on diatoms and carried out on dried samples of six diatom species (Craticula, Cymatopleura, Gomphonema, Gyrosigma, Nitzschia and Stauroneis sp.) (Figure 2) [12]. Cells were collected from a mud sample using an eyelash and placed on a glass slide. For the cleaning and immobilizing of the cells, the slide was rinsed with ethanol. The inner surface of the frustule was also characterized after boiling the shell in sulphuric acid to remove organic material and divide the shell into two halves. Diatoms were imaged in air, and obtained AFM images were found to be comparable with SEM images, and because no metal coating was necessary, AFM proved to be an especially suitable method for studying delicate samples. The imaging of live microalgal cells in their natural hydrated state soon followed [13,14,55,59,64]. Although most studies in the field of microalgal research that have used AFM have been conducted on diatoms, AFM, as a method, has also been used for the morphological study of other microalgal classes, including Chlorophyceae [18,21,22], Chlorodendrophyceae [21,22], Dinophyceae [16,17], Trebouxiophyceae [23] and Porphyridiophyceae [25].
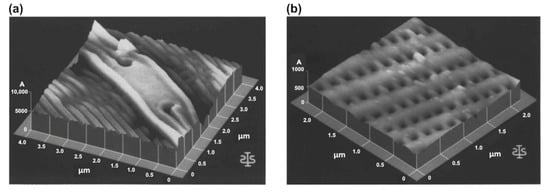
Figure 2.
First AFM images of microalgal cells. Three-dimensional topographical images of diatom from the genus Craticula: central node, scan size 4 µm × 4 µm and vertical scale 1 µm (a); inside view of the frustule, scan size 2.5 µm × 2.5 µm and vertical scale 100 nm (b). Reprinted with permission from [12].
3.1. Nanomorphology of Diatoms
Diatoms (class Bacillariophyceae) are a broad group of microalgae that are abundant in both freshwater and marine environments. Diatoms possess a silicified cell wall that has species-specific patterns and is made up of two overlapping thecae, an epitheca and a smaller hypotheca [56]. Each theca contains a valve and one or more girdle bands. The complete silicified cell wall assembly is known as a frustule, and it consists of silicified components and organic layers. The extracellular organic layer covers the entire cell and can be present as thick mucilaginous capsules and thin, tightly bound organic sheaths or casings [65]. Fixation and dehydration artefacts have made it difficult to perform ultrastructural studies using electron microscopy to examine the interactions between organic layers and their substructures. AFM, as a method, enabled the investigation of the morphological and physical properties of organic surface layers and of silicified components of diatoms. In addition, the nanoscale investigation of diatom silica structures via the AFM imaging of acid-cleaned frustules provided new insights into the biosilification process of diatoms.
3.1.1. Nanostructural Characterization of Living Diatom Cells
The first AFM characterization of the whole cell of a living diatom was performed by Almquist and co-workers in 2001 [13], where the nanostructure and micromechanical properties of the diatom, Navicula pelliculosa, were studied. Diatoms were grown in a liquid medium, mechanically transferred to the silanized (3-aminopropyl-triethoxysilane) mica surface and immediately imaged in air. The complex structure of the pores and poroids was easily resolved via AFM (Figure 3). The valva was found to be split by a raphe and enclosed with central ribs. Between the central ribs and the edge of the valve, rectangular sieve plates or puncta (pores and poroids) form bands of striae, which merge into elongated slits at the margin of the valve.
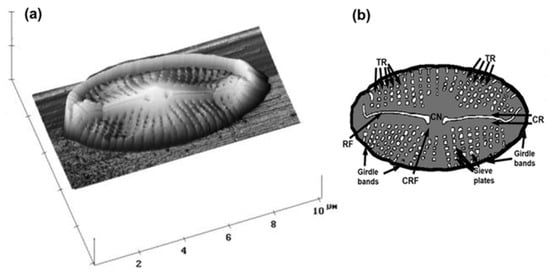
Figure 3.
First AFM image of the whole living microalgal cell. AFM image of diatom, Navicula pellicullosa, presented in 3D topographic view (scan size 10 µm × 10 µm; vertical scale 2 µm) showing raphe fissure and complex structure of pores and poroids (a); schematic drawing of a diatom frustule; CR—central rib, CN—central nodule, TR—transapical rib, RF—raphe fissure, CRF—curve-ended side of raphe fissure (b). Reprinted with permission from [13].
Crawford and co-workers [14] and Higgins and co-workers [55] used AFM to investigate the nanostructure and relationship between siliceous components and organic constituents of the cell walls of diatoms, Pinnularia viridis and Craspedostauros australis, under natural hydrated conditions. For the imaging of the living, motile diatoms in artificial media, cells were deposited on a glass slide modified with an adhesive polymer (poly-L-lysine or polyethylenimine). The AFM imaging of diatoms in contact mode showed that the cells were covered with a thick mucilaginous layer that was not present only around the raphe fissure. The imaging of cells in contact mode with higher forces removed the extracellular polymeric substance (EPS) coating to expose the surface of the silicified wall underneath. By using tapping mode with small amplitudes to reduce forces during scanning, the EPS coating was preserved, and it was shown that the EPS coatings had a different nanostructures specific to each species [64]. While the EPS of C. australis had a grooved surface, the EPS of P. viridis had a spherical particulate structure. Unlike C. australis and P. viridis, N. navis-varingica cells had no surface mucilage layer covering the girdle region. The girdle region of N. navis-varingica was covered by distinct hard spheres with a radius of 50–100 nm [64]. The EPS coating has previously only been observed under a scanning electron microscope as dry, strand-like material via SEM [65]. However, AFM studies revealed that the EPS coating is actually a discrete, structured polymer layer that maintains its integrity and association with the silica frustule. Besides the topography of the diatom frustules, Gebeshuber and co-workers [59] also determined the thickness of the organic coating encasing the siliceous skeleton. After the mechanical removal of the material with increased force during scanning, they found that the thickness of the organic coating for benthic species was about 10 nm.
The nanostructural and nanomechanical properties of different morphotypes (fusiform, triradiate and ovoid) of live Phaeodactylum tricornutum were studied by Francius and co-workers [26]. Using topographic images, they observed that the surface of the ovoid form has greater roughness and is coated with extracellular polymers, while the fusiform and triradiate forms have a smoother and non-structured surface. Besides the differences in morphology, they also investigated the nanomechanical properties of P. tricornutum morphotypes. They revealed that the silicified ovoid form has about a five-times-stiffer cell wall than that of the two non-silicified forms, and the girdle region of fusiform and ovoid forms is five times softer compared to that of the valve. Further, based on the AFM study, a cell wall model of a weakly silicified marine diatom, Cylindrotheca closterium, was proposed by Pletikapić and co-workers [29]. For morphological visualization and characterization, they used AFM contact-mode imaging, which showed the general features of the C. closterium cell (Figure 4). The structure and composition of the cell wall were studied by removing organic material via an acid treatment, after which, silica particles of 15 nm were identified in the valve region. Based on these results Pletikapić and co-workers [29] proposed a model of a cell wall in which individual silica nanoparticles were incorporated into an organic matrix.
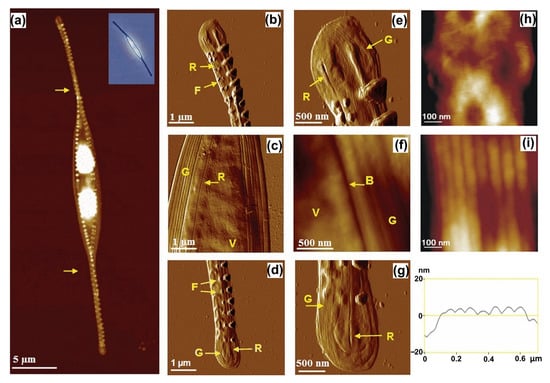
Figure 4.
AFM image of a whole Cylindrotheca closterium cell with inset optical micrograph (a) and its morphological details (b–i). Comparison of valve (h); and girdle band regions (i) at high resolution, with girdle band height profile. Images were acquired using contact mode in air and are presented as height data (a,f,h,i) with vertical scales of 700 nm (a), 250 nm (f), 25 nm (h) and 250 nm (i) and deflection data (b–e,g). Arrows in (a) indicate the spiral twist around the longitudinal cell axis. Labels V, G, R, F and B indicate the following features: valve, girdle band, raphe opening, fibulae and boundary between valve and girdle band, respectively. Reprinted with permission from [29].
