1. Introduction
Cryptosporidium is a common protozoal cause of human acute gastroenteritis in the UK, with
Cryptosporidium hominis and
Cryptosporidium parvum accounting for most laboratory-confirmed cases [
1]. Cryptosporidiosis is a self-limiting illness, but those with underlying immunosuppression may experience complications. Predominant symptoms include watery diarrhoea and abdominal cramps; although the incubation period is reported in communicable disease handbooks to range from one to 12 days with an average of seven days [
2], the biology of the parasite [
3] and human infectivity studies indicate onset of symptoms is more likely after 3 days [
4]. Diarrhoea can last for around two weeks and is generally longer than other infectious causes of gastroenteritis. High numbers of oocysts may be shed in faeces, and very low numbers can cause disease [
5]. Asymptomatic carriage can occur [
1].
In the UK, methods for testing stool samples for
Cryptosporidium at local laboratories vary according to those described in national guidelines [
6], but all laboratory-confirmed cases of cryptosporidiosis are routinely, statutorily notified to the public health agencies [
7]. Subsequently, information is usually forwarded to local environmental health officials, and confirmed cases are interviewed by Environmental Health Officers (EHOs) using a standardised case questionnaire, which includes information on illness onset, symptoms, and detailed exposure information for the two weeks prior to symptoms, including recreational water use [
8]. Infection control advice is also provided, including not to swim for two weeks after symptoms cease [
9]. Completed case questionnaires are usually sent to the local Health Protection Team (HPT) and prospectively entered into local and national systems in order to identify potential links between cases [
8].
In England and Wales, the national Cryptosporidium Reference Unit (CRU), Public Health Wales, Swansea, UK provides confirmatory specialist testing by polymerase chain reaction (PCR) and immunofluorescence microscopy (IFM) where there is a clinically high index of suspicion of illness, but tests are negative locally for
Cryptosporidium. Species identification and subtyping by PCR or PCR sequencing are also provided. All positive specimens identified by local laboratories are requested to be forwarded for species identification, and subtyping is currently carried out as part of outbreak investigations by sequencing part of the
gp60 gene [
10]. There is much variation in the
gp60 gene amongst
C. parvum cases in the UK, but most
C. hominis cases in the UK and northern Europe are
gp60 genotype IbA10G2 [
11].
Transmission of
Cryptosporidium is usually faecal–oral with person-to-person, animal-to-person, foodborne or waterborne spread [
2]. Outbreaks in England and Wales linked to animal contact, environmental contact, and food have been caused by
C. parvum and recreational water outbreaks are predominantly
C. hominis, with few drinking water outbreaks reported in recent years [
10]. In the period 2009 to 2017, 46% of the 178
Cryptosporidium outbreaks involved recreational waters, mostly swimming pools [
10]. Usual pool water disinfection processes such as chlorination are not effective in killing
Cryptosporidium [
12], and filtration with flocculation is required to remove the oocysts [
13]. Outbreak investigations have utilised epidemiological, environmental, and microbiological evidence, including
Cryptosporidium speciation and subtyping to understand links between cases [
10].
Between 26 September 2016 and 17 October 2016, 51 cases of cryptosporidiosis were reported to the Public Health England (PHE) North East Centre in Newcastle. The EHOs’ routine questionnaires initially showed that four cases reported recently swimming at one or both of two local swimming pool facilities (pools A and B) or were a household contact of a user of these pools. Epidemiological, microbiological, and environmental investigations regarding the four cases and the two pools were initiated, and a multi-agency outbreak control team (OCT) was convened on 24 October 2016 to review the investigation, instigate case finding, develop hypotheses regarding potential exposures, and implement control measures.
This outbreak was included in the summary analysis presented previously [
10], but here, we describe the outbreak investigation and the impact and value of a combined approach integrating epidemiology, diagnostic and reference microbiology, and environmental investigations in defining the extent of the outbreak and the likely source and transmission pathways.
2. Materials and Methods
Cryptosporidium notifications were received by the Public Health England (PHE) North East Centre and information was forwarded to local environmental health officials. To identify potential links between
Cryptosporidium cases, completed case questionnaires (
Supplementary material Figure S1) were prospectively entered by the HPT into local and national systems. Cases who described exposure to pool A or B were included in the investigation. Symptomatic household or close contacts identified during case interviews were also asked about relevant exposures. Exposure information was reviewed in order to identify additional shared risk factors or other plausible sources of infection. EHOs, local hospitals, and primary care services in the area were notified of the situation to enhance case finding, ensure that new cases were reported promptly, and reinforce advice to cases about refraining from using swimming pools while ill and for two weeks after symptoms cease [
9].
