Alkylphenols and Chlorophenols Remediation in Vertical Flow Constructed Wetlands: Removal Efficiency and Microbial Community Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Microcosm Experiments
2.3. CWs Sampling
2.4. Analytical Methodologies
2.4.1. Nutrient and COD Analysis
2.4.2. Phenolic Compounds Analysis
2.5. Microbial Communities’ Characterisation
2.5.1. DNA Extraction, PCR Amplification and Sequencing
2.5.2. Bioinformatics Analysis
2.5.3. Diversity and Community Composition of Microbial Community
2.6. Statistical Analysis
3. Results
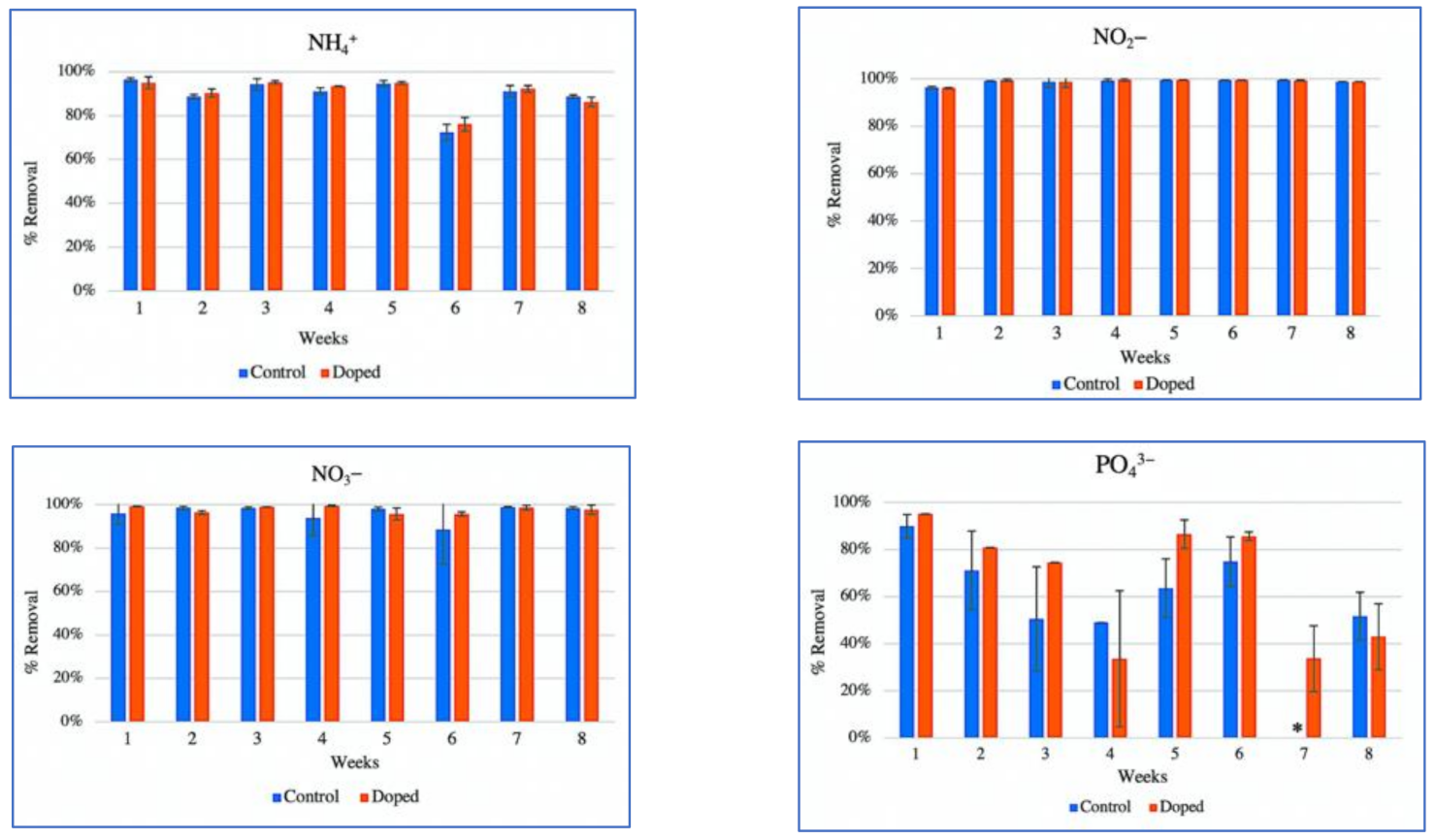
3.1. Removal of a Mixture of Alkylphenol Compounds and Nutrients from Contaminated Water by CWs Microcosms