The AFM imaging of microalgal morphological structures also provided insights into the possible mechanisms of species-specific protection and responses to various environmental pollutants. The effect of silver nanoparticles (Ag NPs) on diatoms, Cylindrotheca closterium and Cylindrotheca fusiformis, and their EPS were studied in the work of Pletikapić and co-workers [36]. Ag NPs-treated cells of both species increased in height due to the degeneration of the chloroplast and showed a deformed surface in both the valve and girdle band regions. Ag NPs were attached to the valve and girdle band regions, and pore-like lesions were found in the valve region. Cells exposed to the Ag NPs had fibrils or a fibrillar network around them, in which Ag NPs were embedded. Further experiments showed that the Ag NPs did not cause fibrils to cross-link or change their height. To better understand the effects of heavy metals on diatoms, Mišić Radić and co-workers [66] studied the morphological features of C. closterium exposed to cadmium. Cd-induced changes were seen as irregular patterns of silica spheres on the more silicified parts of the cell, the girdle band and around the raphe. These changes indicate that C. closterium showed a morphological response, in addition to a physiological response, which were probably caused by the interference of Cd with the diatom biosilification process. To study the effect of nanoplastics on microalgae, Mišić Radić and co-workers [32] used positively charged (amine-modified) and negatively charged (carboxyl-modified) polystyrene nanoplastics (PS NPs) and analyzed the nanostructural and nanomechanical responses of C. closterium. AFM imaging revealed the adsorption of amine-modified PS NPs on the cell surface and the incorporation of both amine-modified and carboxyl-modified PS NPs into the EPS network. In this work, AFM nanomechanical measurements were taken for the first time to investigate the effects of nanoplastics on the mechanical properties of microalgal cells and showed a nanoplastics-induced decrease in cell elasticity. These results suggest that via interactions with microalgal cell walls or with microalgal EPS, nanoplastics may endanger marine microalgae, but also cause higher trophic levels, threatening the health and stability of the marine ecosystem.
AFM imaging also provided an answer to the question of what holds the cells of the diatom, Bacteriastrum jadranum, together in their chain-like colony, in which cells and their setae are not in direct contact. In this study, Bosak and co-workers [37] showed that it is a polysaccharide cell jacket structure that encloses colony cells and setae. The cell jacket was generally visualized via Alcian Blue staining, but the AFM results revealed that the structure of the cell jacket has a form of cross-linked fibrillar network. The network was composed of hexagonally shaped pores that were connected by thicker fibrils and reinforced by branched fibrils, and it was found that the cell jacket is an essential part of the B. jadranum cell.
3.1.2. Nanostructural Characterization of Diatom Frustule
Due to their ability to reproduce complex three-dimensional structures with features controlled at different length scales, diatoms present an excellent model for studying silica biomineralization and the development of biomimetic approaches. Studying diatom silica structures via the AFM imaging of acid-cleaned frustules provided new insights into the biosilification process. Although cells are not alive after the treatment, the acid treatment removes the organic layer and exposes the silica structures more. AFM studies of diatom frustule components showed that they are made out of a conglomerate of spherical silica particles, which was consistent with studies performed using electron microscopy [67]. Crawford and co-workers [14] were the first ones to use AFM to resolve the nanostructure of silica in the cell wall of the diatom. They studied the morphological characteristics of frustules of different diatom species (Pinnularia viridis and Hantzschia amphioxys) and showed that the diatom frustules are composed of packed silica spheres, with significant size differences between the different species. The surface silica of P. viridis had a smooth surface, although when high-resolution imaging was performed, small-scale granularity was observed. The silica in cross-sections of P. viridis consisted of spherical silica particles, with average diameters of 45 nm in the valve and 40 nm in the girdle bands, while in the H. amphioxys cross-section, the average diameters of silica particles were 37 nm and 38 nm in the valve and girdle bands, respectively. This study demonstrates that while silica particles are similar in size for the same frustule component (valve or the girdle bands), there can be large differences in silica particle size between the valves and the girdle bands of the same species. The size range of these particles, as determined via AFM imaging in contact mode, confirmed previous electron microscopy observations that developing diatom frustules are formed by the aggregation of silica particles ranging from 30 nm to 50 nm in diameter [68]. AFM has also been used to study the silica of the large centric diatom, Coscinodiscus granii [69]. Unlike P. viridis and H. amphioxys [14], the surface of C. granii was granular and consisted of fused silica particles, with a diameter ranging from 100 nm to 200 nm. In a different investigation on Coscinodiscus sp., the similar organization of silica on the surface was observed, with smaller particle sizes ranging from 50 nm to 80 nm in diameter [62]. It was concluded that the difference in particle size between the two Coscinodiscus species is probably due to the role of species-specific organics in their formation. The AFM analysis of the nanostructure of silica from the diatom, Ditylum brightwelli, revealed a number of morphological differences between the frustule and the central spine [70]. The fundamental difference between them was a solid, smooth surface in the case of the cell wall and a surface with nanospheres and a central hollow in the central spine. There were other distinctions as well, such as pores in the cell wall, but there were none in the central spine.
The AFM results also provided insights into the assembly concepts involved in diatom silica structure formation, thus contributing to the development of biomimetic approaches. New insights into biosilification processes were provided by Hildebrand and co-workers [71], who studied the synthesis of the silica cell wall in the diatom, Thalassiosira pseudonana. Three different scales of structural organization were recognized, and different silica morphologies were discovered during the creation of different cell wall substructures. The largest scale, called microscale structure formation, is the formation of the overall shape of the valve and girdle bands. The smallest scale, nanoscale structure formation, is the nanostructured morphology of the initial silica polymerization product. The third scale, called mesoscale structure formation, involves the formation of organized substructures within the silica deposition vesicles. This work was important in correlating structure formation with the genes and proteins involved, but it also allowed the modelling of the process and provided a basis for the modification of the structure by genetic or non-genetic means. Hildebrand and co-workers [72] presented an AFM study of the cell walls of 16 diatom species to investigate the design principles involved in the assembly of silica structures and the underlying organics. Samples for AFM imaging were prepared via an acid treatment of diatoms to remove organic layer and imaged in air. The silica structures of diatoms substantially differed for the same features of different species, as well as for different features within a single species and on different faces of the same object (Figure 5). The obtained results suggest that there is no strict relationship between the morphology of nanoscale silica and the type of structure it contains. At the mesoscale, linear structures predominated, suggesting that the assembly or organization of linear organic molecules or subcellular assemblies has an essential and conserved function in structure formation at this scale. The microscale structure has a general influence on the nano-and mesoscale structure, demonstrating that the shaping of the silica deposition vesicle is important in structure formation.
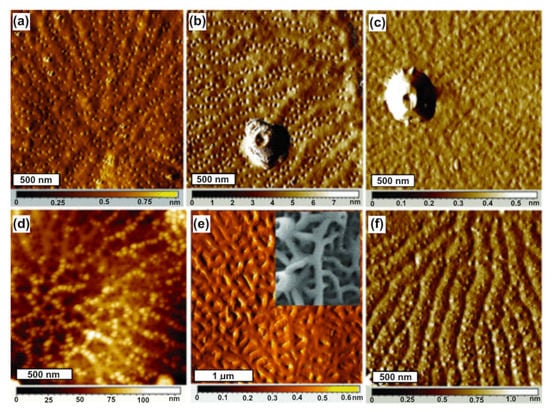
Figure 5.
Similar proximal valve features and distinct distal valve features in three Thalassiosira species. Proximal valve surfaces of T. pseudonana (a); T. weissflogii (b); T. oceanica (c). Distal surfaces of T. pseudonana (d); T. weissflogii with inset SEM image (e); T. oceanica (f). Reprinted with permission from [72].
3.2. Nanostructural Characterization of Other Microalgal Classes
Although most studies performed in the field of microalgae research using AFM have been on diatoms, the AFM method has also been applied for morphological studies of other microalgal classes, including Chlorophyceae [18,21,22], Chlorodendrophyceae [21,22], Dinophyceae [16,17], Trebouxiophyceae [23,24] and Porphyridiophyceae [25].