The local diagnostic microbiology laboratories routinely tested all stools from diarrhoea patients for
Cryptosporidium. Faecal specimens were also requested from any symptomatic household contacts that were identified during case investigations or from individuals who contacted EHOs or PHE to report symptoms following using either pool. For
Cryptosporidium diagnosis, one local laboratory used an automated, plate format enzyme-linked immunosorbent assay, and the other used auramine phenol-stained microscopy [
6]. Faecal specimens from locally diagnosed cryptosporidiosis cases were routinely forwarded to the national Cryptosporidium Reference Unit (CRU) and were especially requested from those who described exposure to pools A or B in the 1 to 12 days prior to developing symptoms. The CRU characterized the
Cryptosporidium species by real-time PCR based on the
Lib13 and
A135 genes [
14]; the PCR comprised two separate amplification assays in the same tube enabling identification of co-infections with
C. hominis and
C. parvum. Subtyping was undertaken by PCR amplification and sequence analysis of part of the 60 kDa glycoprotein (
gp60) gene [
15].
Cryptosporidium-negative stools from patients with a high index of suspicion of cryptosporidiosis were also sent to the CRU for specialist, sensitive testing by immunofluorescence microscopy (Crypto-Cel; TCS Biosciences, UK).
EHOs undertook environmental investigations at the facilities using an environmental risk assessment tool. The tool is part of guidance for the investigation of cases linked to swimming pools [
16], is based on standards published by the Pool Water Treatment Advisory Group [
17], and includes parameters to assess whether the pool is being managed accordingly as well as those pertinent to the control of
Cryptosporidium. This involved reviewing pool policies, management and operating procedures (required under industry and Health and Safety Executive guidance as being essential to safe operation), pool water treatment, routine microbiological sampling results, pre-swim hygiene practices, and pool records. Relevant experts were consulted regarding chemical management. EHOs also obtained advice from the PHE’s Food, Water and Environment (FWE) laboratory. Due to the time elapsed since the exposure of cases, during which multiple turnover periods and backwash cycles had occurred, and the implementation of early control measures, it was thought unlikely that the results of any pool water sampling for
Cryptosporidium would further inform actions being taken. In accordance with national guidance about such situations [
16], pool water sampling for
Cryptosporidium was not undertaken.
Microbiological and environmental findings of the investigation were summarized descriptively. We described the epidemiology of the cases by time, place, and person (date of onset of symptoms, dates/times of pool use, age and sex distribution, and cryptosporidium microbiology) using the following case definitions:
Probable case: An individual who visited, or was a close contact of someone who visited, pool A and/or B on or after 12 September, and who subsequently developed diarrhoea within 1–12 days of attendance, for whom a reference laboratory confirmed diagnosis was not obtained (excluding those who had a local laboratory negative result unless epidemiologically linked to another confirmed case).
Confirmed case: As probable with reference laboratory confirmation of Cryptosporidium.
Probable and confirmed cases were further described as primary or secondary outbreak cases:
Primary outbreak case: An individual who visited pool A and/or B on, or after 12 September, and who subsequently developed diarrhoea within 1–12 days of attendance.
Secondary outbreak case: An individual who developed diarrhoea in the two weeks after contact with a primary case (household or equivalent contact), with no other exposure thought more likely to account for their symptoms.