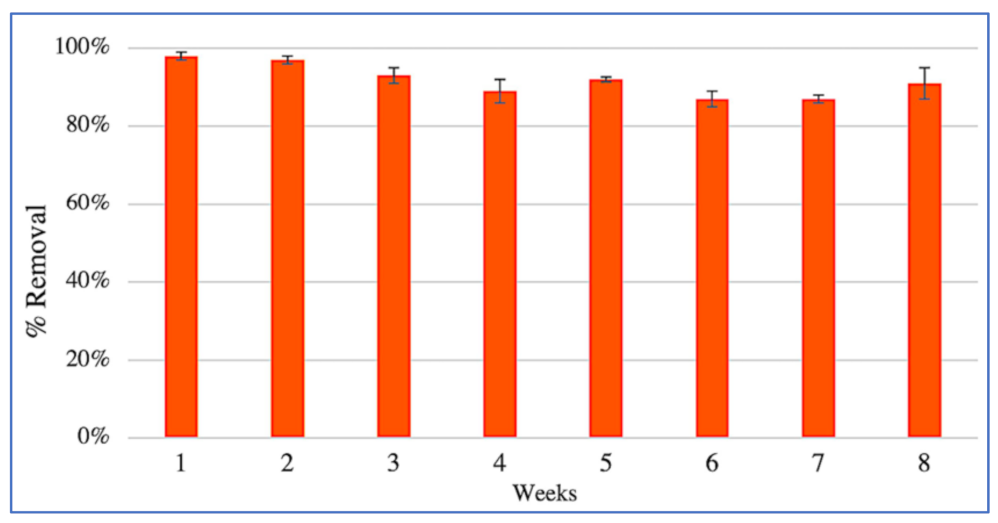
3.2. Removal of PCP and Nutrients from Contaminated Water by CWs Microcosms
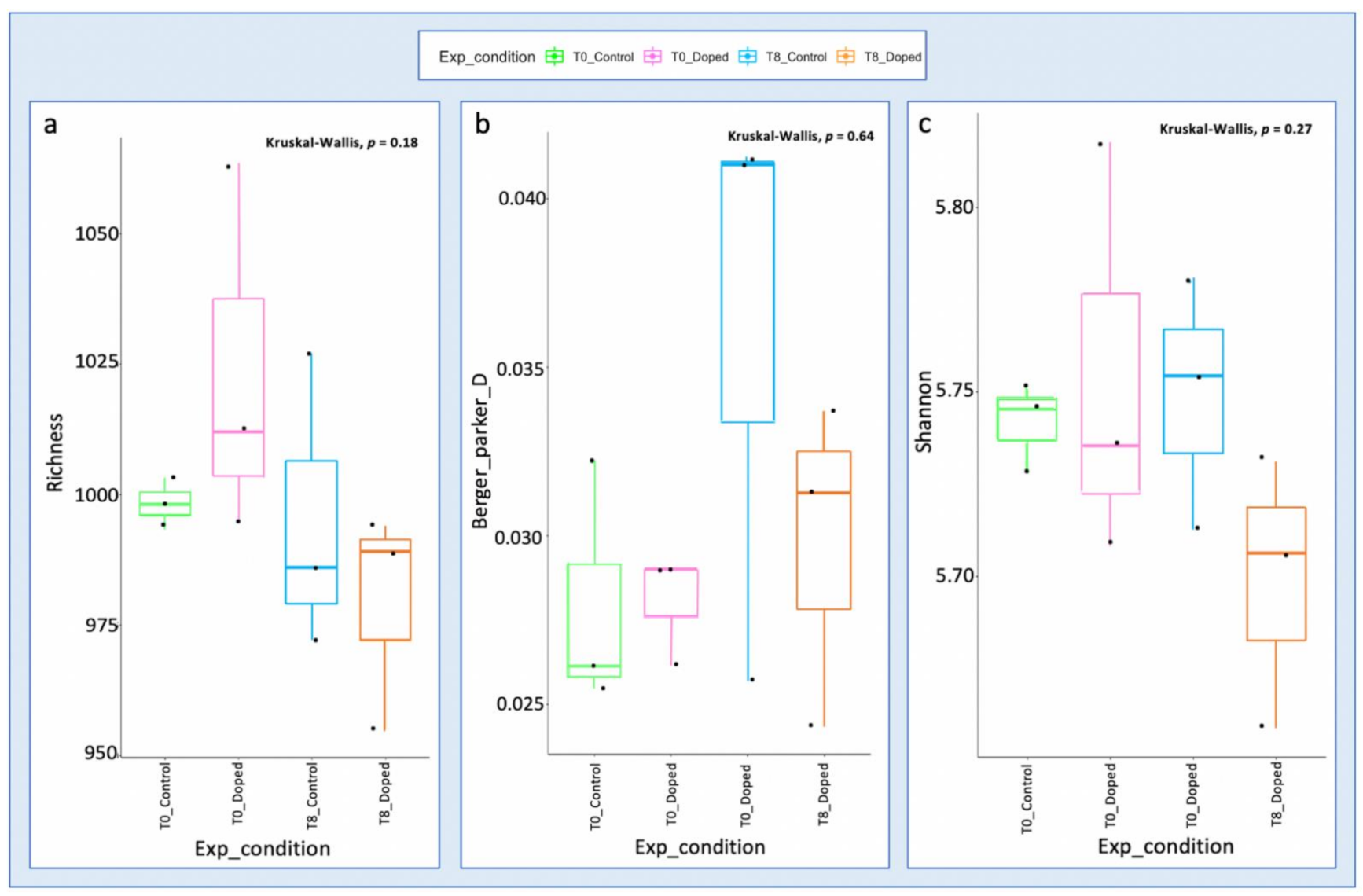
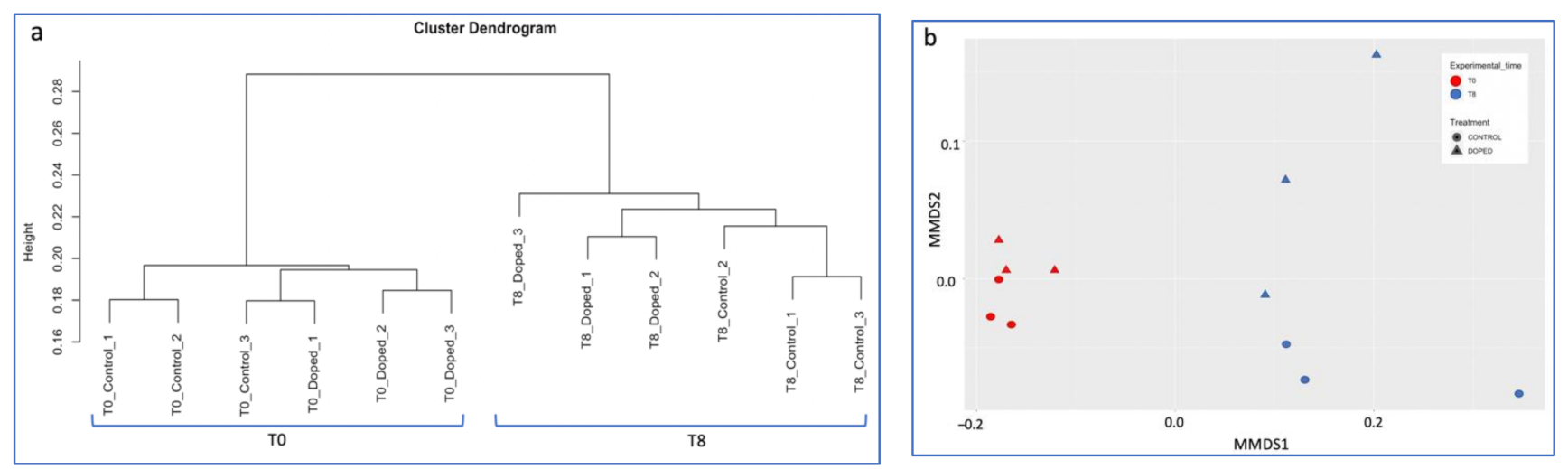
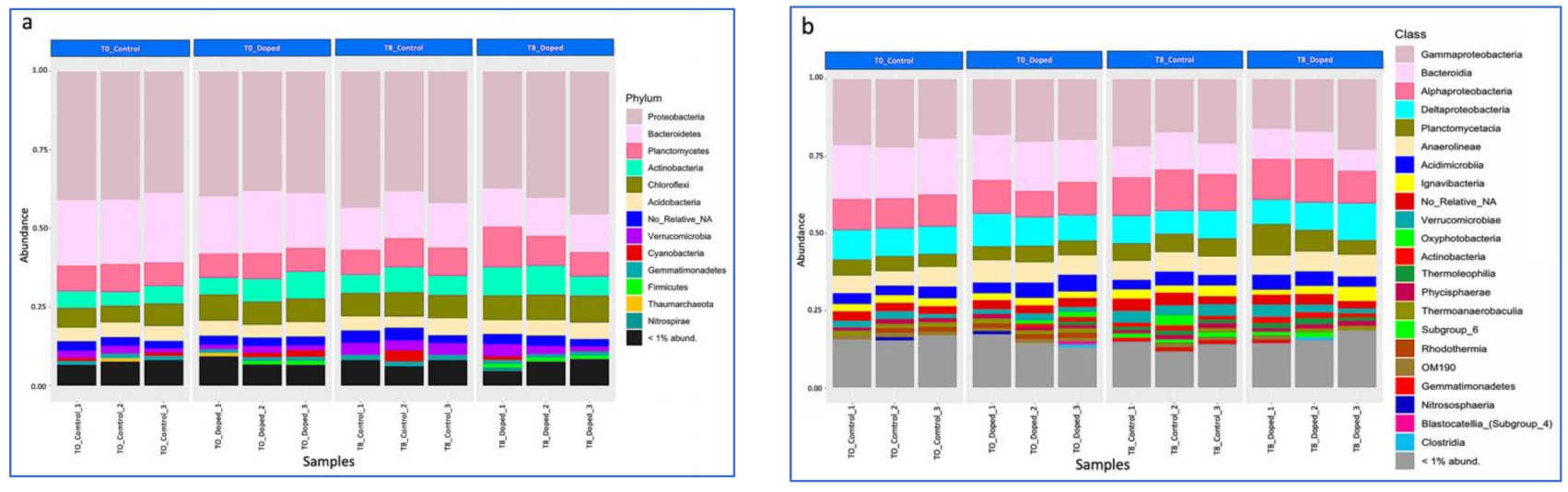
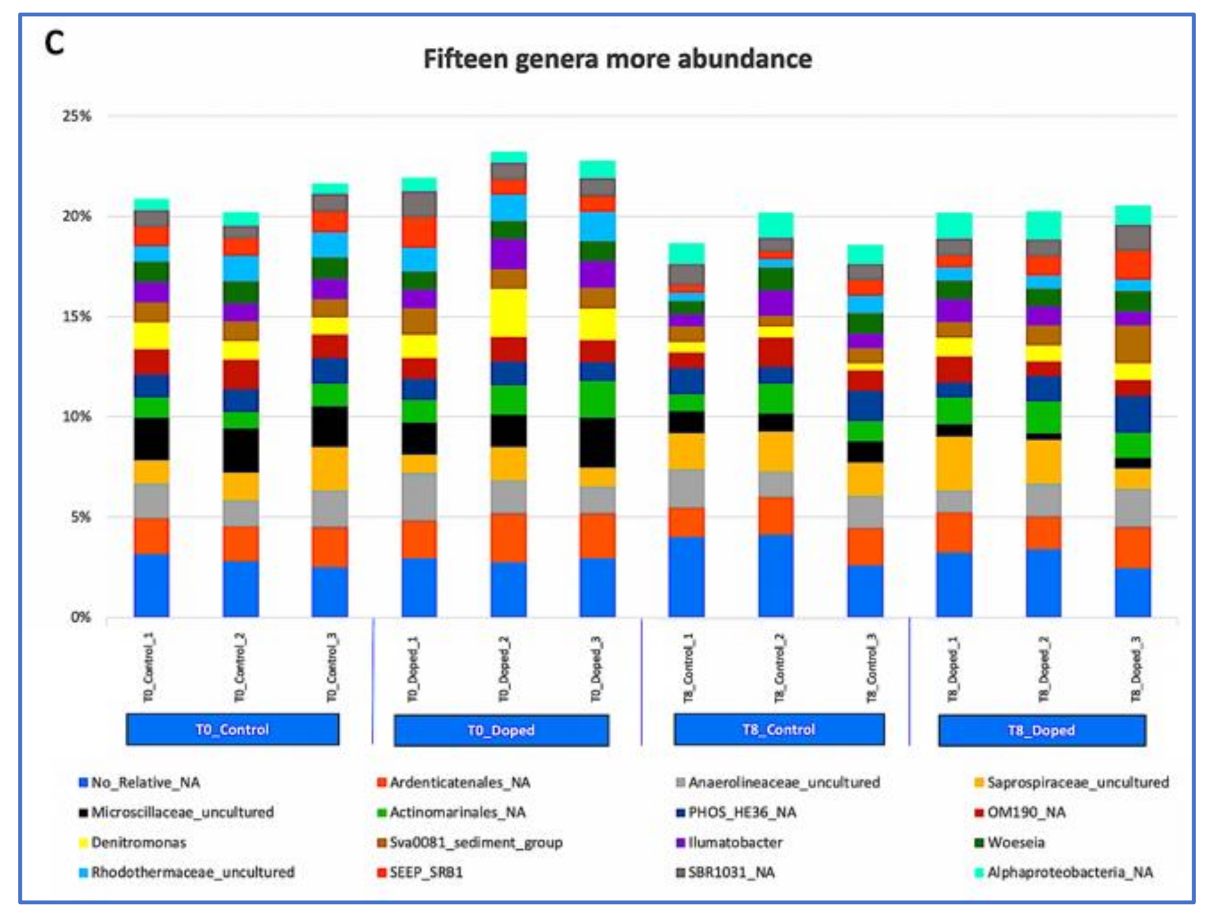
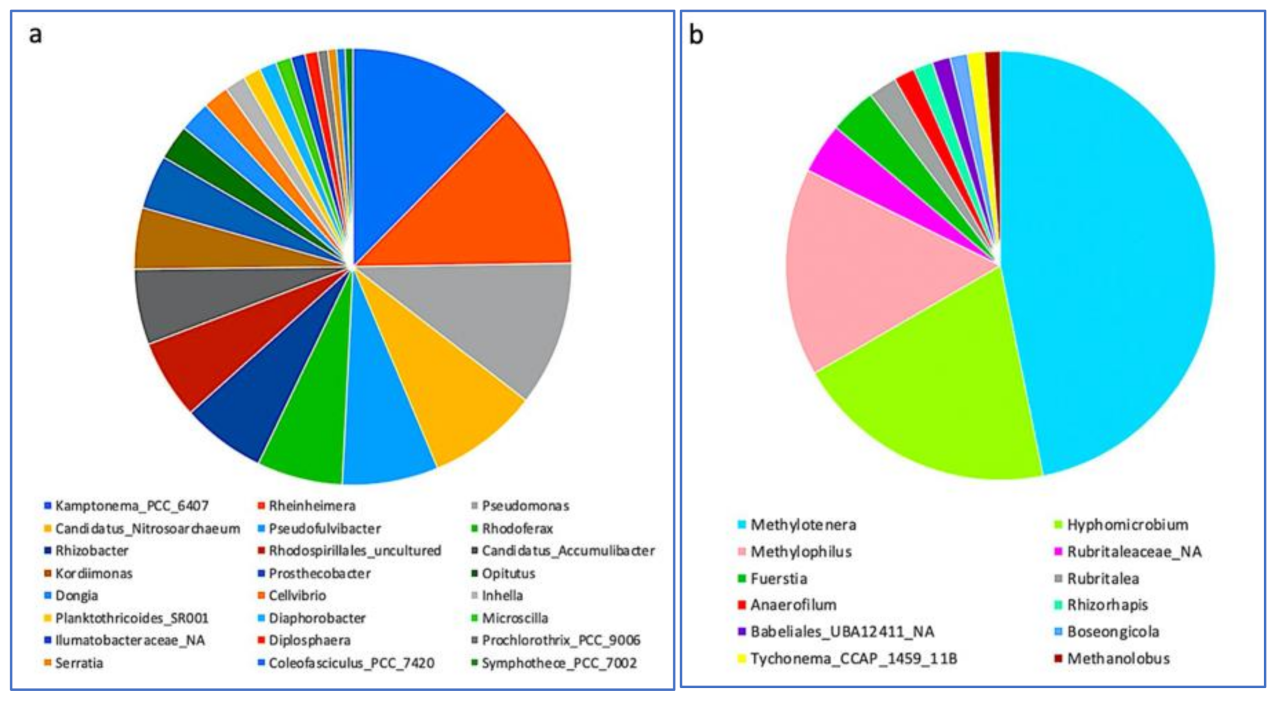
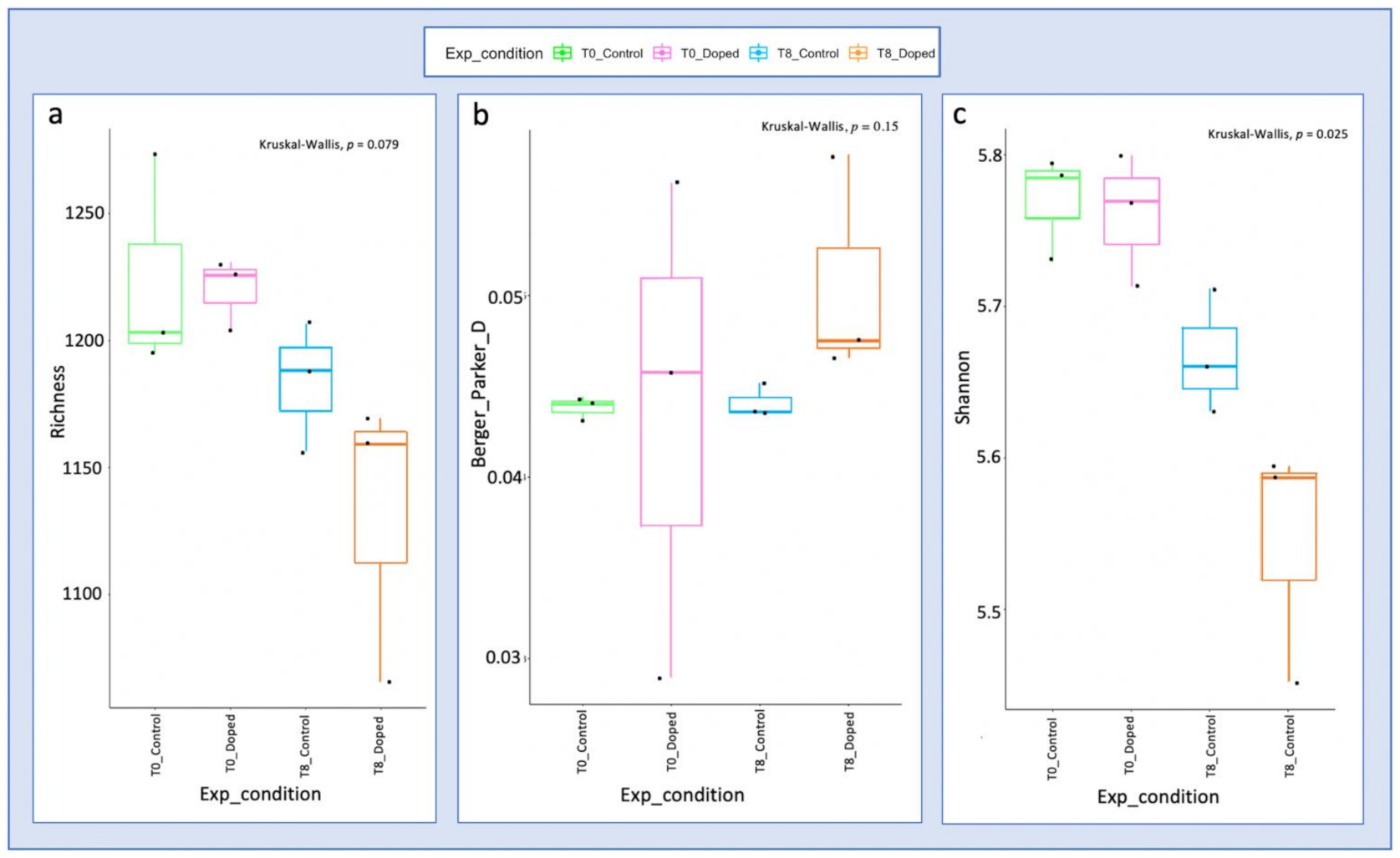
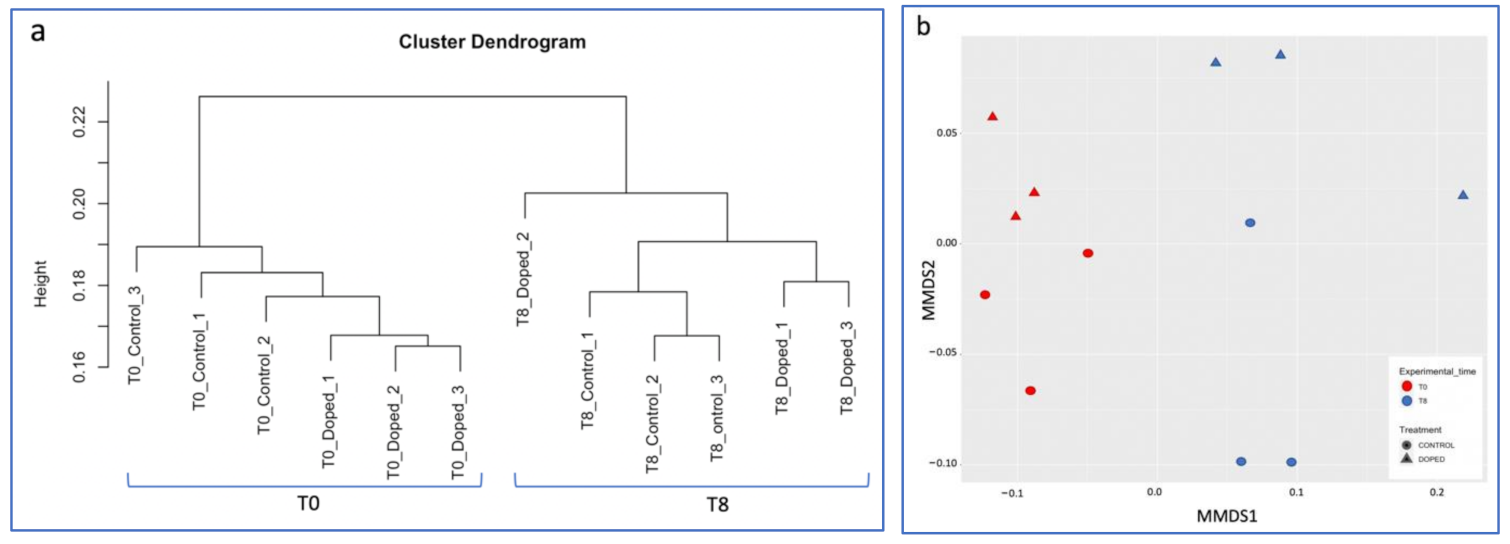
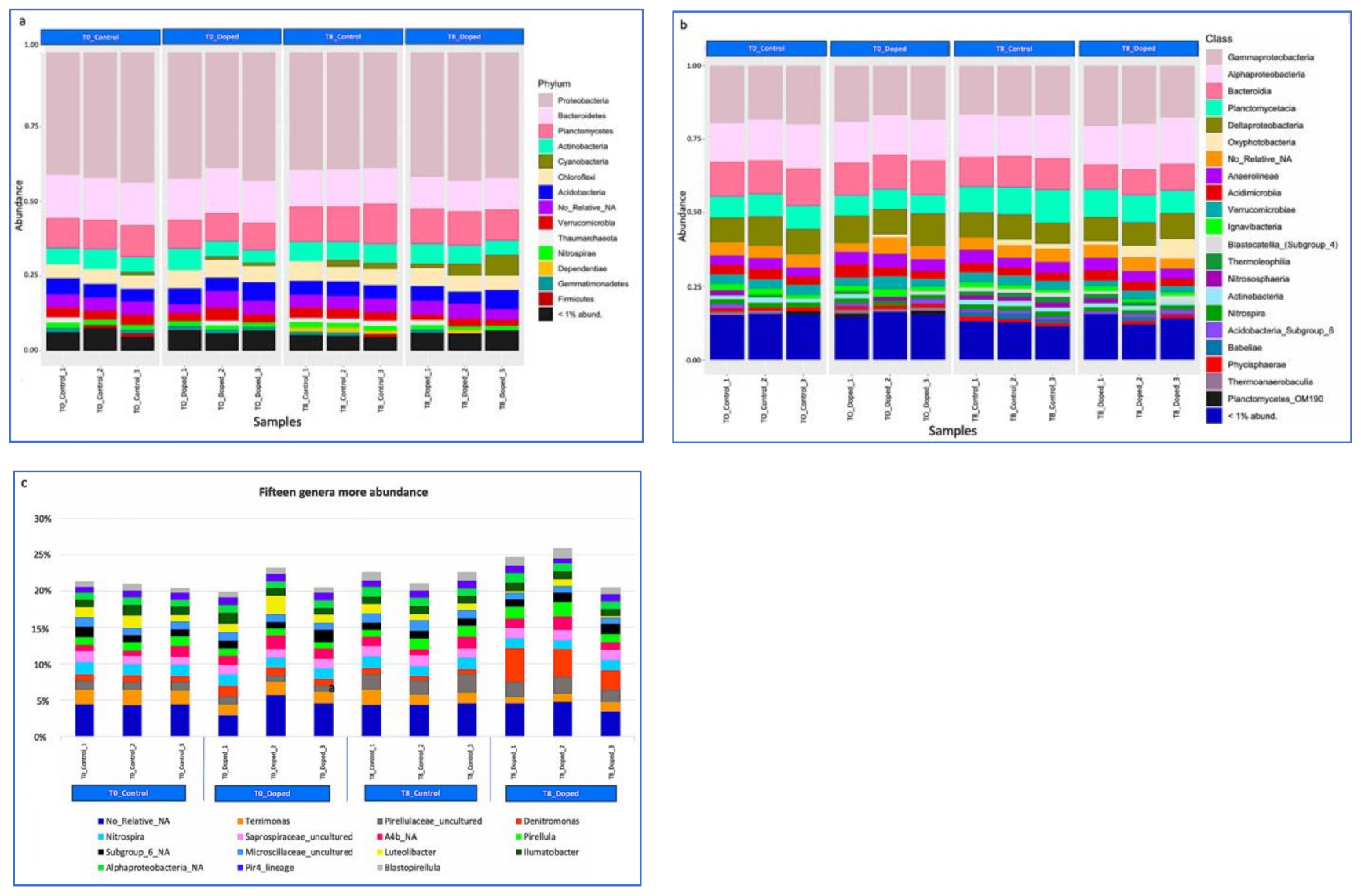
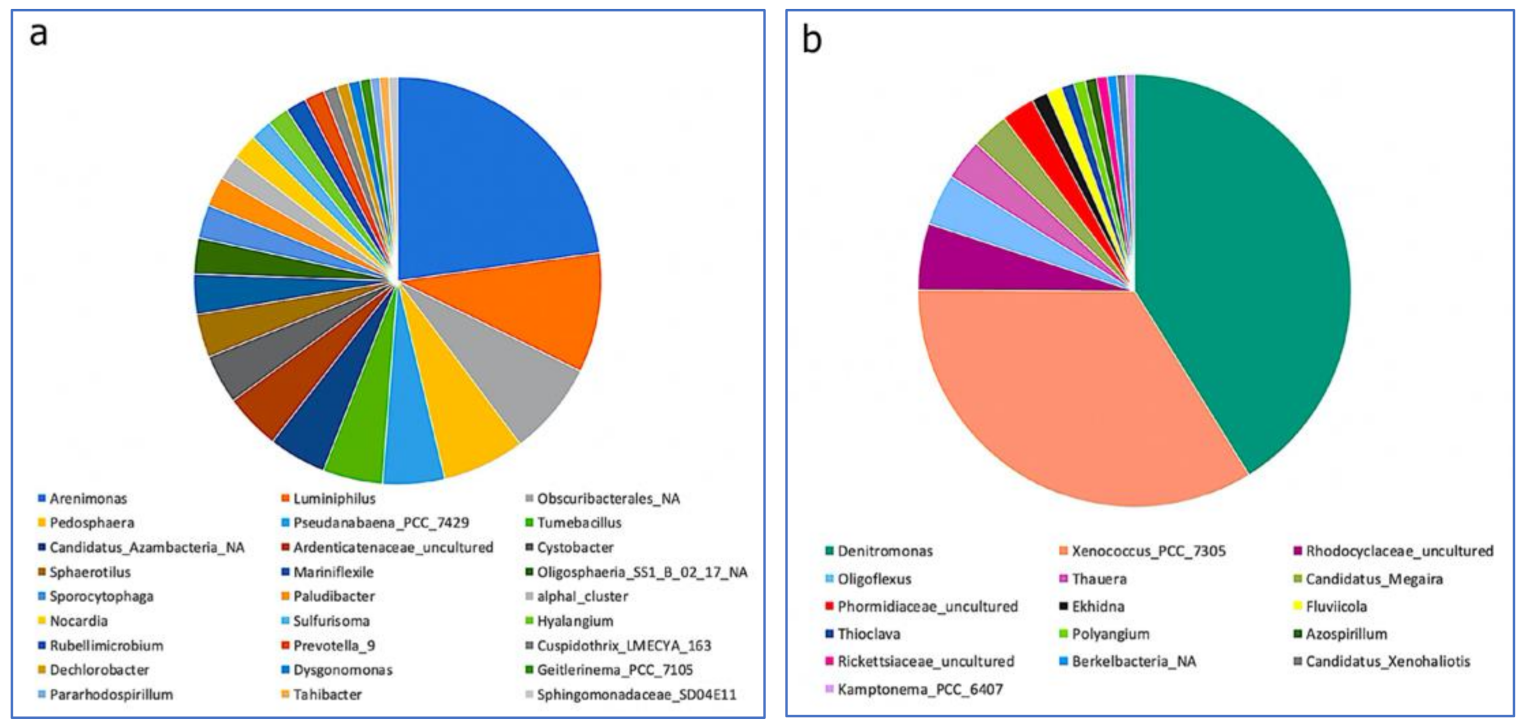
3.3. Impact of a Mixture of Alkylphenol Compounds on CWs Substrate Microbial Community
3.4. Impact of PCP on CWs Substrate Microbial Community
4. Discussion
