Batch Studies on the Biodegradation Potential of Paracetamol, Fluoxetine and 17α-Ethinylestradiol by the Micrococcus yunnanensis Strain TJPT4 Recovered from Marine Organisms
Abstract
:1. Introduction
2. Materials and Methods
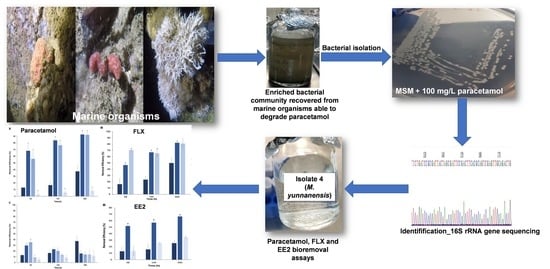
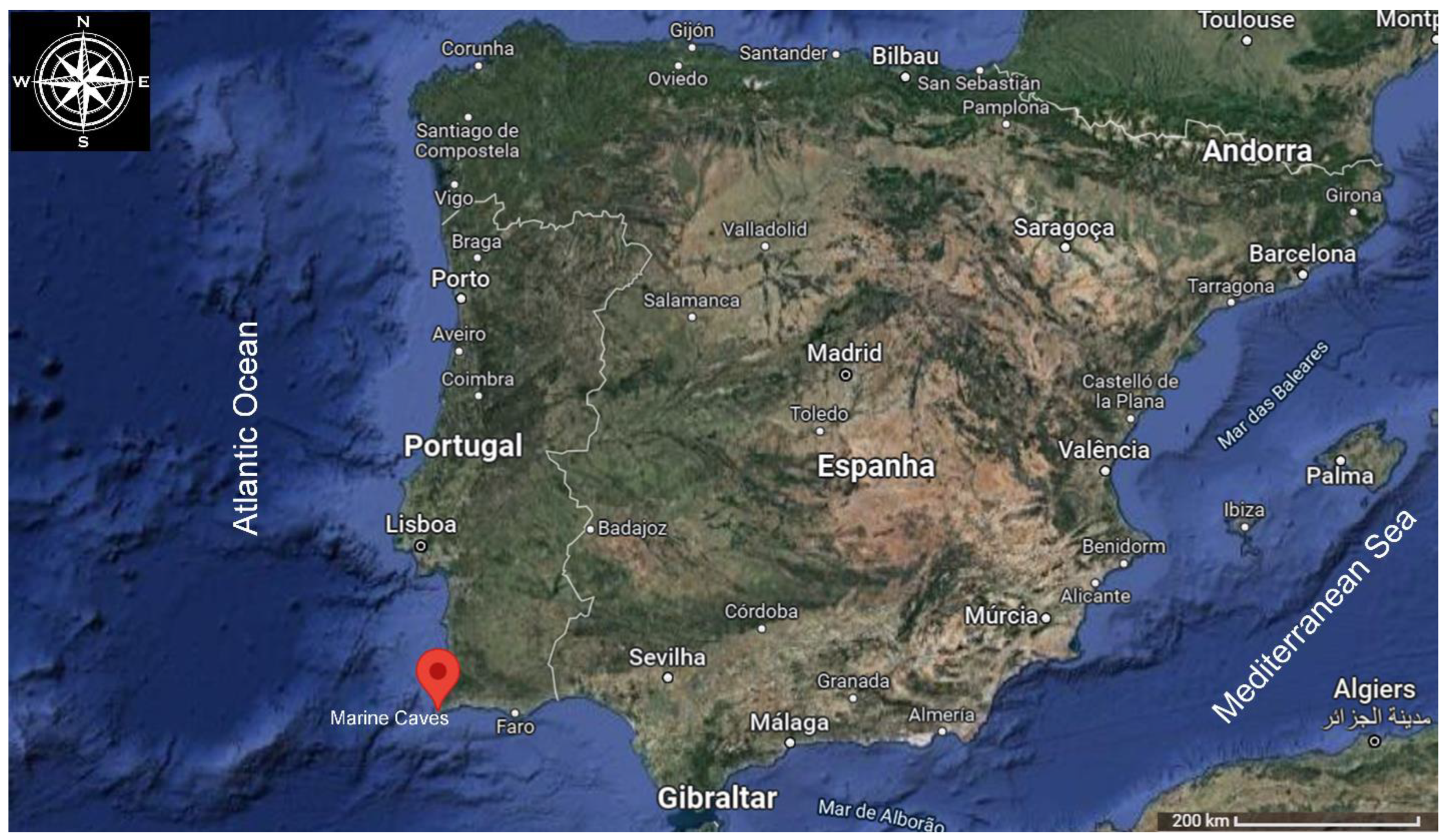
2.1. Inoculum Source
2.2. Recovery of Bacteria from Marine Organisms
2.3. Inoculum Enrichment
2.4. Pharmaceutical Bioremoval Studies Using Bacteria Recovered from Marine Organisms
2.4.1. Bacterial Isolates Preparation
2.4.2. Removal Assays with Bacterial Isolates
2.4.3. Pharmaceutical and Metabolites Analysis
2.5. Statistical Analysis
3. Results/Discussion
3.1. Bacterial isolation from Consortia Recovered from Marine Organisms with the Ability to Degrade Paracetamol
3.2. Bioremoval of Pharmaceuticals by Micrococcus yunnanensis Strain TJPT4
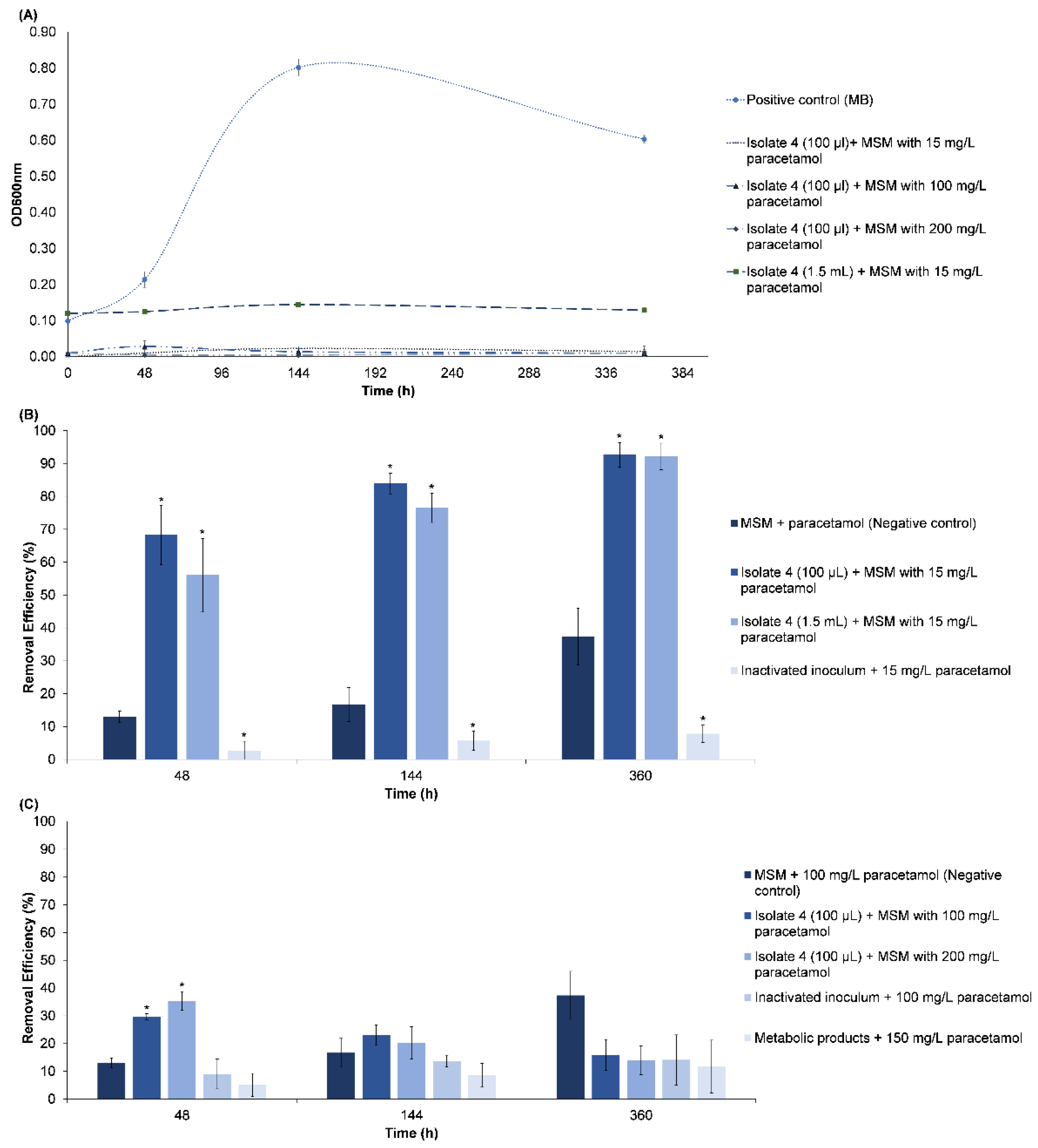
3.2.1. Paracetamol Removal
3.2.2. Fluoxetine Removal
Adsorption of FLX onto the Inactivated Cells
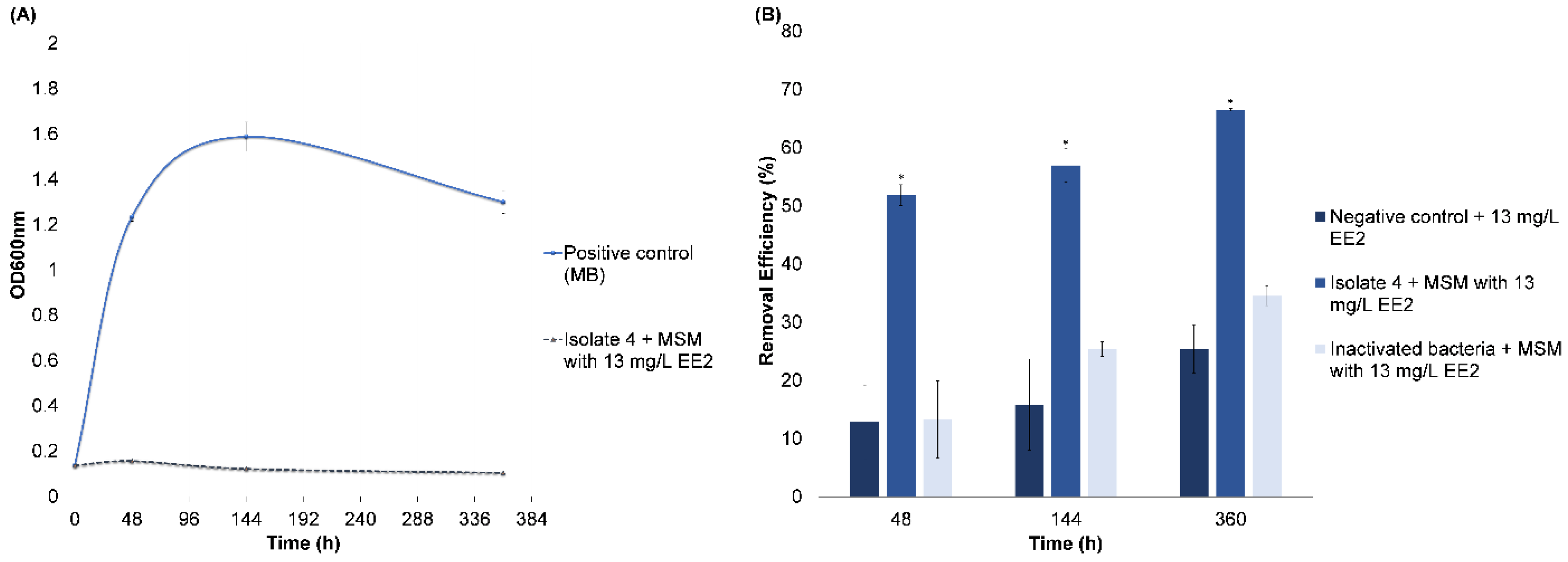
3.2.3. EE2 Removal
3.3. Hypothetical Genetic Potential of M. yunnanensis for Drug Biodegradation Based on Its Encoded Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurnham, C. Limitations of sewage treatment plants in handling industrial wastes. J. Water Pollut. Control Fed. 1960, 32, 211–215. [Google Scholar]
- Gu, Y.; Li, B.; Zhong, X.; Liu, C.; Ma, B. Bacterial community composition and function in a tropical municipal wastewater treatment plant. Water 2022, 14, 1537. [Google Scholar] [CrossRef]
- Martins, M.; Sanches, S.; Pereira, I.A. Anaerobic biodegradation of pharmaceutical compounds: New insights into the pharmaceutical-degrading bacteria. J. Hazard Mater. 2018, 357, 289–297. [Google Scholar] [CrossRef]
- Pólvora, S.; Aníbal, J.; Martins, A. Saline intrusions at Almargem waste water treatment plant in different tidal cycles. In Proceedings of the 2020 2nd International Congress on Engineering and Sustainability in the XXI Century INCREaSE, Faro, Portugal, 9–11 October 2019; pp. 786–798. [Google Scholar] [CrossRef]