3. Results
3.1. Microbiological Investigation
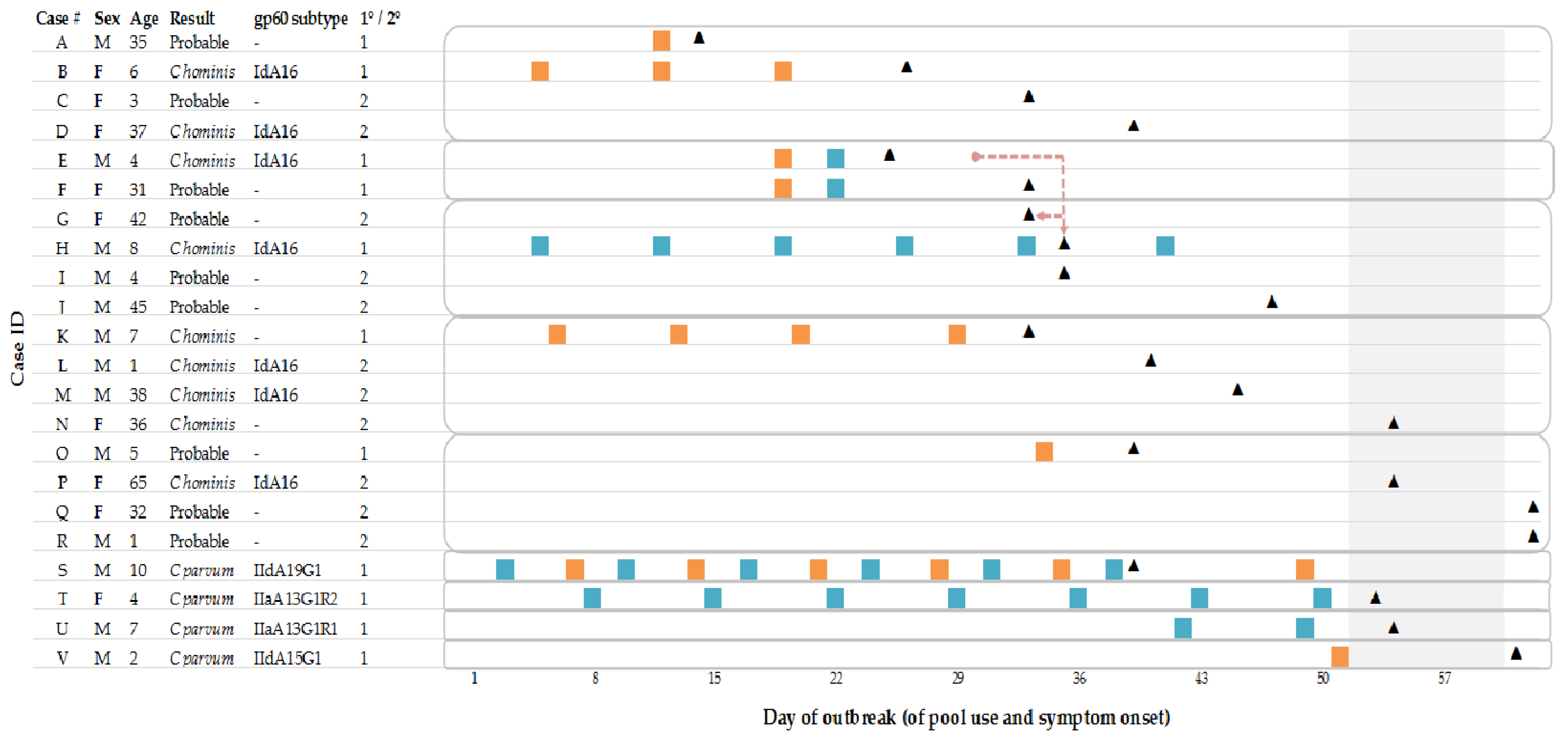
Of the 22 diarrhoea cases, 14 submitted a faecal specimen of which 13 were confirmed positive for Cryptosporidium by the CRU. Of these, one primary and one secondary case had tested negative by auramine phenol microscopy at the local laboratory but were positive by immunofluorescence microscopy at the reference laboratory. The specimen from one symptomatic case who had contact with a confirmed primary case showed a positive reaction in the enzyme immunoassay at the local laboratory but was not confirmed at the reference laboratory so was classified as probable; there were nine probable cases.
Of the 13 confirmed
Cryptosporidium faecal specimens, nine were
C. hominis: four primary and five secondary cases (
Table 1). Four cases were
C. parvum, all primary. Further characterisation by sequencing of the
gp60 gene was possible for eleven of the thirteen confirmed cases (
Table 1). All four
C. parvum cases had different
gp60 subtypes (
Table 1). Seven of the nine
C. hominis cases were subtyped and were identified as IdA16. Three of the IdA16 cases were primary, and four were secondary (
Table 1).