4.1. CW Performance
4.2. Alkylphenols and Chlorophenols Removal in CW Systems
4.3. Microbial Communities’ Characterization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United States Environmental Protection Agency. Special Report on Environmental Endocrine Disruption: An Effects Assessment and Analysis; UESPA: Washington, DC, USA, 1997. [Google Scholar]
- Sharma, V.K.; Anquandah, G.A.K.; Yngard, R.A.; Kim, H.; Fekete, J.; Bouzek, K.; Ray, A.K.; Golovko, D. Nonylphenol, Octylphenol, and Bisphenol-A in the Aquatic Environment: A Review on Occurrence, Fate, and Treatment. J. Environ. Sci. Health Part A Toxic Hazard. Subst. Environ. Eng. 2009, 44, 423–442. [Google Scholar] [CrossRef]
- Orton, F.; Lutz, I.; Kloas, W.; Routledge, E.J. Endocrine Disrupting Effects of Herbicides and Pentachlorophenol: In Vitro and In Vivo Evidence. Environ. Sci. Technol. 2009, 43, 2144–2150. [Google Scholar] [CrossRef] [PubMed]
- Water Framework Directive. Directive of the European Parliament and of the Council 2000/60/EC. Off. J. Eur. Parliam. 2000, 327, 1–82. [Google Scholar]
- United States Environmental Protection Agency. USEPA 2008. US Environmental Protection Agency. In Registration Eligibility Decision for Pentachlorophenol. Available online: http://www.epa.gov/oppsrrd1/REDs/pentachlorophenol_red.pdf (accessed on 30 January 2020).