- Fusi-Schmidhauser, T.; Preston, N.J.; Keller, N.; Gamondi, C. Conservative management of COVID-19 patients-emergency palliative care in action. J. Pain Symptom. Manag. 2020, 60, e27–e30. [Google Scholar] [CrossRef]
- Palma, T.L.; Donaldben, M.N.; Costa, M.C.; Carlier, J.D. Putative role of Flavobacterium, Dokdonella and Methylophilus strains in paracetamol biodegradation. Water Air Soil. Pollut. 2018, 229, 200. [Google Scholar] [CrossRef]
- Brumovský, M.; Bečanová, J.; Kohoutek, J.; Borghini, M.; Nizzetto, L. Contaminants of emerging concern in the open sea waters of the Western Mediterranean. Environ. Pollut. 2017, 229, 976–983. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, J.; Zhu, R.; Zhou, Q.; Chen, J. Degradation of paracetamol by pure bacterial cultures and their microbial consortium. Appl. Microbiol. Biotechnol. 2013, 97, 3687–4369. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, L.; Chen, J. Paracetamol in the environment and its degradation by microorganisms. Appl. Microbiol. Biotechnol. 2012, 96, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Joss, A.; Zabczynski, S.; Göbel, A.; Hofmann, B.; Löfer, D.; McArdell, C.S.; Ternes, T.A.; Thomsen, A.; Siegrist, H. Biological degradation of pharmaceuticals in municipal wastewater treatment: Proposing a classification scheme. Water Res. 2006, 40, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Ferchichi, M.; Dhaouadi, H. Sorption of paracetamol onto biomaterials. Water Sci. Technol. 2016, 74, 287–294. [Google Scholar] [CrossRef]
- Daniel, D.; Nunes, B.; Pinto, E.; Ferreira, I.M.P.L.V.O.; Correia, A.T. Assessment of paracetamol toxic effects under varying seawater pH conditions on the marine polychaete Hediste diversicolor using biochemical endpoints. Biology 2022, 11, 581. Available online: https://www.mdpi.com/2079-7737/11/4/581 (accessed on 3 August 2022). [CrossRef] [PubMed]
- Kwon, J.W.; Armbrust, K.L. Laboratory persistence and fate of fluoxetine in aquatic environments. Environ. Toxicol. Chem. 2006, 25, 2561–2568. [Google Scholar] [CrossRef]
- Kreke, N.; Dietrich, D.R. Physiological endpoints for potential SSRI interactions in fish. Crit. Rev. Toxicol. 2008, 38, 215–247. [Google Scholar] [CrossRef] [Green Version]