3.2. Descriptive Epidemiological Analysis
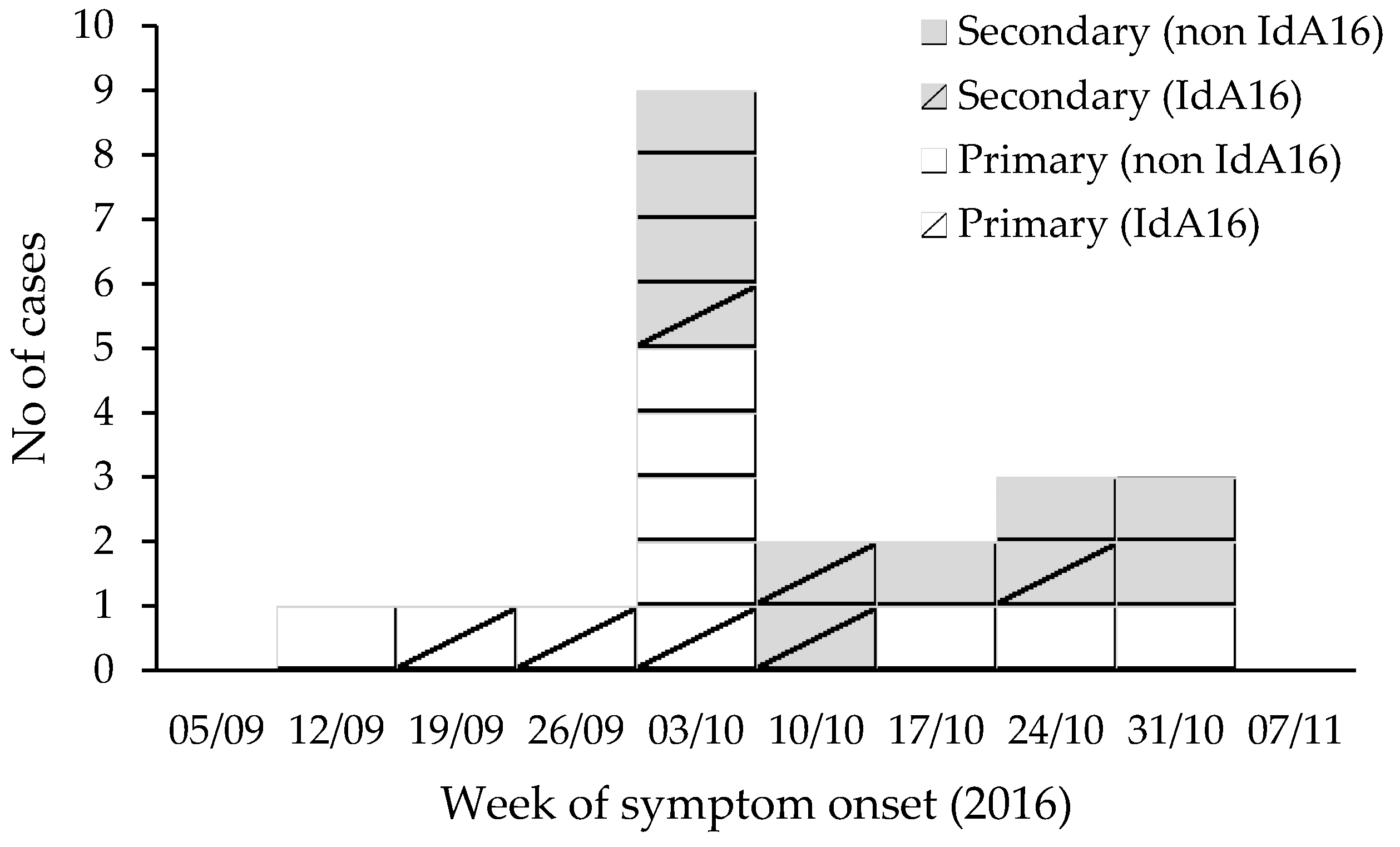
The investigation identified cases in nine households: 11 primary and 11 secondary (
Table 1;
Figure 1). Primary cases’ onset dates ranged from 14 September to 31 October 2016 with pool use up to 21 October 2016. The median age for all cases was seven years (range 1–65 years), and 59% of cases were aged ten years or under (
Figure 2). Fifty-nine percent were male (
Figure 2).
All 22 cases reported diarrhoea, fourteen (64%) reported abdominal pain, ten (45%) reported vomiting, and ten reported other symptoms including fever, nausea, lethargy, and appetite loss. Two children were admitted to hospital. For the ten cases where symptom duration was known, the median duration of symptoms was seven days (range 3 to 10 days). Two male cases, aged 8 and 10 years, described returning to swimming less than two weeks after their reported end of symptoms (
Figure 2).