The function of mucilage in the toxicity mechanism of the dinoflagellate, Ostreopsis cf. ovata., was investigated by Giussani and co-workers [16]. The cell surface of Ostreopsis cf. ovata was characterized at the sub-nanometer resolution via AFM, and the association between mucilage and thecal plates, pores and trychocysts was investigated. Cells were dried and rinsed with ultrapure water, which caused them to disaggregate and break into separate plates. AFM characterization revealed three different cellulose layers of the hypothecal plates, on which scattered pores were observed. Imaging at a higher resolution revealed conical-tubular structures around some pores, suggesting that these are involved in the extrusion mechanism of the trychocyst. The AFM results confirmed the possible active role of filaments in conveying toxicity. The algicidal activity of the fungal strain, Talaromyces purpurogenus, against the dinoflagellate, Prorocentrum donghaiense, was investigated by Shu and co-workers [17]. Increased malondialdehyde levels and ATPase activities indicated that damage occurred to the cell membrane, which was confirmed via AFM. AFM images showed wrinkles and breaks on the cell surface of P. donghaiense, resulting in an increase in roughness. These changes in the cells eventually led to the disintegration of the algal cells due to the changes in physiological metabolism. The consequences of stress on the algal cell membrane were investigated via light microscopy and AFM imaging [18]. The marine alga, Dunaliella tertiolecta, was exposed to hypotonic stress, which resulted in cell swelling, and subsequently, to cell rupture and the formation of plasma membrane vesicles. AFM images of the plasma membrane vesicles showed an average vesicle thickness of ~20 nm, indicating that each membrane bilayer had a thickness of about 10 nm, which is comparable with the estimated cell envelope thickness of 9 nm, as determined with the electron microscope. On the AFM images of vesicles acquired at a higher magnification, proteins were observed as densely packed globules with a height ranging from 5 to 15 nm. Novosel and co-workers [21,22] studied the effect of temperature and salinity stress on three different algal species (Cylindrotheca closterium, Dunaliella tertiolecta and Tetraselmis suecica). The AFM characterization of algal surface morphology showed no specific morphological change in the cell surface with temperature and salinity variations, but a change was observed in the size of the algal species. Cells grown at a higher temperature or at lower salinity were smaller, which is consistent with the commonly known fact that the cell size of microalgae decreases with temperature [73], and also, with decreasing salinity [74]. In addition, Novosel and co-workers were the first ones to visualize micropearls in the cells of Tetraselmis suecica, which had previously been identified via SEM [75]. High-speed AFM was applied to visualize floc formation during electrocoagulation floatation (ECF) in the presence and absence of algae (Chlorella sorokiniana) [23]. It was shown that the floc formed nanometer-sized clumps that aggregated together and had a spheroid shape when isolated. An AFM image of C. sorokiniana after the treatment with ECF showed spherical clumps that were smaller than the clumps from ECF-treated media without cells and were rougher. Liu and co-workers [25] used AFM to directly assess the supramolecular assembly of membrane protein complexes in the membrane of Porphyridium cruentum. They isolated phycobilisomes (PBsomes) and PBsome-containing thylakoid membranes from the unicellular red alga, P. cruentum, and studied the structure of PBsomes and their distribution on the thylakoid membrane of P. cruentum. Different types of PBsome arrangement were observed. On one sample, PBsomes were arranged randomly and tended to form locally clustered motives, whereas on a sample grown under low light, they were organized in more parallel rows. These results suggest that thylakoid membranes adapt to light to increase the energy-harvesting efficiency.
4. Mapping Surface Properties of Microalgal Cells
In addition to the ability to image cells at high resolution, AFM has evolved also as an important tool for deciphering cell surface properties, such as elasticity, adhesion, hydrophobicity, and in recent years, viscoelasticity. Although cell surface properties are crucial for understanding the relationship between form and function, only a few studies have addressed this issue in microalgae, with elasticity being the most commonly studied property.
4.1. Nanomechanical Properties of Microalgal Cells
In recent years, AFM, as a tool, has provided new insights into the mechanical properties of various microalgal cells. In this type of measurement, force–distance curves are acquired and analyzed using different theoretical models to obtain information about a sample’s elasticity (i.e., Young’s modulus) and viscoelasticity (i.e., storage modulus and loss modulus). The first AFM experiments conducted to measure the elasticity of microalgal cells were performed by Almquist and co-workers [13] on the diatom, Navicula pelliculosa. Subsequently, AFM was used to map the elastic properties of various microalgal species [15,20,21,22,26,27,28,29,30,31,32,33]. However, to our knowledge, the viscoelastic properties of microalgal cells have not been previously reported, and in this review, we present new unpublished data of viscoelasticity of the microalga, Dunaliella tertiolecta.
4.1.1. Elasticity of Microalgal Cells
Almquist and co-workers [13] were the first ones to measure the elasticity of microalgal cells using AFM. They showed that the elastic modulus of diatom, Navicula pelliculosa, ranged from seven to hundreds of GPa, which was comparable to values of known silicas. Similar values (1–60 GPa) have been demonstrated for the diatom, Coscinodiscus sp., and attributed to differences in the biomineralization and porosity of the cell wall [13,28]. Francius and co-workers [26] studied the elasticity of the three morphotypes of the diatom, Phaeodactylum tricornutum. The elastic modulus differed from one morphotype to another, with the silicified ovoid form being about five times stiffer (elastic modulus of 500 kPa) than that of the two non-silicified forms (100 kPa). However, all three morphotypes have lower elastic modulus values compared to the values reported for other diatoms. Karp-Boss and co-workers [27] provided the first evidence for changes in local elastic properties during the cell cycle of the marine diatom, Lithodesmium undulatum. They demonstrated that the local elastic modulus is a highly dynamic feature by investigating Young’s elastic modulus and employing a fluorescent dye. The elastic modulus of stained regions was much lower than that of unstained regions, indicating that freshly generated cell wall components are often softer than those inherited from parent cells are. Further, to better understand the association between organic component and the silica structure in diatoms, Pletikapić and co-workers [29] characterized the nanomechanical properties of a weakly silicified marine diatom, Cylindrotheca closterium. For morphological visualization and characterization, they used contact mode, and for measuring the nanomechanical properties, they used Peak Force Tapping AFM. The stiffest and least deformable part of the cell surface were the fibulae, and the valve region was the most deformable and the softest part. Studies on the mechanical properties of microalgal cells have shown that the elastic modulus is species-specific, but it also depends on the growth phase, pH value of the growth medium, exposure to various stresses, etc. Pillet and co-workers [20] used AFM to investigate the nanomechanical properties of Dunaliella tertiolecta cells in different growth phases. They showed that the cells are stiffer in the exponential phase than they are in the stationary phase, implying that the cell membrane undergoes molecular changes throughout aging. Formosa-Dague and co-workers [76] measured the mechanical properties of P. tricornutum cells at different pH values and showed that the elasticity of the cells increased with increasing pH. Further, the nanomechanical characterization of three different algal species (Cylindrotheca closterium, Dunaliella tertiolecta and Tetraselmis suecica) grown at different temperatures and salinities showed that all cells become stiffer at lower temperatures and lower salinities [21,22]. A nanoplastics-induced decrease in cell stiffness was observed by Mišić Radić and co-workers [32] in the study of the effects of nanoplastics on Cylindrotheca closterium cells.