- Peters, J.R.; Granek, E.F.; de Rivera, C.E.; Rollins, M. Prozac in the water: Chronic fluoxetine exposure and predation risk interact to shape behaviors in an estuarine crab. Ecol. Evol. 2017, 7, 9151–9161. [Google Scholar] [CrossRef]
- Vasskog, T.; Anderssen, T.; Pedersen-Bjergaard, T.; Kallenborn, R.; Jensen, E. Occurrence of selective serotonin reuptake inhibitors in sewage and receiving waters at Spitsbergen and in Norway. J. Chromatogr. A 2008, 1185, 194–205. [Google Scholar] [CrossRef]
- Khan, M.F.; Murphy, C.D. Bacterial degradation of the anti-depressant drug fluoxetine produces trifluoroacetic acid and fluoride ion. Appl. Microbiol. Biotechnol. 2021, 105, 9359–9369. [Google Scholar] [CrossRef]
- Das, S.; Ray, N.M.; Wan, J.; Khan, A.; Chakraborty, T.; Ray, M.B. Micropollutants in wastewater: Fate and removal processes in: Physico-chemical wastewater treatment and resource recovery. IntechOpen 2017, 5, 76–107. [Google Scholar] [CrossRef] [Green Version]
- Velazquez, Y.F.; Nacheva, P.M. Biodegradability of fluoxetine, mefenamic acid, and metoprolol using different microbial consortiums. Environ. Sci. Pollut. Res. Int. 2017, 24, 6779–6793. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Vijitkul, P.; Kongsema, M.; Toommakorn, T.; Bullangpoti, V. Investigation of genotoxicity, mutagenicity, and cytotoxicity in erythrocytes of Nile tilapia (Oreochromis niloticus) after fluoxetine exposure. Toxicol Rep. 2022, 29, 588–596. [Google Scholar] [CrossRef]
- Commission Implementing Decision (EU) 2020/1161 of 4 August 2020 Establishing a Watch List of Substances for Union-Wide Monitoring in the Field of Water Policy Pursuant to Directive 2008/105/EC of the European Parliament and of the Council (Notified under Document Number C(2020) 5205). Off. J. Eur. Union 2020, L257, 32–35. Available online: https://op.europa.eu/en/publication-detail/-/publication/5bd350f7-d7b1-11ea-adf7-01aa75ed71a1 (accessed on 7 June 2022).
- Huang, B.; Wang, B.; Ren, D.; Jin, W.; Liu, J.; Peng, J.; Pan, X. Occurrence, removal and bioaccumulation of steroid estrogens in Dianchi lake catchment. China. Environ. Int. 2013, 59, 262–273. [Google Scholar] [CrossRef]