No cases, or households with multiple cases, had multiple
Cryptosporidium species detected. The seven cases with
C. hominis IdA16 (four male and three female) were from five different households, in which other individuals with diarrhoea were identified, although further testing of symptomatic household members was only undertaken in three of the households, with a total of 11 secondary cases identified (
Figure 2). The three primary cases were aged 4, 6, and 8 years, and the secondary cases were aged 1, 37, 38, and 65 years (
Figure 2). Of the primary cases, one reported swimming at pool A, one reported swimming at pool B, and one reported swimming at both pools (
Figure 2). The case who had only swam at pool A and the four secondary cases all had contact with a probable or confirmed primary case who had used pool B. Therefore, all IdA16 cases had either direct or indirect exposure via a primary case to pool B (
Figure 2).
All four
C. parvum cases had different gp60 subtypes; two cases had used pool A only, one had used both pools, and one had used only pool B (
Figure 2).
In addition to pool use, ten cases reported contact with animals, which is also a plausible Cryptosporidium exposure, especially for C. parvum. This contact was mostly with companion animals (dogs, cats, rabbits, tortoise) and reported by C. hominis and C. parvum cases. One child in a family of four (all C. hominis, two known IdA16) also reported visiting a petting farm within 14 days of onset of illness. Three cases from two different households also reported having social contact, outside of the swimming pools, on one occasion involving accidental faecal contact. There were no other reported exposures that linked the cases or appeared more likely to explain their illness.
The outbreak was declared over on 1 December, four weeks after the pools re-opened (on 1 November). The last primary case was referred on 14 November with onset on 31 October and had been swimming prior to the pool closures only. No further reports of illness with exposure to either pool were identified.
3.3. Environmental Investigation
The setting was two public leisure facilities in North East England approximately four kilometres apart. Both pools were owned by the same council and managed by the same team, and the water came from the same two mains (municipal) supplies. Both were monitored for Cryptosporidium, one continuously and the other weekly; there were no detections from either works (Northumbrian Water Limited data). Although pool staff had attended formal pool plant operator training courses, documented evidence was not provided. Both facilities had a main and a learner pool each with separate treatment plants, and they were multi-use facilities accessed by members of the general public for recreational swimming, lessons, and clubs. Pool A was also used by groups using equipment that had been used in open water e.g., sub-Aqua. EHOs first visited the pools on 18 October, and the pools voluntarily closed as a precautionary measure on 21 October in order to undertake control measures.
At both facilities, the pool surrounds were considered to be in a good and clean condition, and the water was clear with no sediment at the bottom of the pools. Calcium hypochlorite was the main disinfectant in use, and manual pH and combined/free chlorine testing was undertaken four times a day. Supplementary UV light treatment was in place for the main pools using slip stream treatment, which was provided by a medium pressure UVC irradiation unit. Medium rate (<25 m/h) sealed sand and gravel filter vessels were in use for both pools at both facilities. Filter backwashing procedures were documented and reported to take place at least twice a week at night after closure. There was a lack of water replacement as the backwash did not go to drain. Poly aluminium chloride was used as a coagulant with continuous dosing. Routine microbiological sampling was undertaken on a monthly basis using an accredited laboratory, and results for the most recent three months (July, August, and September) were reported to be satisfactory. Hand-washing facilities were unavailable at the nappy changing stations. No swim hygiene advice was on display at either facility; although not a legal requirement, it is good practice [
17].
Measured physical and chemical parameters were satisfactory or within range except those mentioned here. In pool A, there was evidence of fluctuating free chlorine levels, ranging from 0.39 to 2.2 mg/l, and combined chlorine levels were frequently greater than half the free chlorine level. This indicated that the water treatment plant was either having difficulty coping with bather pollution or that detergents in the pool water had combined with chlorine. A coagulant dosing pump was indicating that a service was due but was still dosing effectively. A faecal release had been reported to management with details of how it was handled, but had not been recorded in the pool log book. The event occurred more than a week before the onset of any cases; it was a formed stool that was assessed to have been dealt with appropriately at the time, as per guidance [
17,
18], and considered to be lower risk than a liquid stool. However, the lack of recording reduced confidence that there were no other events. No other major operational issues were identified.