- Routt, R.J.; Roberts, J.R. Recognition and Management of Pesticide Poisonings; USEPA: Washington, DC, USA, 1999; p. 236. [Google Scholar]
- Renner, R. European Bans on Surfactant Trigger Transatlantic Debate. Environ. Sci. Technol. 1997, 31, 316–320. [Google Scholar] [CrossRef]
- Visvanathan, C.; Thu, L.N.; Jegatheesan, V.; Anotai, J. Biodegradation of Pentachlorophenol in a Membrane Bioreactor. Desalination 2005, 183, 455–464. [Google Scholar] [CrossRef]
- Yang, S.; Shibata, A.; Yoshida, N.; Katayama, A. Anaerobic Mineralization of Pentachlorophenol (PCP) by Combining PCP-Dechlorinating and Phenol-Degrading Cultures. Biotechnol. Bioeng. 2009, 102, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, T.; Kannan, K.; Giesy, J.P.; Masunaga, S. Contribution of Known Endocrine Disrupting Substances to the Estrogenic Activity in Tama River Water Samples from Japan Using Instrumental Analysis and in Vitro Reporter Gene Assay. Water Res. 2004, 38, 4491–4501. [Google Scholar] [CrossRef]
- Zhang, Z.; Feng, Y.; Gao, P.; Wang, C.; Ren, N. Occurrence and Removal Efficiencies of Eight EDCs and Estrogenicity in a STP. J. Environ. Monit. 2011, 13, 1366–1373. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, H.; Sun, Y.; Wang, C.; Shi, X.-L.; Hu, H.-Y.; Kameya, T.; Fujie, K. Ecological Risk of Estrogenic Endocrine Disrupting Chemicals in Sewage Plant Effluent and Reclaimed Water. Environ. Pollut. 2013, 180, 339–344. [Google Scholar] [CrossRef]
- Leech, D.M.; Snyder, M.T.; Wetzel, R.G. Natural Organic Matter and Sunlight Accelerate the Degradation of 17ß-Estradiol in Water. Sci. Total Environ. 2009, 407, 2087–2092. [Google Scholar] [CrossRef]
- Yang, X.; Flowers, R.C.; Weinberg, H.S.; Singer, P.C. Occurrence and Removal of Pharmaceuticals and Personal Care Products (PPCPs) in an Advanced Wastewater Reclamation Plant. Water Res. 2011, 45, 5218–5228. [Google Scholar] [CrossRef]
- Zhang, D.Q.; Gersberg, R.M.; Hua, T.; Zhu, J.; Tuan, N.A.; Tan, S.K. Pharmaceutical Removal in Tropical Subsurface Flow Constructed Wetlands at Varying Hydraulic Loading Rates. Chemosphere 2012, 87, 273–277. [Google Scholar] [CrossRef]
- Matamoros, V.; Bayona, J.M. Behavior of emerging pollutants in constructed wetlands. In Emerging Contaminants from Industrial and Municipal Waste; Barceló, D., Petrovic, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 5, pp. 199–214. [Google Scholar] [CrossRef]
- Wirasnita, R.; Mori, K.; Toyama, T. Effect of Activated Carbon on Removal of Four Phenolic Endocrine-Disrupting Compounds, Bisphenol A, Bisphenol F, Bisphenol S, and 4-Tert-Butylphenol in Constructed Wetlands. Chemosphere 2018, 210, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Fujii, D.A.D.; Soda, S.; Machimura, T.; Ike, M. Removal of Phenol, Bisphenol A, and 4-Tert-Butylphenol from Synthetic Landfill Leachate by Vertical Flow Constructed Wetlands. Sci. Total Environ. 2017, 578, 566–576. [Google Scholar] [CrossRef]
- Toro-Vélez, A.F.; Madera-Parra, C.A.; Peña-Varón, M.R.; Lee, W.Y.; Bezares-Cruz, J.C.; Walker, W.S.; Cárdenas-Henao, H.; Quesada-Calderón, S.; García-Hernández, H.; Lens, P.N.L. BPA and NP Removal from Municipal Wastewater by Tropical Horizontal Subsurface Constructed Wetlands. Sci. Total Environ. 2016, 542, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Papaevangelou, V.A.; Gikas, G.D.; Tsihrintzis, V.A.; Antonopoulou, M.; Konstantinou, I.K. Removal of Endocrine Disrupting Chemicals in HSF and VF Pilot-Scale Constructed Wetlands. Chem. Eng. J. 2016, 294, 146–156. [Google Scholar] [CrossRef]
- Song, H.-L.; Nakano, K.; Taniguchi, T.; Nomura, M.; Nishimura, O. Estrogen Removal from Treated Municipal Effluent in Small-Scale Constructed Wetland with Different Depth. Bioresour. Technol. 2009, 100, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-L.; Yang, X.-L.; Nakano, K.; Nomura, M.; Nishimura, O.; Li, X.-N. Elimination of Estrogens and Estrogenic Activity from Sewage Treatment Works Effluents in Subsurface and Surface Flow Constructed Wetlands. Int. J. Environ. Anal. Chem. 2011, 91, 600–614. [Google Scholar] [CrossRef]
- Carvalho, P.N.; Araújo, J.L.; Mucha, A.P.; Basto, M.C.P.; Almeida, C.M.R. Potential of Constructed Wetlands Microcosms for the Removal of Veterinary Pharmaceuticals from Livestock Wastewater. Bioresour. Technol. 2013, 134, 412–416. [Google Scholar] [CrossRef]
- Gorito, A.M.; Ribeiro, A.R.; Gomes, C.R.; Almeida, C.M.R.; Silva, A.M.T. Constructed Wetland Microcosms for the Removal of Organic Micropollutants from Freshwater Aquaculture Effluents. Sci. Total Environ. 2018, 644, 1171–1180. [Google Scholar] [CrossRef]
- Gorito, A.M.; Ribeiro, A.R.; Almeida, C.M.R.; Silva, A.M.T. A Review on the Application of Constructed Wetlands for the Removal of Priority Substances and Contaminants of Emerging Concern Listed in Recently Launched EU Legislation. Environ. Pollut. 2017, 227, 428–443. [Google Scholar] [CrossRef] [PubMed]
- Grasshoff, K.; Ehrhardt, M.; Kremling, K. Methods of Seawater Analysis. Second, Revised and Extended Edition. Chemical Oceanography; Academia Press: London, UK; New York, NY, USA, 1983. [Google Scholar]
- Jones, M.N. Nitrate Reduction by Shaking with Cadmium: Alternative to Cadmium Columns. Water Res. 1984, 18, 643–646. [Google Scholar] [CrossRef]
- De Morais, P.; Stoichev, T.; Basto, M.C.P.; Carvalho, P.N.; Vasconcelos, M.T.S.D. A Headspace SPME-GC-ECD Method Suitable for Determination of Chlorophenols in Water Samples. Anal. Bioanal. Chem. 2011, 399, 2531–2538. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every Base Matters: Assessing Small Subunit RRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Bragança, I.; Mucha, A.P.; Tomasino, M.P.; Santos, F.; Lemos, P.C.; Delerue-Matos, C.; Domingues, V.F. Deltamethrin Impact in a Cabbage Planted Soil: Degradation and Effect on Microbial Community Structure. Chemosphere 2019, 220, 1179–1186. [Google Scholar] [CrossRef]
- Calheiros, C.S.C.; Carecho, J.; Tomasino, M.P.; Almeida, C.M.R.; Mucha, A.P. Floating Wetland Islands Implementation and Biodiversity Assessment in a Port Marina. Water 2020, 12, 3273. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- R Core Team. R Core Team, 2008a language and environment for statistical computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 30 January 2020).
- Oksanen, J.F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Peter, R.; Minchin, R.B.; O’Hara, G.L.; Simpson, P.; et al. Vegan: Community Ecology Package (R package Version 2.5-5). Available online: https://CRAN.R-project.org/package=vegan (accessed on 30 January 2020).
- Wickham, H. Getting started with Qplot. In Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; pp. 9–26. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Chazdon, R.L.; Chao, A.; Colwell, R.K.; Lin, S.-Y.; Norden, N.; Letcher, S.G.; Clark, D.B.; Finegan, B.; Arroyo, J.P. A Novel Statistical Method for Classifying Habitat Generalists and Specialists. Ecology 2011, 92, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Y.; Chen, X.; Li, Y. Effects of Carbon Sources on Sludge Performance and Microbial Community for 4-Chlorophenol Wastewater Treatment in Sequencing Batch Reactors. Bioresour. Technol. 2018, 255, 22–28. [Google Scholar] [CrossRef]
- Cooper, P. A Review of the Design and Performance of Vertical-Flow and Hybrid Reed Bed Treatment Systems. Water Sci. Technol. 1999, 40, 1–9. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Biswas, D.K.; Nian, Y.; Jiang, G. Potential of Constructed Wetlands in Treating the Eutrophic Water: Evidence from Taihu Lake of China. Bioresour. Technol. 2008, 99, 1656–1663. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Li, X.; Fu, C.; Shi, T.; Yan, P. Effects of Influent Nitrogen Loads on Nitrogen and COD Removal in Horizontal Subsurface Flow Constructed Wetlands during Different Growth Periods of Phragmites Australis. Sci. Total Environ. 2018, 635, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Vymazal, J. Removal of Nutrients in Various Types of Constructed Wetlands. Sci. Total Environ. 2007, 380, 48–65. [Google Scholar] [CrossRef]