- Klaic, M.; Jirsa, F. 17α-Ethinylestradiol (EE2): Concentrations in the environment and methods for wastewater treatment—An update. RSC Adv. 2022, 12, 12794–12805. [Google Scholar] [CrossRef]
- Ternes, T.; Stumpf, M.; Mueller, J.; Haberer, K.; Wilken, R.-D.; Servos, M. Behavior and occurrence of estrogens in municipal sewage treatment plants-I. investigations in Germany, Canada and Brazil. Sci. Total Environ. 1999, 225, 81–90. [Google Scholar] [CrossRef]
- Bhandari, R.K.; Deem, S.L.; Holliday, D.K.; Jandegian, C.M.; Kassotis, C.D.; Nagel, S.C.; Tillitt, D.E.; Vom Saal, F.S.; Rosenfeld, C.S. Effects of the environmental estrogenic contaminants bisphenol A and 17α-ethinyl estradiol on sexual development and adult behaviors in aquatic wildlife species. Gen. Comp. Endocrinol. 2015, 214, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Schröder, P.; Helmreich, B.; Skrbic, B.; Carballa, M.; Papa, M.; Pastore, C.; Emre, Z.; Oehmen, A.; Langenhoff, A.; Molinos, M.; et al. Status of hormones and painkillers in wastewater effluents across several European states-considerations for the EU watch list concerning estradiols and diclofenac. Environ. Sci. Pollut. Res. Int. 2016, 23, 12835–12866. [Google Scholar] [CrossRef] [PubMed]
- Palma, T.L.; Shylova, A.; Costa, M.C. Isolation and characterization of bacteria from activated sludge capable of degrading 17α-ethinylestradiol, a contaminant of high environmental concern. Microbiology 2021, 167, 4. [Google Scholar] [CrossRef] [PubMed]
- Haiyan, R.; Shulan, J.; ud din Ahmad, N.; Dao, W.; Chengwu, C. Degradation characteristics and metabolic pathway of 17-alphaethynylestradiol by Sphingobacterium sp. JCR5. Chemosphere 2007, 66, 340–346. [Google Scholar] [CrossRef]
- Wojnarowski, K.; Podobiński, P.; Cholewińska, P.; Smoliński, J.; Dorobisz, K. Impact of estrogens present in environment on health and welfare of animals. Animals 2021, 11, 2152. [Google Scholar] [CrossRef]