Pool B reported that high pH values of 7.6–7.8 were regularly recorded, which would have reduced the effectiveness of the coagulant and thus filtration, and also of the chlorine disinfection. A faecal release prior to the outbreak was reported during the pool inspection, outside the incubation period of the first case, but no further details were available. The paperwork for recording chemical checks did not indicate when action was required. There were occasions when backwashing may have occurred while the pool was in use. The water from the showers on the pool side, which were used both pre-swim and post-swim, ran into a deck-level drain.
3.4. Outbreak Control Measures
At both pools, further control measures were implemented on 21 October including a backwash and subsequent replacement of one-third of the pool water, increased coagulant dosing, replacement of 150 mm filter medium (as contaminants might penetrate to below 100 mm), and then circulating the water for 24 h, providing greater than six turnover periods at correct disinfection and filtration parameters. This was to minimise any potential risk should Cryptosporidium oocysts remain in the pool. The pools provided verbal updates to visitors and improved signage regarding exclusion advice following diarrhoeal illness. Staff training on pool water management focussed on managing chlorine and pH levels. Staff deep cleaned the pool surrounds. The backwashing procedure was remedied so that the backwash was discharged into the drain.
Other general hygiene measures included providing handwashing advice and the additional provision of sanitising hand wipes at the nappy changing stations; although these were of limited use for such purposes and not effective against Cryptosporidium, they were a temporary measure as there were no sinks in the immediate vicinity of these stations.
Pool management issued letters to parents of children attending swimming lessons and groups that used the pools informing them of the situation and highlighting swimming exclusion advice. Both facilities introduced reimbursement or credit for any swimming lessons missed due to diarrhoeal illness and advised swimmers not to swim for two weeks following diarrhoeal illness.
3.5. Material Deposited
One
C. hominis gp60 sequence, UKH96, representing the seven identical IdA16 subtypes, was previously placed on Genbank with the accession number MK391442 when the outbreak was included in a published summary of
Cryptosporidium outbreaks [
10]. The four
C. parvum sequences were placed on Genbank, accession numbers MZ596421 through to MZ596424.
4. Discussion
This report describes a complex prolonged cryptosporidiosis outbreak of 22 confirmed and probable cases that were most likely linked epidemiologically in time and place to two swimming pools. Although Cryptosporidium is a notifiable agent in the UK, the full extent of the outbreak may have been underestimated, as some symptomatic cases may not have sought medical attention or were not tested. It is not known whether the probable cases had cryptosporidiosis nor whether any asymptomatic contacts were infected. There may have been recall bias towards cases reporting recreational water-related exposures, once there was awareness of the outbreak, and under reporting of other exposures. As some cases had multiple exposures to one or both of the pools and contact with other cases prior to symptom onset, it was not always possible to identify whether a case was primary or secondary or to calculate incubation periods. The epidemiology and timing of symptom onset were used to assess this, although this may not have been correct and was not possible to verify.
The outbreak coincided with the seasonal cryptosporidiosis increase [
19], so a rapid and consistent investigation of new cases was important in identifying common exposures. Those carrying out surveillance should remain alert to identifying and investigating common exposures, but this is not always done, and practice in England and Wales is variable [
8]. It is also important that other possible exposures are considered when cases occur in a close geographical area, as swimming is a common activity. There was no indication in the
Cryptosporidium case surveillance data for the area supplied by the water treatment works that cases might be linked to mains water consumption, and there had been no detections in the treated water. A child in one family reported visiting a petting farm, but all cases were
C. hominis, and it would be highly unusual to acquire
C. hominis from animals [
10]. The distinctly different
C. parvum subtypes indicated that despite being epidemiologically linked, the cases were likely to be sporadic cases of incidental finding without a single common source. This may reflect that swimming is a common exposure, particularly amongst the affected age group. Although they were investigated in the same way, none of the
C. parvum cases were linked to secondary spread, which has been reported to occur more frequently with
C. hominis [
19,
20].
An analytical epidemiological study was considered but was not carried out due to the statistical limitations of the small number of cases initially notified and because control measures had already been implemented. However, investigators should consider carrying out analytical studies in larger outbreaks in order to identify risk factors associated with cases [
21].