- Sirivedhin, T.; Gray, K.A. Factors Affecting Denitrification Rates in Experimental Wetlands: Field and Laboratory Studies. Ecol. Eng. 2006, 26, 167–181. [Google Scholar] [CrossRef]
- Kang, Y.; Xie, H.; Li, B.; Zhang, J.; Hao Ngo, H.; Guo, W.; Guo, Z.; Kong, Q.; Liang, S.; Liu, J.; et al. Performance of Constructed Wetlands and Associated Mechanisms of PAHs Removal with Mussels. Chem. Eng. J. 2019, 357, 280–287. [Google Scholar] [CrossRef]
- Martín, M.; Gargallo, S.; Hernández-Crespo, C.; Oliver, N. Phosphorus and Nitrogen Removal from Tertiary Treated Urban Wastewaters by a Vertical Flow Constructed Wetland. Ecol. Eng. 2013, 61, 34–42. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, X.; Yang, G.; Wang, D.; Wu, Y.; Li, Y.; Huang, X.; Fu, Q.; Wang, Q.; Liu, Y.; et al. Norfloxacin-Induced Effect on Enhanced Biological Phosphorus Removal from Wastewater after Long-Term Exposure. J. Hazard. Mater. 2020, 392, 122336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yue, Q.; Yang, K.; Zhao, P.; Gao, B. Enhanced Phosphorus and Ciprofloxacin Removal in a Modified BAF System by Configuring Fe-C Micro Electrolysis: Investigation on Pollutants Removal and Degradation Mechanisms. J. Hazard. Mater. 2018, 342, 705–714. [Google Scholar] [CrossRef]
- Belmont, M.A.; Metcalfe, C.D. Feasibility of Using Ornamental Plants (Zantedeschia Aethiopica) in Subsurface Flow Treatment Wetlands to Remove Nitrogen, Chemical Oxygen Demand and Nonylphenol Ethoxylate Surfactants—A Laboratory-Scale Study. Ecol. Eng. 2003, 21, 233–247. [Google Scholar] [CrossRef]
- Dai, Y.; Tao, R.; Tai, Y.; Tam, N.F.; Dan, A.; Yang, Y. Application of a Full-Scale Newly Developed Stacked Constructed Wetland and an Assembled Bio-Filter for Reducing Phenolic Endocrine Disrupting Chemicals from Secondary Effluent. Ecol. Eng. 2017, 99, 496–503. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Liaw, E.T.; Fan, K.M. Removal of Veterinary Antibiotics, Alkylphenolic Compounds, and Estrogens from the Wuluo Constructed Wetland in Southern Taiwan. J. Environ. Sci. Health Part A 2015, 50, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.K.; Kumar, S.; Sharma, C. Removal of Chlorophenolics from Pulp and Paper Mill Wastewater through Constructed Wetland. Water Environ. Res. Res. Public Water Environ. Fed. 2013, 85, 54–62. [Google Scholar] [CrossRef]
- David, A.; Fenet, H.; Gomez, E. Alkylphenols in Marine Environments: Distribution Monitoring Strategies and Detection Considerations. Mar. Pollut. Bull. 2009, 58, 953–960. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Igbinosa, E.O. Chlorophenols and Other Related Derivatives of Environmental Concern: Properties, Distribution and Microbial Degradation Processes. Chemosphere 2011, 83, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Connaughton, D.F.; Stedinger, J.R.; Lion, L.W.; Shuler, M.L. Description of Time-Varying Desorption Kinetics: Release of Naphthalene from Contaminated Soils. Environ. Sci. Technol. 1993, 27, 2397–2403. [Google Scholar] [CrossRef]
- Zhou, J.L. Sorption and Remobilization Behavior of 4-Tert-Octylphenol in Aquatic Systems. Environ. Sci. Technol. 2006, 40, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Pardue, J.H.; Moe, W.M.; Valsaraj, K.T. Mineralization of Desorption-Resistant 1,4-Dichlorobenzene in Wetland Soils. Environ. Toxicol. Chem. 2003, 22, 2312–2322. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, E.; Scow, K.M. Repeated Inoculation as a Strategy for the Remediation of Low Concentrations of Phenanthrene in Soil. Biodegradation 2001, 12, 201–207. [Google Scholar] [CrossRef]
- Allard, A.-S.; Remberger, M.; Neilson, A.H. The Negative Impact of Aging on the Loss of PAH Components in a Creosote-Contaminated Soil. Int. Biodeterior. Biodegrad. 2000, 46, 43–49. [Google Scholar] [CrossRef]
- Wick, L.Y.; Colangelo, T.; Harms, H. Kinetics of Mass Transfer-Limited Bacterial Growth on Solid PAHs. Environ. Sci. Technol. 2001, 35, 354–361. [Google Scholar] [CrossRef]
- Hamker, J.W.; Thompsom, J.M. Adsorption. In Organic Chemicals in Soil Environment; Goring, C., Hamaker, J., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 1971; Volume 1. [Google Scholar]
- Chien, S.-W.C.; Chen, S.-H.; Li, C.-J. Effect of Soil PH and Organic Matter on the Adsorption and Desorption of Pentachlorophenol. Environ. Sci. Pollut. Res. Int. 2018, 25, 5269–5279. [Google Scholar] [CrossRef]
- Braeckevelt, M.; Rokadia, H.; Imfeld, G.; Stelzer, N.; Paschke, H.; Kuschk, P.; Kastner, M.; Richnow, H.H.; Weber, S. Assessment of in Situ Biodegradation of Monochlorobenzene in Contaminated Groundwater Treated in a Constructed Wetland. Environ. Pollut. 2007, 148, 428–437. [Google Scholar] [CrossRef]
- Braeckevelt, M.; Mirschel, G.; Wiessner, A.; Rueckert, M.; Reiche, N.; Vogt, C.; Schultz, A.; Paschke, H.; Kuschk, P.; Kaestner, M. Treatment of Chlorobenzene-Contaminated Groundwater in a Pilot-Scale Constructed Wetland. Ecol. Eng. 2008, 33, 45–53. [Google Scholar] [CrossRef]
- Ma, X.; Havelka, M.M. Phytotoxicity of Chlorinated Benzenes to Typha Angustifolia and Phragmites Communis. Environ. Toxicol. 2009, 24, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, Y.; Liu, Y.; Chang, H.; Li, Z.; Xue, J. Uptake and Translocation of Organic Pollutants in Plants: A Review. J. Integr. Agric. 2017, 16, 1659–1668. [Google Scholar] [CrossRef]
- Zhao, L.; Zhu, C.; Gao, C.; Jiang, J.; Yang, J.; Yang, S. Phytoremediation of Pentachlorophenol-Contaminated Sediments by Aquatic Macrophytes. Environ. Earth Sci. 2011, 64, 581–588. [Google Scholar] [CrossRef]
- Hübner, T.M.; Tischer, S.; Tanneberg, H.; Kuschk, P. Influence of Phenol and Phenanthrene on the Growth of Phalaris Arundinacea and Phragmites Australis. Int. J. Phytoremediation 2000, 2, 331–342. [Google Scholar] [CrossRef]
- Schröder, P.; Collins, C. Conjugating Enzymes Involved in Xenobiotic Metabolism of Organic Xenobiotics in Plants. Int. J. Phytoremediation 2002, 4, 247–265. [Google Scholar] [CrossRef]
- Liste, H.-H.; Alexander, M. Plant-Promoted Pyrene Degradation in Soil. Chemosphere 2000, 40, 7–10. [Google Scholar] [CrossRef]
- Yang, C.; Yulai, W.; Wang, M.; Chen, H. Plant Species Influence Microbial Metabolic Activity and Butachlor Biodegradation in a Riparian Soil from Chongming Island, China. Geoderma 2013, 193, 165–171. [Google Scholar] [CrossRef]
- Hechmi, N.; Aissa, N.B.; Abdenaceur, H.; Jedidi, N. Evaluating the Phytoremediation Potential of Phragmites Australis Grown in Pentachlorophenol and Cadmium Co-Contaminated Soils. Environ. Sci. Pollut. Res. Int. 2014, 21, 1304–1313. [Google Scholar] [CrossRef]
- Hechmi, N.; Ben Aissa, N.; Abdenaceur, H.; Jedidi, N. Uptake and Bioaccumulation of Pentachlorophenol by Emergent Wetland Plant Phragmites Australis (Common Reed) in Cadmium Co-Contaminated Soil. Int. J. Phytoremediation 2015, 17, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhang, S.; Christie, P.; Wang, S.; Xie, M. Behavior of Decabromodiphenyl Ether (BDE-209) in the Soil-Plant System: Uptake, Translocation, and Metabolism in Plants and Dissipation in Soil. Environ. Sci. Technol. 2010, 44, 663–667. [Google Scholar] [CrossRef]
- Ekelund, R.; Granmo, Å.; Magnusson, K.; Berggren, M.; Bergman, Å. Biodegradation of 4-Nonylphenol in Seawater and Sediment. Environ. Pollut. 1993, 79, 59–61. [Google Scholar] [CrossRef]
- Nishio, E.; Ichiki, Y.; Tamura, H.; Morita, S.; Watanabe, K.; Yoshikawa, H. Isolation of Bacterial Strains That Produce the Endocrine Disruptor, Octylphenol Diethoxylates, in Paddy Fields. Biosci. Biotechnol. Biochem. 2002, 66, 1792–1798. [Google Scholar] [CrossRef]
- Ying, G.-G.; Williams, B.; Kookana, R. Environmental Fate of Alkylphenols and Alkylphenol Ethoxylates—A Review. Environ. Int. 2002, 28, 215–226. [Google Scholar] [CrossRef]