- Allen, E.E.; Bartlett, D.H. Piezophiles: Microbial adaptation to the deep-sea environment. In Extremophiles; Eolss Publishers Co., Ltd.: Oxford, UK, 2002; pp. 1–26. [Google Scholar]
- Palma, T.L.; Shylova, A.; Carlier, J.D.; Costa, M.C. An autochthonous aerobic bacterial community and its cultivable isolates capable of degrading fluoxetine. J. Chem. Technol. Biotechnol. 2021, 96, 10. [Google Scholar] [CrossRef]
- Palma, T.L.; Magno, G.; Costa, M.C. Biodegradation of paracetamol by some gram positive bacterial isolates. Curr. Microbiol. 2021, 78, 2774–2786. [Google Scholar] [CrossRef]
- Palma, T.L.; Costa, M.C. Anaerobic biodegradation of fluoxetine using a high performance bacterial community. Anaerobe 2021, 68, 102356. [Google Scholar] [CrossRef]
- Ho, Y.; McKay, G. Pseudo-second order model for sorption processes. Process Biochem. 1999, 34, 451–465. [Google Scholar] [CrossRef]
- Çeçen, F.; Gül, G. Biodegradation of five pharmaceuticals: Estimation by predictive Models and comparison with activated sludge data. Int. J. Environ. Sci. Technol. 2021, 18, 327–340. [Google Scholar] [CrossRef]
- Betts, G. Other spoilage bacteria 23. In Food Spoilage Microorganisms; Woodhead Publishing Series in Food Science, Technology and Nutrition; Blackburn, C.d.W., Ed.; Woodhead Publishing; Elsevier; Unilever Research Colworth: Bedford, UK, 2006; pp. 668–693. [Google Scholar] [CrossRef]
- Zhao, G.Z.; Li, J.; Qin, S.; Zhang, Y.Q.; Zhu, W.Y.; Jiang, C.L.; Xu, L.H.; Li, W.J. Micrococcus yunnanensis sp. nov., a novel actinobacterium isolated from surface-sterilized Polyspora axillaris roots. Int. J. Syst. Evol. Microbiol. 2009, 59, 2383–2387. [Google Scholar] [CrossRef] [PubMed]
- Palma, T.L.; Vieira, B.; Nunes, J.; Lourenço, J.P.; Monteiro, O.C.; Costa, M.C. Photodegradation of chloramphenicol and paracetamol using PbS/TiO2 nanocomposites produced by green synthesis. J. Iran. Chem. Soc. 2020, 17, 2013–2031. [Google Scholar] [CrossRef]
- El-Naas, M.H.; Alhaija, M.A. Modeling of Adsorption Processes. In Mathematical Modelling; Brennan, C.R., Ed.; Nova Publishers Inc.: Hauppauge, NY, USA, 2011; pp. 1–22. [Google Scholar]