In line with usual public health practice, the case definition included those who developed diarrhoea within 1–12 days of attendance at either pool [
2]. However, symptoms are unlikely to develop before three days post exposure to
C. hominis [
4], and recommendations for the public health management of gastrointestinal infections show that the usual range is 3–12 days and the median range is five to seven days [
9]. However, the first identified confirmed case had an onset 2 days after using pool B, indicating either an unusually short incubation period or mis-recall of their swim date. Including case definitions with a broad time interval may be helpful in such circumstances and where cases of other pathogens with shorter incubation periods might also occur.
The species identification and
gp60 subtyping results added value to the interpretation of the epidemiology by identifying cases that were likely to have a common source of infection. Seven cases had a rare
C. hominis subtype, IdA16, supporting the hypothesis that they were likely to be associated with a single local source of exposure (one pool) followed by wider household and community spread. The hypothesis was also supported by existing knowledge;
C. parvum is more commonly acquired from zoonotic transmission than human, and outbreaks linked to swimming pools are more commonly caused by
C. hominis [
10]. Prior to this outbreak, only three isolates of
C. hominis IdA16 had been identified by the CRU in different regions of England in 2008 and 2015, and no other cases had been identified in the UK (verified by consultation with counterparts in Scotland; Claire Alexander, personal communication). One of the UK cases was published previously [
22]. Globally, other cases of
C. hominis IdA16 were identified at the time from the Basic Local Alignment Search Tool database and PubMed, from Australia [
23,
24], Bangladesh [
25], Egypt (GenBank), India [
26], and Tasmania [
27] and in wastewater samples in China [
28]. More recently, cases have been reported in arctic Canada [
29], Israel [
30], and Sweden, although these were linked to travel to China and Sri Lanka [
31]. Nevertheless, IdA16 remains a rare subtype.
Speciation and
gp60 subtyping results are not always available for all cases in an outbreak, as samples may not be sent to the CRU or the
gp60 PCR may not be positive. This could pose an additional limitation to case finding if case definitions are too restrictive. In this investigation, the application of specialist tests to two cases that tested negative locally but where there was a high suspicion of cryptosporidiosis due to epidemiological links to other confirmed cases was fruitful in providing a diagnosis and species but not subtype. The diagnostic tests used for
Cryptosporidium in the UK vary in sensitivity, and comparison has shown the value of using specialist tests such as immunofluorescence microscopy [
32,
33].
As not all cases were confirmed, it is not known whether cases in households were infected with multiple species or genotypes. Given that some cases had multiple exposures and background sporadic cases were also identified, this is possible. The PCR to identify species would have a high likelihood of detecting if both
C. hominis and
C. parvum were present in a specimen, but the Sanger sequencing technology for analysis of the gp60 gene would be unlikely to pick up multiple infections unless additional analysis is undertaken. More recently, we were able to re-investigate the
C. parvum sequences for previously obscured mixed infections using the Tracking of Indels by DEcomposition (TIDE) algorithm to analyse shifts in target sequences [
34]. This post hoc analysis identified an additional underlying
Cryptosporidium sequence in the sample from case U and also probably case S. This indicated the likelihood of multiplicity of infection in these cases, which is especially relevant for case S, who swam on multiple occasions at both pools. The TIDE analysis was unsuccessful for case V and identified only underlying sequence stutter in the sample from case T. Improved reference chromatograms are needed for routine application of the TIDE algorithm, and reference sequences are needed to extend its use to
C. hominis subtypes.
In some households, the case with the earliest onset and with pool exposure was not confirmed, but subsequent symptomatic cases were. This is expected, as public and health professional awareness of a situation increases and subsequent cases may be more likely to attend healthcare and submit a sample.
From a pool operator and management and from a public health perspective, control measures at swimming pools need to be implemented quickly [
18,
21], and these are often based on a hypothesized exposure rather than confirmed and before diagnosis. However, the sooner diagnostic and reference laboratory results are available, the sooner the OCT can be reassured that the control measures put in place are appropriate. Investigations should apply a precautionary approach where there is a suggested, but not confirmed, pathway of exposure. In this outbreak, individuals were known to be swimming while potentially infectious, chemical values were out of range, there were reported and potentially unreported faecal accidents, and multiple epidemiologically linked cases had been identified with no other more likely exposure.