- Fujii, K.; Urano, N.; Kimura, S.; Nomura, Y.; Karube, I. Microbial Degradation of Nonylphenol in Some Aquatic Environments. Fish. Sci. 2000, 66, 44–48. [Google Scholar] [CrossRef]
- Ushiba, Y.; Takahara, Y.; Ohta, H. Sphingobium amiense sp. Nov., a Novel Nonylphenol-Degrading Bacterium Isolated from a River Sediment. Int. J. Syst. Evol. Microbiol. 2003, 53, 2045–2048. [Google Scholar] [CrossRef]
- Toyama, T.; Ojima, T.; Tanaka, Y.; Mori, K.; Morikawa, M. Sustainable Biodegradation of Phenolic Endocrine-Disrupting Chemicals by Phragmites Australis–Rhizosphere Bacteria Association. Water Sci. Technol. 2013, 68, 522–529. [Google Scholar] [CrossRef]
- D’Angelo, E.M.; Reddy, K.R. Aerobic and Anaerobic Transformations of Pentachlorophenol in Wetland Soils. Soil Sci. Soc. Am. J. 2000, 64, 933–943. [Google Scholar] [CrossRef]
- Zeng, G.; Yu, Z.; Chen, Y.; Zhang, J.; Li, H.; Yu, M.; Zhao, M. Response of Compost Maturity and Microbial Community Composition to Pentachlorophenol (PCP)-Contaminated Soil during Composting. Bioresour. Technol. 2011, 102, 5905–5911. [Google Scholar] [CrossRef]
- Liu, H.; Li, G.; Li, X.; Chen, J. Molecular Characterization of Bacterial Community in Aerobic Granular Sludge Stressed by Pentachlorophenol. J. Environ. Sci. 2008, 20, 1243–1249. [Google Scholar] [CrossRef]
- Beaulieu, M.; Bécaert, V.; Deschênes, L.; Villemur, R. Evolution of Bacterial Diversity during Enrichment of PCP-Degrading Activated Soils. Microb. Ecol. 2000, 40, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; He, Y.; Tang, X.; Brookes, P.C.; Xu, J. Reconstruction of Microbial Community Structures as Evidences for Soil Redox Coupled Reductive Dechlorination of PCP in a Mangrove Soil. Sci. Total Environ. 2017, 596, 147–157. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; He, T.; Xie, S. Change of Microbial Community Structure and Functional Gene Abundance in Nonylphenol-Degrading Sediment. Appl. Microbiol. Biotechnol. 2015, 99, 3259–3268. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, Y.; Zhou, J.; Zhou, Q.; Vymazal, J.; Kuschk, P. Transformation of Chloroform in Model Treatment Wetlands: From Mass Balance to Microbial Analysis. Environ. Sci. Technol. 2015, 49, 6198–6205. [Google Scholar] [CrossRef]
- Guan, W.; Yin, M.; He, T.; Xie, S. Influence of Substrate Type on Microbial Community Structure in Vertical-Flow Constructed Wetland Treating Polluted River Water. Environ. Sci. Pollut. Res. Int. 2015, 22, 16202–16209. [Google Scholar] [CrossRef]
- He, T.; Guan, W.; Luan, Z.; Xie, S. Spatiotemporal Variation of Bacterial and Archaeal Communities in a Pilot-Scale Constructed Wetland for Surface Water Treatment. Appl. Microbiol. Biotechnol. 2016, 100, 1479–1488. [Google Scholar] [CrossRef]
- Lang, X.; Chen, X.; Xu, A.; Song, Z.; Wang, X.; Wang, H. Variation of Bacterial and Archaeal Community Structures in a Full-Scale Constructed Wetlands for Wastewater Treatment. Archaea 2018, 2018, 9319345. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, B.; Zou, R.; Dai, Y.; Xie, S.; Yuan, B. Biodegradation of Antibiotic Ciprofloxacin: Pathways, Influential Factors, and Bacterial Community Structure. Environ. Sci. Pollut. Res. 2016, 23, 7911–7918. [Google Scholar] [CrossRef]
- Zhang, S.; Courtois, S.; Gitungo, S.; Raczko, R.F.; Dyksen, J.E.; Li, M.; Axe, L. Microbial Community Analysis in Biologically Active Filters Exhibiting Efficient Removal of Emerging Contaminants and Impact of Operational Conditions. Sci. Total Environ. 2018, 640–641, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Smalla, K.; Wieland, G.; Buchner, A.; Zock, A.; Parzy, J.; Kaiser, S.; Roskot, N.; Heuer, H.; Berg, G. Bulk and Rhizosphere Soil Bacterial Communities Studied by Denaturing Gradient Gel Electrophoresis: Plant-Dependent Enrichment and Seasonal Shifts Revealed. Appl. Environ. Microbiol. 2001, 67, 4742–4751. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant Species and Soil Type Cooperatively Shape the Structure and Function of Microbial Communities in the Rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Calheiros, C.S.C.; Duque, A.F.; Moura, A.; Henriques, I.S.; Correia, A.; Rangel, A.O.S.S.; Castro, P.M.L. Changes in the Bacterial Community Structure in Two-Stage Constructed Wetlands with Different Plants for Industrial Wastewater Treatment. Bioresour. Technol. 2009, 100, 3228–3235. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation Aspects of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Dai, Y.; Xie, S. Anaerobic Biodegradation of Nonylphenol in River Sediment under Nitrate- or Sulfate-Reducing Conditions and Associated Bacterial Community. J. Hazard. Mater. 2015, 286, 306–314. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.; Xia, S.; Zhao, J. Study of 4-t-Octylphenol Degradation and Microbial Community in Granular Sludge. J. Environ. Sci. 2008, 20, 167–171. [Google Scholar] [CrossRef]
- Li, H.; Shen, T.-T.; Wang, X.-L.; Lin, K.-F.; Liu, Y.-D.; Lu, S.-G.; Gu, J.-D.; Wang, P.; Lu, Q.; Du, X.-M. Biodegradation of Perchloroethylene and Chlorophenol Co-Contamination and Toxic Effect on Activated Sludge Performance. Bioresour. Technol. 2013, 137, 286–293. [Google Scholar] [CrossRef]
- Yu, H.; Feng, C.; Liu, X.; Yi, X.; Ren, Y.; Wei, C. Enhanced Anaerobic Dechlorination of Polychlorinated Biphenyl in Sediments by Bioanode Stimulation. Environ. Pollut. 2016, 211, 81–89. [Google Scholar] [CrossRef]
- De Weert, J.; Viñas, M.; Grotenhuis, T.; Rijnaarts, H.; Langenhoff, A. Aerobic Nonylphenol Degradation and Nitro-Nonylphenol Formation by Microbial Cultures from Sediments. Appl. Microbiol. Biotechnol. 2010, 86, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Jontofsohn, M.; Stoffels, M.; Hartmann, A.; Pfister, G.; Jüttner, I.; Severin-Edmair, G.; Schramm, K.-W.; Schloter, M. Influence of Nonylphenol on the Microbial Community of Lake Sediments in Microcosms. Sci. Total Environ. 2002, 285, 3–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Sei, K.; Toyama, T.; Ike, M.; Zhang, J.; Yang, M.; Kamagata, Y. Changes of Catabolic Genes and Microbial Community Structures during Biodegradation of Nonylphenol Ethoxylates and Nonylphenol in Natural Water Microcosms. Biochem. Eng. J. 2008, 39, 288–296. [Google Scholar] [CrossRef]
- Stenrød, M.; Klemsdal, S.S.; Norli, H.R.; Eklo, O.M. Effects of Picoxystrobin and 4-n-Nonylphenol on Soil Microbial Community Structure and Respiration Activity. PLoS ONE 2013, 8, e66989. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, Y.; Sun, W.; Xie, S.; Liu, Y. Nonylphenol Biodegradation in River Sediment and Associated Shifts in Community Structures of Bacteria and Ammonia-Oxidizing Microorganisms. Ecotoxicol. Environ. Saf. 2014, 106, 1–5. [Google Scholar] [CrossRef]
- Domene, X.; Chelinho, S.; Sousa, J.P. Effects of Nonylphenol on a Soil Community Using Microcosms. J. Soils Sediments 2010, 10, 556–567. [Google Scholar] [CrossRef]
- Rodríguez, J.; Gallampois, C.M.J.; Timonen, S.; Andersson, A.; Sinkko, H.; Haglund, P.; Berglund, Å.M.M.; Ripszam, M.; Figueroa, D.; Tysklind, M.; et al. Effects of Organic Pollutants on Bacterial Communities Under Future Climate Change Scenarios. Front. Microbiol. 2018, 9, 2926. [Google Scholar] [CrossRef]
- Cai, T.; Qian, L.; Cai, S.; Chen, L. Biodegradation of Benazolin-Ethyl by Strain Methyloversatilis Sp. Cd-1 Isolated from Activated Sludge. Curr. Microbiol. 2011, 62, 570–577. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Doronina, N.V.; Khmelenina, V.N. Biotechnological Potential of Aerobic Methylotrophic Bacteria: A Review of Current State and Future Prospects. Appl. Biochem. Microbiol. 2005, 41, 433–441. [Google Scholar] [CrossRef]
- Lu, C.; Hong, Y.; Liu, J.; Gao, Y.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. A PAH-Degrading Bacterial Community Enriched with Contaminated Agricultural Soil and Its Utility for Microbial Bioremediation. Environ. Pollut. 2019, 251, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Yagi, J.M.; Sims, D.; Brettin, T.; Bruce, D.; Madsen, E.L. The Genome of Polaromonas Naphthalenivorans Strain CJ2, Isolated from Coal Tar-Contaminated Sediment, Reveals Physiological and Metabolic Versatility and Evolution through Extensive Horizontal Gene Transfer. Environ. Microbiol. 2009, 11, 2253–2270. [Google Scholar] [CrossRef] [PubMed]