- Kumar, K.V.; Gadipelli, S.; Wood, B.; Ramisetty, K.A.; Stewart, A.A.; Howard, C.A.; Brett, D.J.L.; Rodriguez-Reinoso, F. Characterization of the adsorption site energies and heterogeneous surfaces of porous materials. J. Mater. Chem. 2019, 7, 10104–10137. [Google Scholar] [CrossRef] [Green Version]
- Chopra, S.; Kumar, D. Characterization, optimization and kinetics study of acetaminophen degradation by Bacillus drentensis strain S1 and waste water degradation analysis. Bioresour. Bioprocess. 2020, 7, 9. [Google Scholar] [CrossRef]
- Żur, J.; Wojcieszyńska, D.; Hupert-Kocurek, K.; Marchlewicz, A.; Guzik, U. Paracetamol-toxicity and microbial utilization. Pseudomonas moorei KB4 as a case study for exploring degradation pathway. Chemosphere 2018, 206, 192–202. [Google Scholar] [CrossRef]
- Sharma, K.; Kaushik, G.; Thotakura, N.; Raza, K.; Sharma, N.; Nimesh, S. Enhancement effects of process optimization technique while elucidating the degradation pathways of drugs present in pharmaceutical industry wastewater using Micrococcus yunnanensis. Chemosphere 2020, 238, 124689. [Google Scholar] [CrossRef]
- Shammas, N.K.; Wang, L.K.; Pereira, N.C.; Hung, Y. Biological Treatment Processes. In The Handbook of Environmental Engineering; Shammas, N.K., Wang, L.K., Pereira, N.C., Hung, Y., Eds.; The Humana Press: Towona, NJ, USA, 2009; pp. 513–538. [Google Scholar]
- Ejhed, H.; Fång, J.; Hansen, K.; Graae, L.; Rahmberg, M.; Magnér, J.; Dorgeloh, E.; Plaza, G. The effect of hydraulic retention time in onsite wastewater treatment and removal of pharmaceuticals, hormones and phenolic utility substances. Sci. Total Environ. 2018, 618, 250–261. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. Application of microorganisms in bioremediation-review. J. Environ. Microbiol. 2017, 1, 02–09. [Google Scholar]
- PATRIC 3.6.12. Available online: https://www.patricbrc.org/view/Genome/566027.4 (accessed on 1 August 2022).