The fluctuating pH identified in pool B, which was often greater than the recommended maximum value of 7.4 [
17], would have played a contributory role in reducing the effectiveness of coagulation, reducing filter efficiency for removing
Cryptosporidium [
17,
35]. Although UV is effective at treating
Cryptosporidium [
36], this was not delivered full-stream, contrary to recommendations [
17]. The OCT hypothesized that an unidentified faecal event or serial contamination may have occurred at pool B, which either overloaded the filtration system or resulted in persistent environmental contamination. A lack of water replacement subsequent to backwashing filters meant that one mechanism for diluting contaminants was missing, and recycling backwash may have perpetuated the contamination. Results of the routine microbiological monitoring showed that the chlorine disinfection was effective but do not provide a good indicator for
Cryptosporidium. The decision not to sample the pool water for
Cryptosporidium was based on the likelihood that oocysts would not be found following multiple turnovers and backwash water cycles and some water replacement, and that some intervention measures had already been initiated.
The evidence from this outbreak suggests that
C. hominis IdA16 may be a relatively infectious subtype with apparent higher transmissibility than
C. parvum. Although both species are transmissible,
C. hominis cases have been reported to have higher excretion levels [
37] and are more often associated with family or household clusters [
19]. When cycle threshold values (which can estimate the number of oocysts present in a sample and therefore indicate the magnitude of the infection) from the real-time PCR used to identify
C. hominis were compared, the outbreak IdA16 cases were similar to those of contemporaneous background cases of
C. hominis IbA10G2 cases predominant at the time (CRU data, not shown) and are regarded as virulent [
38]. There were insufficient local
C. parvum cases at the time for similar comparisons to be made.
Some swimmers reported returning to the pool less than two weeks after the end of symptoms and may have returned to swimming while still infectious. Cryptosporidiosis cases should be reminded not to use pools for 14 days after their symptoms resolve, as per guidance on the prevention of person-to-person transmission [
9]. It has been reported previously that swimmers can be reluctant to adhere to the advice, and it seems difficult to prevent cases returning [
21]. The reimbursement and credits provided by the pool facilities may help to mitigate this.
5. Conclusions
Although this outbreak occurred in 2016, swimming pool-related outbreaks of cryptosporidiosis continue to occur and can be complex as shown by the investigation described here. While pool operating procedures should be sufficient to cope with low-level contamination [
39], failings may occur, and significant expertise is required to identify and address these [
40]. Pool management teams should ensure that operational guidance is followed [
17] and reinforce and maintain swim hygiene messages [
16]. These are especially important during peak use periods [
39] and during outbreaks, as there will be returning swimmers, and they may also use other pools. Messaging should also be reinforced beyond the pool facilities, such as during healthcare consultations and via information leaflets/posters. There should be a general availability of resources (posters/leaflets etc.) for pools to use in order to advise users about exclusion periods; see for example those provided by the Pool Water Treatment Advisory Group (PWTAG) available at
https://www.pwtag.org/posters/ (accessed on 4 November 2021). Public health professionals investigating cases of
Cryptosporidium linked to swimming pools should refer to local outbreak plans and the published guidance [
16]. This includes the legal framework, normal pool water treatment, operating parameters and standards, key indicators of good pool management, establishing and investigating a link between cases and pools, emergency actions for public health protection, public health messages, independent sources of advice, and the inspection of swimming pools for which a checklist is provided [
16]. Cryptosporidiosis cases should be reminded not to use pools for 14 days after their symptoms resolve, as per guidance on the prevention of person-to-person transmission [
9].
The Cryptosporidium species identification and gp60 subtyping results added value to the interpretation of the epidemiology by identifying cases that were likely to have a common source of infection. This outbreak investigation demonstrated the added value of using gp60 subtyping in helping to support the hypothesis, particularly as some cases had multiple links and exposures. Subtyping distinguished between cases that were likely to have a common exposure or were likely to be sporadic. The rarity of the C. hominis subtype identified also supported the hypothesis that the C. hominis cases had been exposed, directly or indirectly, to a single common local source.
While routine detailed epidemiological information should always be obtained in outbreak investigations, this should be supplemented by characterization of the species and subtype. This would add value to investigations by helping to support or refute the hypothesis, particularly in swimming pool outbreaks where exposure histories can be complex and multiple sources of infection are suspected. Awareness of the availability and value of subtyping to laboratories should be raised so that more investigations benefit from this service in the future and in order to further understanding of cryptosporidium molecular epidemiology.