4.1.2. Viscoelasticity of Microalgal Cells
Despite the widespread use of AFM for the mechanical characterization of biological samples, including cells, most prior research has used the standard Hertzian contact mechanics model and measured the apparent elastic modulus of the cells, while ignoring viscoelastic effects. In the field of microalgae research, to our knowledge, there are no published data on the viscoelasticity of microalgal cells; so, in this article, we present our preliminary unpublished data on the viscoelasticity of microalga, Dunaliella tertiolecta, in the exponential growth phase (Figure 6). Measurements were taken using JPK NanoWizard 4 XP (Bruker, Berlin, Germany) coupled to an Olympus IX73 microscope (Olympus, Tokyo, Japan) using the JPK CellMech Package (Bruker, Berlin, Germany). Silicon nitride probes (MLCT-BIO, cantilever C, nominal frequency 7 kHz, spring constant 0.01 N/m; Bruker, Camarillo, CA, USA) were used and calibrated on a clean Petri dish in filtered seawater (0.2 µm) before the measurement. A plastic Petri dish coated with 0.1% polyethylenimine (PEI), with filtered seawater as an imaging buffer, was used. For cell imaging quantitative imaging (QITM), the following settings were applied: setpoint 1.5 nN, scan size 10 µm × 10 µm, z-length 2000 nm, z-speed 75 µm/s and 128 pix resolution (Figure 6a—inset). The parameters used for acquiring viscoelastic data were: (i) extend segment—z-length: 8 µm, z-speed: 8 µm/s, sample rate: 2048 Hz, and setpoint: 1.5 nN; (ii) pause segment—time: 1 s, sample rate: 2048 Hz; (iii) modulation segment—frequency: 10 Hz, amplitude: 10 nm, and sample rate: 2048 Hz; (iv) retract segment—z-length: 8 µm, z-speed: 8 µm/s, and sample rate: 2048 Hz (Figure 6b). JPK data processing software with JPK CellMech Package (JPKSPM Data Processing Software, version 7.0.165, Bruker, Berlin, Germany) was used for data processing.
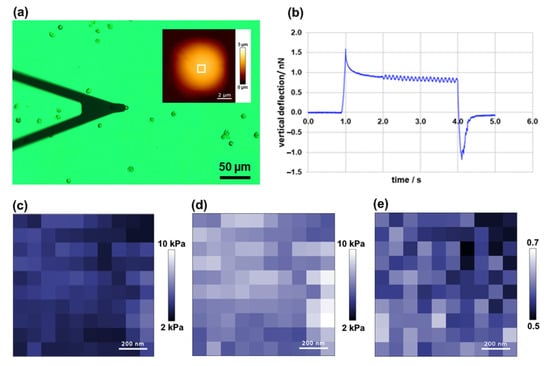
Figure 6.
Viscoelasticity of microalga Dunaliella tertiolecta. Optical micrograph of AFM cantilever and D. tertiolecta cells (inset shows AFM height image of the cell acquired in quantitative imaging (QI) mode) (a); representative vertical deflection vs. time curve (b); loss modulus map (c); storage modulus map (d); loss tangent map (e). Maps were acquired on the selected cell area (1 µm × 1 µm) shown on AFM inset in (a).
Representative maps of loss and storage moduli obtained of a D. tertiolecta cell (area: 1 µm × 1 µm) are shown in Figure 6c,d, respectively. The mean value of the loss (viscous) modulus was 3.75 ± 0.66 kPa (Figure 6c), and the mean value of the storage (elastic) modulus was 6.56 ± 1.14 kPa (Figure 6d). The value of the loss tangent was 0.57 ± 0.03 (Figure 6e), indicating that D. tertiolecta cells exhibit a relatively high level of energy dissipation when they are being deformed, which is typical for cells and biomaterials. However, the presented results are preliminary, representing one cell analyzed at one indentation frequency (10 Hz), and further measurements are required to understand the frequency dependent mechanical response of algal cells better.
4.2. Hydrophobicity of Microalgal Cells
Although measuring the interaction between an uncoated tip (usually silicon nitride) and a sample offers valuable data on the nanomechanical properties of the sample, using chemical force microscopy enables the study of other surface properties and interactions. In CFM, modified AFM tips are used to monitor specific interaction forces, thus enabling the mapping of the spatial distribution of chemical groups on living cells [34]. By using AFM tips modified with hydrophilic (OH) or hydrophobic (CH3) functional groups, AFM can provide important data on cell surface hydrophobicity [77]. Despite the opportunity provided by CFM to study cell hydrophobicity, only a few studies have been performed to investigate these interactions in microalgal cells.
Pillet and co-workers [20] used CFM with a hydrophobic methyl-terminated tip and mapped the hydrophobicity of a living Dunaliella tertiolecta cell in different growth phases (Figure 7). They showed that the hydrophobicity of cells depends on the growth phase, with cells in the exponential growth phase being more hydrophobic than the cells in the stationary growth phase are.
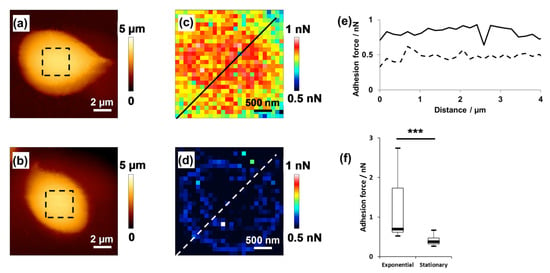
Figure 7.
Hydrophobicity of alga D. tertiolecta. Height image of an algal cell in the exponential phase (a); height image of an algal cell in the stationary phase (b); adhesion force map (c) measured by the interaction between the hydrophobic AFM tip and the selected cell area shown in (a); adhesion force map (d) of the selected cell area shown in (b); cross-sections from the adhesion force maps in the exponential phase (full line) and stationary phase (dashed line) (e); statistical analysis of hydrophobicity (f). Significant differences were observed (***). Reprinted with permission from [20].
Lavieale and co-workers [78] investigated the distribution of hydrophobic forces at the molecular level on the cell surface of Nitzschia palea. Using hydrophobic and hydrophilic tips, they showed that the EPS on N. palea’s surface is organized as adhesive patches and is accumulated more in the center of the cell than it is at the apexes. Novosel and co-workers [21,22] showed that the change in hydrophobicity of cells is caused by both temperature and salinity. At elevated temperatures, D. tertiolecta and T. suecica were extremely hydrophilic, whereas C. closterium was the most hydrophobic, and at lower salinity, all three species tested had a more hydrophobic character. Demir and co-workers [79] developed an interesting approach to study cell hydrophobicity using air bubbles. They developed a novel approach to studying the interactions between air bubbles and cell surface using Fluidic Force Microscopy (FluidFM) technology, which combines AFM with microfluidics. They used AFM cantilevers with an integrated micro-sized channel connected to a pressure controller that generates air bubbles. They showed that the generated bubbles can be utilized to investigate the interactions with hydrophobic samples. Overall, these studies show that chemical force microscopy can provide important information about the hydrophobic properties of microalgae at the subcellular level, complementing nanomechanical properties of the cell.
4.3. Adhesion Studies of Microalgal Cells
Understanding the mechanism of cell adhesion is a major challenge. These interactions are complex and include specific and non-specific interactions. Traditionally used macroscopic assays, which are used to study the mechanism of microbial adhesion and biofilm formation [11], can be complemented with AFM force spectroscopy, which can provide quantitative information on adhesion forces. In addition, the use of cellular probes, in which AFM cantilevers are functionalized with the cell, enables researchers to measure the interactions between the cell and the surface at a single-cell level. Over the past two decades, the number of cell adhesion studies based on single-cell force spectroscopy (SCFS) has increased [80,81]. Bowen and co-workers [82] were the first ones to use a single, live, immobilized cell as a probe to study adhesion. After this study, a large number of cell probes from different microorganisms were used. Despite the great amount of interest on this approach to studying cell–surface interactions, there are only a few studies that focused on the adhesion of microalgal cells, especially diatoms. Since diatoms are the major players of biofilms in aquatic environments, learning about the mechanisms that control their adhesion is important for developing new strategies against biofouling.
Arce and co-workers [35] were the first ones to use SCFS in studies of microalgal adhesion. They investigated the adhesion of the freshwater diatom, Navicula sp., to different hydrophobic and hydrophilic surfaces by using a cantilever functionalized with an Navicula sp. cell (Figure 8). The force–distance curves showed similar adhesion strengths on a hydrophobic silicone elastomer that inhibits biofilm formation (Intersleek) and on mica (hydrophilic), suggesting that the EPS of Navicula sp. has both hydrophobic and hydrophilic properties.
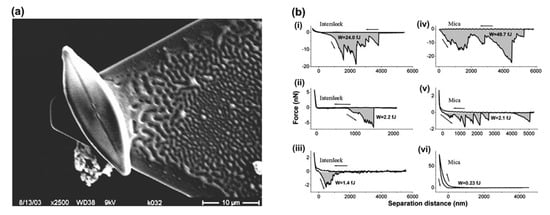
Figure 8.