- Rios-Miguel, A.B.; Smith, G.J.; Cremers, G.; van Alen, T.; Jetten, M.S.M.; Op den Camp, H.J.M.; Welte, C.U. Microbial paracetamol degradation involves a high diversity of novel amidase enzyme candidates. Water Res. 2022, 16, X. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.W.Y.; Chakrabarti, N.; Ing, C.; Halgas, O.; To, T.K.W.; Wälti, M.; Petit, A.-P.C.; Tran, C.; Savchenko, A.; Yakunin, A.F.; et al. Defluorination capability of l-2-haloacid dehalogenases in the HAD-like hydrolase superfamily correlates with active site compactness. ChemBioChem 2022, 23, e202100414. [Google Scholar] [CrossRef]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012, 335, 233–235. Available online: https://www.science.org/doi/10.1126/science.1215063 (accessed on 8 August 2022). [CrossRef] [Green Version]
- Chen, Y.L.; Yu, C.P.; Lee, T.H.; Goh, K.S.; Chu, K.H.; Wang, P.H.; Ismail, W.; Shih, C.J.; Chiang, Y.R. Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 2017, 24, 712–724.e7. [Google Scholar] [CrossRef] [PubMed]


| Pharmaceutical Compound/Sole Carbon Source | Study | Process | Strain/ Microbial Consortium | Respiratory Condition | Biodegradation | Adsorption | Extra Carbon Source | Putative Metabolic Pathway/ Degradation Products |
|---|---|---|---|---|---|---|---|---|
| Paracetamol | This work | Batch | Bacterial communities/ M. yunnanensis strain TJPT4 | Aerobic | >60% of 15 mg/L after 48 h | N.S. | No |  |
| Palma et al. [33] | Batch | Bacillus cereus group; Corynebacterium nuruki; Enterococcus faecium | Aerobic | >90% of 200 mg/L after 48 h | N.S. | No | 4-Aminophenol; hydroquinone; 2-hexenoic acid | |
| Chopra and Kumar [42] | 20 L batch reactor | Bacillus drentensis strain S1 | Anaerobic | >90% of 50–1200 mg/L after 48 h | N.D. | No | Hydroquinone; 2-isopropyl-5-methylcyclohexanone; oxalic acid and lactic acid; formic acid; carbon dioxide | |
| Zur et al. [43] | Batch | Pseudomonas moorei KB4 | Aerobic | 50 mg/L—Complete | N.D. | Glucose | 4-Aminophenol; hydroquinone; p-aminophenol and p-nitrophenol; carboxylic acids | |
| Sharma et al. [44] | Batch | M. yunnanensis KGP04 | Aerobic | >80% after 6 h | N.D. | Dextrose/peptone | N.D. | |
| Fluoxetine | This work | Batch | M. yunnanensis strain TJPT4 | Aerobic | ≈50% after 48 h | >50% after 48 h | No | Biodegradation/adsorption mechanisms |