- Tuan, N.N.; Hsieh, H.-C.; Lin, Y.-W.; Huang, S.-L. Analysis of Bacterial Degradation Pathways for Long-Chain Alkylphenols Involving Phenol Hydroxylase, Alkylphenol Monooxygenase and Catechol Dioxygenase Genes. Bioresour. Technol. 2011, 102, 4232–4240. [Google Scholar] [CrossRef]
- Ma, W.; Nie, C.; Chen, B.; Cheng, X.; Lun, X.; Zeng, F. Adsorption and Biodegradation of Three Selected Endocrine Disrupting Chemicals in River-Based Artificial Groundwater Recharge with Reclaimed Municipal Wastewater. J. Environ. Sci. 2015, 31, 154–163. [Google Scholar] [CrossRef]
- Ma, W.; Yan, Y.; Ma, M.; Zhang, Y.; Nie, C.; Lun, X. Effect of Biochar on Migration and Biodegradation of 4-n-Nonylphenol (NP) during River-Based Groundwater Recharge with Reclaimed Water. Desalin. Water Treat. 2016, 57, 29316–29327. [Google Scholar] [CrossRef]
- Yang, C.-W.; Chang, B.-V.; Chen, L.-Y.; Tang, S.-L. Removal of Nonylphenol by Earthworms and Bacterial Community Change. Int. Biodeterior. Biodegrad. 2014, 96, 9–17. [Google Scholar] [CrossRef]
- Táncsics, A.; Szalay, A.R.; Farkas, M.; Benedek, T.; Szoboszlay, S.; Szabó, I.; Lueders, T. Stable Isotope Probing of Hypoxic Toluene Degradation at the Siklós Aquifer Reveals Prominent Role of Rhodocyclaceae. FEMS Microbiol. Ecol. 2018, 94, 88. [Google Scholar] [CrossRef] [PubMed]
- Kasai, Y.; Takahata, Y.; Manefield, M.; Watanabe, K. RNA-Based Stable Isotope Probing and Isolation of Anaerobic Benzene-Degrading Bacteria from Gasoline-Contaminated Groundwater. Appl. Environ. Microbiol. 2006, 72, 3586–3592. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tian, H.; Huang, F.; Long, W.; Zhang, Q.; Wang, J.; Zhu, Y.; Wu, X.; Chen, G.; Zhao, L.; et al. Time-Resolved Analysis of a Denitrifying Bacterial Community Revealed a Core Microbiome Responsible for the Anaerobic Degradation of Quinoline. Sci. Rep. 2017, 7, 14778. [Google Scholar] [CrossRef]
- Gutierrez, T.; Singleton, D.R.; Aitken, M.D.; Semple, K.T. Stable Isotope Probing of an Algal Bloom to Identify Uncultivated Members of the Rhodobacteraceae Associated with Low-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation. Appl. Environ. Microbiol. 2011, 77, 7856–7860. [Google Scholar] [CrossRef]
- Corteselli, E.M.; Aitken, M.D.; Singleton, D.R. Rugosibacter Aromaticivorans Gen. Nov., Sp. Nov., a Bacterium within the Family Rhodocyclaceae, Isolated from Contaminated Soil, Capable of Degrading Aromatic Compounds. Int. J. Syst. Evol. Microbiol. 2017, 67, 311–318. [Google Scholar] [CrossRef]
- Singleton, D.R.; Hu, J.; Aitken, M.D. Heterologous Expression of Polycyclic Aromatic Hydrocarbon Ring-Hydroxylating Dioxygenase Genes from a Novel Pyrene-Degrading Betaproteobacterium. Appl. Environ. Microbiol. 2012, 78, 3552–3559. [Google Scholar] [CrossRef]
- Regonne, R.K.; Martin, F.; Mbawala, A.; Ngassoum, M.B.; Jouanneau, Y. Identification of Soil Bacteria Able to Degrade Phenanthrene Bound to a Hydrophobic Sorbent in Situ. Environ. Pollut. 2013, 180, 145–151. [Google Scholar] [CrossRef]
- Salinero, K.K.; Keller, K.; Feil, W.S.; Feil, H.; Trong, S.; Di Bartolo, G.; Lapidus, A. Metabolic Analysis of the Soil Microbe Dechloromonas Aromatica Str. RCB: Indications of a Surprisingly Complex Life-Style and Cryptic Anaerobic Pathways for Aromatic Degradation. BMC Genom. 2009, 10, 351. [Google Scholar] [CrossRef]
- Li, Z.-L.; Nan, J.; Huang, C.; Liang, B.; Liu, W.-Z.; Cheng, H.-Y.; Zhang, C.; Zhang, D.; Kong, D.; Kanamaru, K.; et al. Spatial Abundance and Distribution of Potential Microbes and Functional Genes Associated with Anaerobic Mineralization of Pentachlorophenol in a Cylindrical Reactor. Sci. Rep. 2016, 6, 19015. [Google Scholar] [CrossRef]
- Harwood, C.S.; Burchhardt, G.; Herrmann, H.; Fuchs, G. Anaerobic Metabolism of Aromatic Compounds via the Benzoyl-CoA Pathway. FEMS Microbiol. Rev. 1998, 22, 439–458. [Google Scholar] [CrossRef]
- Tong, H.; Liu, C.; Li, F.; Luo, C.; Chen, M.; Hu, M. The Key Microorganisms for Anaerobic Degradation of Pentachlorophenol in Paddy Soil as Revealed by Stable Isotope Probing. J. Hazard. Mater. 2015, 298, 252–260. [Google Scholar] [CrossRef]
- McAllister, K.A.; Lee, H.; Trevors, J.T. Microbial Degradation of Pentachlorophenol. Biodegradation 1996, 7, 1–40. [Google Scholar] [CrossRef]
- Mohn, W.W.; Kennedy, K.J. Reductive Dehalogenation of Chlorophenols by Desulfomonile Tiedjei DCB-1. Appl. Environ. Microbiol. 1992, 58, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Coates, J.D. Hydroxylation and Carboxylation–Two Crucial Steps of Anaerobic Benzene Degradation by Dechloromonas Strain RCB. Appl. Environ. Microbiol. 2005, 71, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, Y.; Sakai, Y.; Uenishi, H.; Uchihashi, Y.; Hiraishi, A.; Yukawa, H.; Yurimoto, H.; Kato, N. Aerobic and Anaerobic Toluene Degradation by a Newly Isolated Denitrifying Bacterium, Thauera Sp. Strain DNT-1. Appl. Environ. Microbiol. 2004, 70, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, A.E.; Nalewajko, C.; Fulthorpe, R.R. The Effects of Cyanobacterial Exudates on Bacterial Growth and Biodegradation of Organic Contaminants. Microb. Ecol. 2006, 51, 4–12. [Google Scholar] [CrossRef] [PubMed]

| Week | CW Influent (mg·L−1) | CW Effluent | |||
|---|---|---|---|---|---|
| Control (mg·L−1) | % of Removal | Doped (mg·L−1) | % of Removal | ||
| 1 | 45 | 114 ± 7 | n.r. | 112 ± 12 | n.r. |
| 2 | 83 | 86 ± 42 | n.r. | 70 ± 61 | 15 ± 74 |
| 3 | 43 | 18 ± 8 | 58 ± 18 | 36 ± 42 | 16 ± 97 |
| 4 | 59 | 23 ± 10 | 61 ± 18 | 61 ± 20 | n.r. |
| 5 | 15 | 4.0 ± 0.1 | 73.0 ± 0.1 | 7 ± 5 | 53 ± 35 |
| 6 | 27 | 13 ± 2 | 52 ± 7 | 6 ± 1 | 78 ± 4 |
| 7 | 34 | 95 ± 12 | n.r. | 64 ± 13 | n.r. |
| 8 | 46 | 14 ± 2 | 69 ± 5 | 8 ± 7 | 83 ± 15 |
| PCP (μg·L−1) | |||
|---|---|---|---|
| Week | Influent | Effluent | |
| Control | Doped | ||
| 1 | <4 × 10−5 * | <4 × 10−5 * | 2.5 ± 0.5 |
| 2 | 3.1 ± 0.6 | ||
| 3 | 7 ± 2 | ||
| 4 | 11 ± 4 | ||
| 5 | 8 ± 2 | ||
| 6 | 13 ± 2 | ||
| 7 | 13 ± 1 | ||
| 8 | 9 ± 4 | ||
| Week | CW Influent (mg·L−1) | CW Effluent | |||
|---|---|---|---|---|---|
| Control (mg·L−1) | % of Removal | Doped (mg·L−1) | % of Removal | ||
| 1 | 23 | 22 ± 6 | 3 ± 25 | 15 ± 3 | 35 ± 15 |
| 2 | 42 | 35 ± 1 | 16 ± 3 | 39 ± 1 | 8 ± 2 |
| 3 | 55 | 36 ± 4 | 35 ± 6 | 29 ± 1 | 48 ± 2 |
| 4 | 46 | 39 ± 5 | 16 ± 10 | 36 ± 4 | 22 ± 8 |
| 5 | 28 | 21 ± 5 | 26 ± 18 | 21 ± 2 | 24 ± 5 |
| 6 | 22 | 27 ± 3 | n.r. | 30 ± 1 | n.r. |
| 7 | 22 | 16 ± 2 | 29 ± 9 | 19 ± 7 | 14 ± 32 |
| 8 | 19 | 24 ± 4 | n.r. | 26 ± 8 | n.r. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montenegro, I.P.F.M.; Mucha, A.P.; Tomasino, M.P.; Gomes, C.R.; Almeida, C.M.R. Alkylphenols and Chlorophenols Remediation in Vertical Flow Constructed Wetlands: Removal Efficiency and Microbial Community Response. Water 2021, 13, 715. https://doi.org/10.3390/w13050715
Montenegro IPFM, Mucha AP, Tomasino MP, Gomes CR, Almeida CMR. Alkylphenols and Chlorophenols Remediation in Vertical Flow Constructed Wetlands: Removal Efficiency and Microbial Community Response. Water. 2021; 13(5):715. https://doi.org/10.3390/w13050715
Chicago/Turabian StyleMontenegro, Inês P. F. M., Ana P. Mucha, Maria Paola Tomasino, Carlos Rocha Gomes, and Cristina Marisa R. Almeida. 2021. "Alkylphenols and Chlorophenols Remediation in Vertical Flow Constructed Wetlands: Removal Efficiency and Microbial Community Response" Water 13, no. 5: 715. https://doi.org/10.3390/w13050715
APA StyleMontenegro, I. P. F. M., Mucha, A. P., Tomasino, M. P., Gomes, C. R., & Almeida, C. M. R. (2021). Alkylphenols and Chlorophenols Remediation in Vertical Flow Constructed Wetlands: Removal Efficiency and Microbial Community Response. Water, 13(5), 715. https://doi.org/10.3390/w13050715