Cell probe experiments. SEM micrograph of a single diatom cell attached with epoxy glue to an AFM tipless cantilever (a); representative force–distance curves obtained with bioprobe diatoms in the stationary phase on Intersleek (i–iii) and mica (iv–vi) surfaces (b). The work of detachment, W, is given in fJ units (10−15 J) for each curve. The arrow represents the approach and retraction directions. Reproduced with permission from [35].
The adhesion of the freshwater diatom, Nitzschia palea, cells to different surfaces was investigated by Laviale and co-workers [78] using SCFS. They used tipless cantilevers modified with polydopamine to attach N. palea cells to the cantilever. The results showed that N. palea cells strongly adhere to hydrophobic surfaces and that adhesion involves the unfolding of surface biopolymers. On the contrary, the adhesion to the hydrophilic surface was lower and appeared to involve polymers of a different nature (without unfoldable domains). Chemical force spectroscopy with hydrophobic tips also showed that adhesion is controlled by hydrophobic EPS distributed in patches on the cells’ surface.
The presented SCFS studies on diatoms show that AFM with a living bioprobe can be successfully used to characterize the adhesion of microalgal cells to various surfaces and that AFM is a valuable tool for the evaluation of diatom attachment to antifouling materials and complements the conventional methods used to characterize the surface properties of diatoms (adhesion tests and electron microscopy). Therefore, using SCFS on other microalgae can provide more insights into surface colonization and biofilm formation in aquatic environments.
5. Future Perspectives
The studies presented in this review show that AFM has great potential in the field of microalgal research (brief summary is given in Supplementary Material, Table S1). The AFM imaging of diatoms allowed the study of the morphological and physical characterization of silicified frustule components and organic layers, as well as living diatoms. In addition, the nanoscale investigation of diatom silica structures via AFM imaging of acid-cleaned frustules provided a new understanding of the design and assembly principles during the formation of the diatom silica structure, contributing to the development of biomimetic approaches. In addition to its ability to image cells at a high resolution, AFM in force spectroscopy mode has also proven to be a valuable tool for deciphering cell surface properties, such as elasticity, adhesion, hydrophobicity, and in recent years, viscoelasticity. Studies on the mechanical properties of microalgal cells have shown that the elastic modulus is species-specific, and also, depends on the growth phase of the cell, pH of the growth medium, exposure to various stress factors, etc. However, to our knowledge, the viscoelastic properties of microalgal cells have not been previously reported, and in this review, we present new unpublished data on the viscoelasticity of microalga, Dunaliella tertiolecta. Since cells generally display both elastic and viscous behaviors, assessing the viscoelastic properties of microalgal cells is important for gaining more insight into cell properties and understanding cell complexity. The application of AFM in single-cell force spectroscopy mode, where cantilevers are functionalized with cells, can provide new insights into the physicochemical properties that govern interactions at the interface between microalgal cells and their environment. The presented SCFS studies on diatoms demonstrate that AFM with a living bioprobe can be successfully used to characterize the adhesion of microalgal cells to various surfaces and that AFM is a promising method for the rapid assessment of diatom attachment to antifouling materials. Recent technological advances in AFM instrumentation have made AFM an even more powerful tool for biological applications. The implementation of the high-speed AFM mode (HS-AFM) enables the following of dynamic processes on the cell surface, which could provide new insights into understanding the behavior of microalgae when they are exposed to stress. Further, by inserting a long, needle-like nanoprobe into a living mammal cell, so-called 3D nanoendoscopy AFM measurements were obtained [83]. Since microalgae have either a rigid shell (such as diatoms) or a thin membrane, it remains to be explored whether these experiments could be performed on microalgal cells, but if so, they would certainly provide more information on the basic mechanisms of cellular functions in microalgal cells. Although AFM provides high sensitivity and resolution in imaging and studying cells, it does not provide other important information, such as information about cellular components and chemical composition. Therefore, combining AFM with other complementary techniques could provide a more comprehensive insight into microalgae. The correlation of AFM with optical microscopy, such as a simple optical microscope, fluorescence microscope, laser scanning confocal microscope or super-resolution microscope, allows the correlation of organelles and chemical components with cell morphology and cell surface properties. In addition, combining AFM with infrared (IR) or Raman spectroscopy, also known as AFM-IR and tip-enhanced Raman spectroscopy (TERS), can provide information on the nanoscale chemical characterization of samples, allowing a correlation between morphology and composition.
6. Conclusions
The cell surface represents the boundary between cells and their environment, and therefore, has a number of crucial roles, including determining cell shape, as well as mediating cellular interactions (cell adhesion, cell aggregation, etc.). Therefore, knowledge about the morphology and surface properties of microalgae is essential for a detailed understanding of their function, behavior, fate and role in aquatic systems. A variety of characterization methods have been used to achieve this goal, including the separation and chemical analysis of cell membrane components, infrared spectroscopy and electron microscopy. These methods frequently involve the manipulation of cells before studying, including drying and staining, and often provide average information about a large number of cells. However, despite numerous investigations that have been conducted to date using various analytical techniques and approaches, many aspects of the unique field of microalgae remain unexplored. Atomic force microscopy, with its multiparametric and multifunctional characterization, i.e., the imaging of the native microalgal cell surface at the single cell level in its natural physiological environment with nano- and sub-nanometric scale resolution and the mapping of surface properties, such as elasticity, viscoelasticity, hydrophobicity and adhesion, can answer many unanswered questions in the microalgal field. In addition, combined systems of AFM with other complementary techniques, such as optical microscopy or Raman and infrared spectroscopy, and others, are continuously evolving and opening new opportunities for the study of biological samples, including microalgae. In conclusion, AFM, as a tool, will continue to play an important role in microalgal research by expanding our knowledge on the microalgal cells’ surface structure and properties, and also, contributing to a better understanding of the mechanisms involved in microalgal behavior.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w15111983/s1. Table S1: Microalgal species and parameters studied via AFM.
Author Contributions
Conceptualization, T.M.R.; methodology, T.M.R., P.V., A.Č. and A.D.; investigation, T.M.R., P.V., A.Č. and A.D.; writing—original draft preparation, T.M.R., P.V., A.Č. and A.D.; writing—review and editing, T.M.R., P.V., A.Č. and A.D.; supervision, T.M.R.; funding acquisition, T.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the project “Research network V4-Croatia for the development of novel drug carriers from algae” (No. 22220115) funded through the International Visegrad Fund.
Data Availability Statement
Not applicable.