| Palma et al. [32] | Batch | Community/Pseudomonas putida; Enterobacter ludwigii; P. nitritireducens; Alcaligenes faecalis; P. aeruginosa; P. nitroreducens | Aerobic | >50% after 48 h | N.D. | Residual carbon sources existing in the raw wastewater/only FLX | N.D. | |
| Khan and Murphy [17] | Batch | P. knackmussii B-13 | Aerobic | 5 mM—Complete after 72 h | N.D. | No | 4-(trifluoromethyl)phenol (TFMP) and 3-(methylamino)-1-phenylpropan-1-ol; 2-hydroxy-6-oxo-7,7,7-trifuorohepta-2,4-dienoic acid (7-TFHOD) | |
| EE2 | This work | Batch | M. yunnanensis strain TJPT4 | Aerobic | >50% of 13 mg/L after 48 h | <50% after 48 h | No |  |
| Palma et al. [28] | Batch | Acinetobacter bouvetii; Acinetobacter kokii; Pantoe agglomerans; Shinella zoogloeoides | Aerobic | >50% of 13 mg/L after 48 h | N.D. | No | Estrone (E1); γ-lactone compounds, 2-pentanedioic acid and 2-butenedioic acid, an intermediate metabolite of the TCA cycle |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, T.; Valentine, J.; Gomes, V.; Faleiro, M.; Costa, M. Batch Studies on the Biodegradation Potential of Paracetamol, Fluoxetine and 17α-Ethinylestradiol by the Micrococcus yunnanensis Strain TJPT4 Recovered from Marine Organisms. Water 2022, 14, 3365. https://doi.org/10.3390/w14213365
Palma T, Valentine J, Gomes V, Faleiro M, Costa M. Batch Studies on the Biodegradation Potential of Paracetamol, Fluoxetine and 17α-Ethinylestradiol by the Micrococcus yunnanensis Strain TJPT4 Recovered from Marine Organisms. Water. 2022; 14(21):3365. https://doi.org/10.3390/w14213365
Chicago/Turabian StylePalma, Tânia, Julia Valentine, Vera Gomes, Maria Faleiro, and Maria Costa. 2022. "Batch Studies on the Biodegradation Potential of Paracetamol, Fluoxetine and 17α-Ethinylestradiol by the Micrococcus yunnanensis Strain TJPT4 Recovered from Marine Organisms" Water 14, no. 21: 3365. https://doi.org/10.3390/w14213365
APA StylePalma, T., Valentine, J., Gomes, V., Faleiro, M., & Costa, M. (2022). Batch Studies on the Biodegradation Potential of Paracetamol, Fluoxetine and 17α-Ethinylestradiol by the Micrococcus yunnanensis Strain TJPT4 Recovered from Marine Organisms. Water, 14(21), 3365. https://doi.org/10.3390/w14213365