Acknowledgments
Permissions for Figures: Figure 2 is reprinted from Ultramicroscopy, 1992, Linder, A.; Colchero, J.; Apell, H.-J.; Marti, O.; Mlynek, J., Scanning Force Microscopy of Diatom Shells, 42–44: 329–332, Copyright (2023), with permission from Elsevier. Figure 3 is reprinted from Journal of Microscopy, 2001, Almqvist, N.; Delamo, Y.; Smith, B.L.; Thomson, N.H.; Bartholdson, A.; Lal, R.; Brzezinski, M.; Hansma, P.K., Micromechanical and Structural Properties of a Pennate Diatom Investigated by Atomic Force Microscopy, 202: 518–532, Copyright (2023), with permission from John Wiley and Sons. Figure 4 is reprinted from Journal of Phycology, 2012, Pletikapić, G.; Berquand, A.; Radić, T.M.; Svetličić, V., Quantitative Nanomechanical Mapping of Marine Diatom in Seawater Using Peak Force Tapping Atomic Force Microscopy, 48: 174–185, Copyright (2023), with permission from John Wiley and Sons. Figure 5 is reprinted from Journal of Microscopy, 2009, Hildebrand, M.; Holton, G.; Joy, D. C.; Doktycz, M. J.; Allison, D. P., Diverse and Conserved Nano- and Mesoscale Structures of Diatom Silica Revealed by Atomic Force Microscopy, 235: 172–187, Copyright (2023), with permission from John Wiley and Sons. Figure 7 is reprinted from Bioelectrochemistry, 2019, Pillet, F.; Dague, E.; Pečar Ilić, J.; Ružić, I.; Rols, M.P.; Ivošević DeNardis, N. Changes in Nanomechanical Properties and Adhesion Dynamics of Algal Cells during Their Growth., 127: 154–162, Copyright (2023), with permission from Elsevier. Figure 8 is reprinted from Biophysical journal, 2004, Arce, F.T.; Avci, R.; Beech, I.B.; Cooksey, K.E.; Wigglesworth-Cooksey, B., A Live Bioprobe for Studying Diatom-Surface Interactions, 87: 4284–4297, Copyright (2023), with permission from Elsevier.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Eaton, P.; West, P. Atomic Force Microscopy; Oxford University Press: New York, NY, USA, 2010; ISBN 978-0-19-957045-4. [Google Scholar]
- Dufrêne, Y.F. Using Nanotechniques to Explore Microbial Surfaces. Nat. Rev. Microbiol. 2004, 2, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging Modes of Atomic Force Microscopy for Application in Molecular and Cell Biology. Nat. Nanotech. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Allison, D.P.; Mortensen, N.P.; Sullivan, C.J.; Doktycz, M.J. Atomic Force Microscopy of Biological Samples. WIREs Nanomed. Nanobiotechnol. 2010, 2, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Lilledahl, M.B.; Stokke, B.T. Novel Imaging Technologies for Characterization of Microbial Extracellular Polysaccharides. Front. Microbiol. 2015, 6, 525. [Google Scholar] [CrossRef] [PubMed]
- De Stefano, M.; Marino, D. Morphology and Taxonomy of Amphicocconeis Gen. Nov. (Achnanthales, Bacillariophyceae, Bacillariophyta) with Considerations on Its Relationship to Other Monoraphid Diatom Genera. Eur. J. Phycol. 2003, 38, 361–370. [Google Scholar] [CrossRef][Green Version]
- Fresnel, J.; Probert, I. The Ultrastructure and Life Cycle of the Coastal Coccolithophorid Ochrosphaera Neapolitana (Prymnesiophyceae). Eur. J. Phycol. 2005, 40, 105–122. [Google Scholar] [CrossRef]
- Morales, E.A.; Edlund, M.B.; Spaulding, S.A. Description and Ultrastructure of Araphid Diatom Species (Bacillariophyceae) Morphologically Similar to Pseudostaurosira Elliptica (Schumann) Edlund et al. Phycol. Res. 2010, 58, 97–107. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic Force Microscopy as a Multifunctional Molecular Toolbox in Nanobiotechnology. Nat. Nanotech. 2008, 3, 261–269. [Google Scholar] [CrossRef]
- Dupres, V.; Alsteens, D.; Andre, G.; Dufrêne, Y.F. Microbial Nanoscopy: A Closer Look at Microbial Cell Surfaces. Trends Microbiol. 2010, 18, 397–405. [Google Scholar] [CrossRef]
- Linder, A.; Colchero, J.; Apell, H.-J.; Marti, O.; Mlynek, J. Scanning Force Microscopy of Diatom Shells. Ultramicroscopy 1992, 42–44, 329–332. [Google Scholar] [CrossRef]
- Almqvist, N.; Delamo, Y.; Smith, B.L.; Thomson, N.H.; Bartholdson, A.; Lal, R.; Brzezinski, M.; Hansma, P.K. Micromechanical and Structural Properties of a Pennate Diatom Investigated by Atomic Force Microscopy. J. Microsc. 2001, 202, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.A.; Higgins, M.J.; Mulvaney, P.; Wetherbee, R. Nanostructure of the Diatom Frustule as Revealed by Atomic Force and Scanning Electron Microscopy. J. Phycol. 2001, 37, 543–554. [Google Scholar] [CrossRef]
- Luís, A.T.; Hlúbiková, D.; Vaché, V.; Choquet, P.; Hoffmann, L.; Ector, L. Atomic Force Microscopy (AFM) Application to Diatom Study: Review and Perspectives. J. Appl. Phycol. 2017, 29, 2989–3001. [Google Scholar] [CrossRef]
- Giussani, V.; Sbrana, F.; Asnaghi, V.; Vassalli, M.; Faimali, M.; Casabianca, S.; Penna, A.; Ciminiello, P.; Dell’Aversano, C.; Tartaglione, L.; et al. Active Role of the Mucilage in the Toxicity Mechanism of the Harmful Benthic Dinoflagellate Ostreopsis Cf. Ovata. Harmful Algae 2015, 44, 46–53. [Google Scholar] [CrossRef]
- Shu, W.; Zhao, L.; Hou, S.; Yu, Q.J.; Tan, S.; Yin, P. Toxic Effect on the Membrane System and Cell Proliferation of Prorocentrum Donghaiense Caused by the Novel Algicidal Fungus Talaromyces Purpurogenus YL13. J. Appl. Phycol. 2017, 29, 275–284. [Google Scholar] [CrossRef]
- DeNardis, N.I.; Ilić, J.P.; Ružić, I.; Pletikapić, G. Cell Adhesion and Spreading at a Charged Interface: Insight into the Mechanism Using Surface Techniques and Mathematical Modelling. Electrochim. Acta 2015, 176, 743–754. [Google Scholar] [CrossRef]
- Ivošević DeNardis, N.; Pečar Ilić, J.; Ružić, I.; Novosel, N.; Mišić Radić, T.; Weber, A.; Kasum, D.; Pavlinska, Z.; Balogh, R.K.; Hajdu, B.; et al. Algal Cell Response to Laboratory-Induced Cadmium Stress: A Multimethod Approach. Eur. Biophys. J. 2019, 48, 231–248. [Google Scholar] [CrossRef]
- Pillet, F.; Dague, E.; Pečar Ilić, J.; Ružić, I.; Rols, M.-P.; Ivošević DeNardis, N. Changes in Nanomechanical Properties and Adhesion Dynamics of Algal Cells during Their Growth. Bioelectrochemistry 2019, 127, 154–162. [Google Scholar] [CrossRef]
- Novosel, N.; Mišić Radić, T.; Zemla, J.; Lekka, M.; Čačković, A.; Kasum, D.; Legović, T.; Žutinić, P.; Gligora Udovič, M.; Ivošević DeNardis, N. Temperature-Induced Response in Algal Cell Surface Properties and Behaviour: An Experimental Approach. J. Appl. Phycol. 2022, 34, 243–259. [Google Scholar] [CrossRef]
- Novosel, N.; Mišić Radić, T.; Levak Zorinc, M.; Zemla, J.; Lekka, M.; Vrana, I.; Gašparović, B.; Horvat, L.; Kasum, D.; Legović, T.; et al. Salinity-Induced Chemical, Mechanical, and Behavioral Changes in Marine Microalgae. J. Appl. Phycol. 2022, 34, 1293–1309. [Google Scholar] [CrossRef]
- Landels, A.; Beacham, T.A.; Evans, C.T.; Carnovale, G.; Raikova, S.; Cole, I.S.; Goddard, P.; Chuck, C.; Allen, M.J. Improving Electrocoagulation Floatation for Harvesting Microalgae. Algal Res. 2019, 39, 101446. [Google Scholar] [CrossRef] [PubMed]
- Demir, I.; Blockx, J.; Dague, E.; Guiraud, P.; Thielemans, W.; Muylaert, K.; Formosa-Dague, C. Nanoscale Evidence Unravels Microalgae Flocculation Mechanism Induced by Chitosan. ACS Appl. Bio Mater. 2020, 3, 8446–8459. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-N.; Aartsma, T.J.; Thomas, J.-C.; Lamers, G.E.M.; Zhou, B.-C.; Zhang, Y.-Z. Watching the Native Supramolecular Architecture of Photosynthetic Membrane in Red Algae. J. Biol. Chem. 2008, 283, 34946–34953. [Google Scholar] [CrossRef]
- Francius, G.; Tesson, B.; Dague, E.; Martin-Jézéquel, V.; Dufrêne, Y.F. Nanostructure and Nanomechanics of Live Phaeodactylum Tricornutum Morphotypes. Environ. Microbiol. 2008, 10, 1344–1356. [Google Scholar] [CrossRef]
- Karp-Boss, L.; Gueta, R.; Rousso, I. Judging Diatoms by Their Cover: Variability in Local Elasticity of Lithodesmium Undulatum Undergoing Cell Division. PLoS ONE 2014, 9, e109089. [Google Scholar] [CrossRef]
- Losic, D.; Short, K.; Mitchell, J.G.; Lal, R.; Voelcker, N.H. AFM Nanoindentations of Diatom Biosilica Surfaces. Langmuir 2007, 23, 5014–5021. [Google Scholar] [CrossRef]
- Pletikapić, G.; Berquand, A.; Radić, T.M.; Svetličić, V. Quantitative Nanomechanical Mapping of Marine Diatom in Seawater Using Peak Force Tapping Atomic Force Microscopy. J. Phycol. 2012, 48, 174–185. [Google Scholar] [CrossRef]
- Warren, K.M.; Mpagazehe, J.N.; LeDuc, P.R.; Higgs, C.F. Probing the Elastic Response of Microalga Scenedesmus Dimorphus in Dry and Aqueous Environments through Atomic Force Microscopy. Appl. Phys. Lett. 2014, 105, 163701. [Google Scholar] [CrossRef]
- Yap, B.H.J.; Crawford, S.A.; Dagastine, R.R.; Scales, P.J.; Martin, G.J.O. Nitrogen Deprivation of Microalgae: Effect on Cell Size, Cell Wall Thickness, Cell Strength, and Resistance to Mechanical Disruption. J. Ind. Microbiol. Biotech. 2016, 43, 1671–1680. [Google Scholar] [CrossRef]
- Mišić Radić, T.; Vukosav, P.; Komazec, B.; Formosa-Dague, C.; Domazet Jurašin, D.; Peharec Štefanić, P.; Čačković, A.; Juraić, K.; Ivošević DeNardis, N. Nanoplastic-Induced Nanostructural, Nanomechanical, and Antioxidant Response of Marine Diatom Cylindrotheca Closterium. Water 2022, 14, 2163. [Google Scholar] [CrossRef]
- Demir-Yilmaz, I.; Guiraud, P.; Formosa-Dague, C. The Contribution of Atomic Force Microscopy (AFM) in Microalgae Studies: A Review. Algal Res. 2021, 60, 102506. [Google Scholar] [CrossRef]
- Dague, E.; Alsteens, D.; Latgé, J.-P.; Verbelen, C.; Raze, D.; Baulard, A.R.; Dufrêne, Y.F. Chemical Force Microscopy of Single Live Cells. Nano Lett. 2007, 7, 3026–3030. [Google Scholar] [CrossRef] [PubMed]
- Arce, F.T.; Avci, R.; Beech, I.B.; Cooksey, K.E.; Wigglesworth-Cooksey, B. A Live Bioprobe for Studying Diatom-Surface Interactions. Biophys. J. 2004, 87, 4284–4297. [Google Scholar] [CrossRef]
- Pletikapić, G.; Žutić, V.; Vrček, I.V.; Svetličić, V. Atomic Force Microscopy Characterization of Silver Nanoparticles Interactions with Marine Diatom Cells and Extracellular Polymeric Substance. J. Mol. Recognit. 2012, 25, 309–317. [Google Scholar] [CrossRef]
- Bosak, S.; Pletikapić, G.; Hozić, A.; Svetličić, V.; Sarno, D.; Viličić, D. A Novel Type of Colony Formation in Marine Planktonic Diatoms Revealed by Atomic Force Microscopy. PLoS ONE 2012, 7, e44851. [Google Scholar] [CrossRef]
- Lee, A.K.; Lewis, D.M.; Ashman, P.J. Force and Energy Requirement for Microalgal Cell Disruption: An Atomic Force Microscope Evaluation. Bioresour. Technol. 2013, 128, 199–206. [Google Scholar] [CrossRef]
- Binning, G.; Rohrer, H.; Gerber, C.; Weibel, E. Surface Studies by Scanning Tunneling Microscopy. In Scanning Tunneling Microscopy; Neddermeyer, H., Ed.; Perspectives in Condensed Matter Physics; Springer: Dordrecht, The Netherlands, 1982; Volume 6, pp. 31–35. ISBN 978-0-7923-2065-4. [Google Scholar]
- Morris, V.J.; Gunning, A.P.; Kirby, A.R. Atomic Force Microscopy for Biologists, 1st ed.; Imperial College Press: London, UK, 1999; ISBN 1-86094-199-0. [Google Scholar]
- Putman, C.A.J.; Van der Werf, K.O.; De Grooth, B.G.; Van Hulst, N.F.; Greve, J. Tapping Mode Atomic Force Microscopy in Liquid. Appl. Phys. Lett. 1994, 64, 2454–2456. [Google Scholar] [CrossRef]
- Radmacher, M. Measuring the Elastic Properties of Biological Samples with the AFM. IEEE Eng. Med. Biol. Mag. 1997, 16, 47–57. [Google Scholar] [CrossRef]
- Jena, B.P.; Hörber, J.K.H. Atomic Force Microscopy in Cell Biology; Academic Press: San Diego, CA, USA, 2002; ISBN 978-0-12-544171-1. [Google Scholar]
- Butt, H.-J.; Jaschke, M.; Ducker, W. Measuring Surface Forces in Aqueous Electrolyte Solution with the Atomic Force Microscope. Bioelectrochem. Bioenerg. 1995, 38, 191–201. [Google Scholar] [CrossRef]
- Heinz, W.F.; Hoh, J.H. Spatially Resolved Force Spectroscopy of Biological Surfaces Using the Atomic Force Microscope. Trends Biotechnol. 1999, 17, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hertz, H. Ueber die Berührung fester elastischer Körper. J. Für Die Reine Und Angew. Math. 1882, 1882, 156–171. [Google Scholar] [CrossRef]
- Kasas, S.; Dietler, G. Probing The Nanomechanical Properties of Viruses, Cells and Cellular Structures. In Life at the Nanoscale; Jenny Stanford Publishing: Singapore, 2011; pp. 335–353. ISBN 981-4267-96-1. [Google Scholar]
- Sneddon, I.N. The Relation between Load and Penetration in the Axisymmetric Boussinesq Problem for a Punch of Arbitrary Profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Cappella, B.; Dietler, G. Force-Distance Curves by Atomic Force Microscopy. Surf. Sci. Rep. 1999, 34, 5–104. [Google Scholar] [CrossRef]
- Alcaraz, J.; Buscemi, L.; Grabulosa, M.; Trepat, X.; Fabry, B.; Farré, R.; Navajas, D. Microrheology of Human Lung Epithelial Cells Measured by Atomic Force Microscopy. Biophys. J. 2003, 84, 2071–2079. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Bao, G.; Wang, N. Cell Mechanics: Mechanical Response, Cell Adhesion, and Molecular Deformation. Annu. Rev. Biomed. Eng. 2000, 2, 189–226. [Google Scholar] [CrossRef]
- Fabry, B.; Maksym, G.N.; Butler, J.P.; Glogauer, M.; Navajas, D.; Fredberg, J.J. Scaling the Microrheology of Living Cells. Phys. Rev. Lett. 2001, 87, 148102. [Google Scholar] [CrossRef]
- Rother, J.; Nöding, H.; Mey, I.; Janshoff, A. Atomic Force Microscopy-Based Microrheology Reveals Significant Differences in the Viscoelastic Response between Malign and Benign Cell Lines. Open Biol. 2014, 4, 140046. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Okajima, T.; Raman, A. Measuring Viscoelasticity of Soft Biological Samples Using Atomic Force Microscopy. Soft Matter 2020, 16, 64–81. [Google Scholar] [CrossRef]
- Higgins, M.J.; Crawford, S.A.; Mulvaney, P.; Wetherbee, R. Characterization of the Adhesive Mucilages Secreted by Live Diatom Cells Using Atomic Force Microscopy. Protist 2002, 153, 25–38. [Google Scholar] [CrossRef]
- Pickett-Heaps, J.; Schmid, A.M.M.; Edgar, L.A. The Cell Biology of Diatom Valve Formation. In Progress in Phycological Research; Biopress Ltd.: Bristol, UK, 1990. [Google Scholar]
- Pletikapić, G.; Mišić Radić, T.; Zimmermann, A.H.; Svetličić, V.; Pfannkuchen, M.; Marić, D.; Godrijan, J.; Žutić, V. AFM Imaging of Extracellular Polymer Release by Marine Diatom Cylindrotheca Closterium (Ehrenberg) Reiman & J.C. Lewin. J. Mol. Recognit. 2011, 24, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Mišić Radić, T.; Svetličić, V.; Žutić, V.; Boulgaropoulos, B. Seawater at the Nanoscale: Marine Gel Imaged by Atomic Force Microscopy. J. Mol. Recognit. 2011, 24, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, I.C.; Kindt, J.H.; Thompson, J.B.; Del Amo, Y.; Stachelberger, H.; Brzezinski, M.A.; Stucky, G.D.; Morse, D.E.; Hansma, P.K. Atomic Force Microscopy Study of Living Diatoms in Ambient Conditions. J. Microsc. 2003, 212, 292–299. [Google Scholar] [CrossRef]
- Evans, C.T.; Baldock, S.J.; Hardy, J.G.; Payton, O.; Picco, L.; Allen, M.J. A Non-Destructive, Tuneable Method to Isolate Live Cells for High-Speed AFM Analysis. Microorganisms 2021, 9, 680. [Google Scholar] [CrossRef]
- Losic, D.; Rosengarten, G.; Mitchell, J.G.; Voelcker, N.H. Pore Architecture of Diatom Frustules: Potential Nanostructured Membranes for Molecular and Particle Separations. J. Nanosci. Nanotech. 2006, 6, 982–989. [Google Scholar] [CrossRef]
- Losic, D.; Pillar, R.J.; Dilger, T.; Mitchell, J.G.; Voelcker, N.H. Atomic Force Microscopy (AFM) Characterisation of the Porous Silica Nanostructure of Two Centric Diatoms. J. Porous Mater. 2007, 14, 61–69. [Google Scholar] [CrossRef]
- Hildebrand, M.; Doktycz, M.J.; Allison, D.P. Application of AFM in Understanding Biomineral Formation in Diatoms. Pflugers Arch. Eur. J. Physiol. 2008, 456, 127–137. [Google Scholar] [CrossRef]
- Higgins, M.J.; Sader, J.E.; Mulvaney, P.; Wetherbee, R. Probing The Surface of Living Diatoms with Atomic Force Microscopy: The Nanostructure and Nanomechanical Properties of the Mucilage Layer. J. Phycol. 2003, 39, 722–734. [Google Scholar] [CrossRef]
- Hoagland, K.D.; Rosowski, J.R.; Gretz, M.R.; Roemer, S.C. Diatom Extracellular Polymeric Substances: Function, Fine Structure, Chemistry, and Physiology. J. Phycol. 1993, 29, 537–566. [Google Scholar] [CrossRef]
- Mišić Radić, T.; Čačković, A.; Penezić, A.; Dautović, J.; Lončar, J.; Omanović, D.; Juraić, K.; Ljubešić, Z. Physiological and Morphological Response of Marine Diatom Cylindrotheca Closterium (Bacillariophyceae) Exposed to Cadmium. Eur. J. Phycol. 2021, 56, 24–36. [Google Scholar] [CrossRef]
- Schmid, A.-M.M.; Schulz, D. Wall Morphogenesis in Diatoms: Deposition of Silica by Cytoplasmic Vesicles. Protoplasma 1979, 100, 267–288. [Google Scholar] [CrossRef]
- Chiappino, M.L.; Volcani, B.E. Studies on the Biochemistry and Fine Structure of Silicia Shell Formation in Diatoms VII. Sequential Cell Wall Development in the PennateNavicula Pelliculosa. Protoplasma 1977, 93, 205–221. [Google Scholar] [CrossRef]
- Noll, F.; Sumper, M.; Hampp, N. Nanostructure of Diatom Silica Surfaces and of Biomimetic Analogues. Nano Lett. 2002, 2, 91–95. [Google Scholar] [CrossRef]
- Heredia, A.; Silva, S.; Santos, C.; Delgadillo, I.; Vrieling, E.G. Analysis of Cross-Sections of Ditylum Brightwelli Biosilica by Tapping Mode Atomic Force Microscopy and Scanning Electron Microscopy. J. Scanning Probe Microsc. 2008, 3, 19–24. [Google Scholar] [CrossRef]
- Hildebrand, M.; York, E.; Kelz, J.I.; Davis, A.K.; Frigeri, L.G.; Allison, D.P.; Doktycz, M.J. Nanoscale Control of Silica Morphology and Three-Dimensional Structure during Diatom Cell Wall Formation. J. Mat. Res. 2006, 21, 2689–2698. [Google Scholar] [CrossRef]
- Hildebrand, M.; Holton, G.; Joy, D.C.; Doktycz, M.J.; Allison, D.P. Diverse and Conserved Nano- and Mesoscale Structures of Diatom Silica Revealed by Atomic Force Microscopy. J. Microsc. 2009, 235, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, D.; Ciotti, B.J.; Montagnes, D.J.S. Protists Decrease in Size Linearly with Temperature: Ca. 2.5% °C−1. Proc. R. Soc. London. Ser. B Biol. Sci. 2003, 270, 2605–2611. [Google Scholar] [CrossRef]
- Litchman, E.; Klausmeier, C.A.; Yoshiyama, K. Contrasting Size Evolution in Marine and Freshwater Diatoms. Proc. Natl. Acad. Sci. USA 2009, 106, 2665–2670. [Google Scholar] [CrossRef]
- Martignier, A.; Filella, M.; Pollok, K.; Melkonian, M.; Bensimon, M.; Barja, F.; Langenhorst, F.; Jaquet, J.-M.; Ariztegui, D. Marine and Freshwater Micropearls: Biomineralization Producing Strontium-Rich Amorphous Calcium Carbonate Inclusions Is Widespread in the Genus Tetraselmis (Chlorophyta). Biogeosciences 2018, 15, 6591–6605. [Google Scholar] [CrossRef]
- Formosa-Dague, C.; Gernigon, V.; Castelain, M.; Daboussi, F.; Guiraud, P. Towards a Better Understanding of the Flocculation/Flotation Mechanism of the Marine Microalgae Phaeodactylum Tricornutum under Increased PH Using Atomic Force Microscopy. Algal Res. 2018, 33, 369–378. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Direct Characterization of the Physicochemical Properties of Fungal Spores Using Functionalized AFM Probes. Biophys. J. 2000, 78, 3286–3291. [Google Scholar] [CrossRef] [PubMed]
- Laviale, M.; Beaussart, A.; Allen, J.; Quilès, F.; El-Kirat-Chatel, S. Probing the Adhesion of the Common Freshwater Diatom Nitzschia Palea at Nanoscale. ACS Appl. Mater. Interfaces 2019, 11, 48574–48582. [Google Scholar] [CrossRef] [PubMed]
- Demir, I.; Lüchtefeld, I.; Lemen, C.; Dague, E.; Guiraud, P.; Zambelli, T.; Formosa-Dague, C. Probing the Interactions between Air Bubbles and (Bio)Interfaces at the Nanoscale Using FluidFM Technology. J. Colloid Interface Sci. 2021, 604, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Beaussart, A.; El-Kirat-Chatel, S.; Sullan, R.M.A.; Dufrêne, Y.F. Atomic Force Microscopy: A New Look at Pathogens. PLOS Pathog. 2013, 9, e1003516. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Sticky Microbes: Forces in Microbial Cell Adhesion. Trends Microbiol. 2015, 23, 376–382. [Google Scholar] [CrossRef]
- Bowen, W.R.; Hilal, N.; Lovitt, R.W.; Wright, C.J. Direct Measurement of the Force of Adhesion of a Single Biological Cell Using an Atomic Force Microscope. Colloids Surf. A Physicochem. Eng. Asp. 1998, 136, 231–234. [Google Scholar] [CrossRef]
- Penedo, M.; Miyazawa, K.; Okano, N.; Furusho, H.; Ichikawa, T.; Alam, M.S.; Miyata, K.; Nakamura, C.; Fukuma, T. Visualizing Intracellular Nanostructures of Living Cells by Nanoendoscopy-AFM. Sci. Adv. 2021, 7, eabj4990. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).