Optimizing ADM1 Calibration and Input Characterization for Effective Co-Digestion Modelling
Abstract
:1. Introduction
2. Materials and Methods
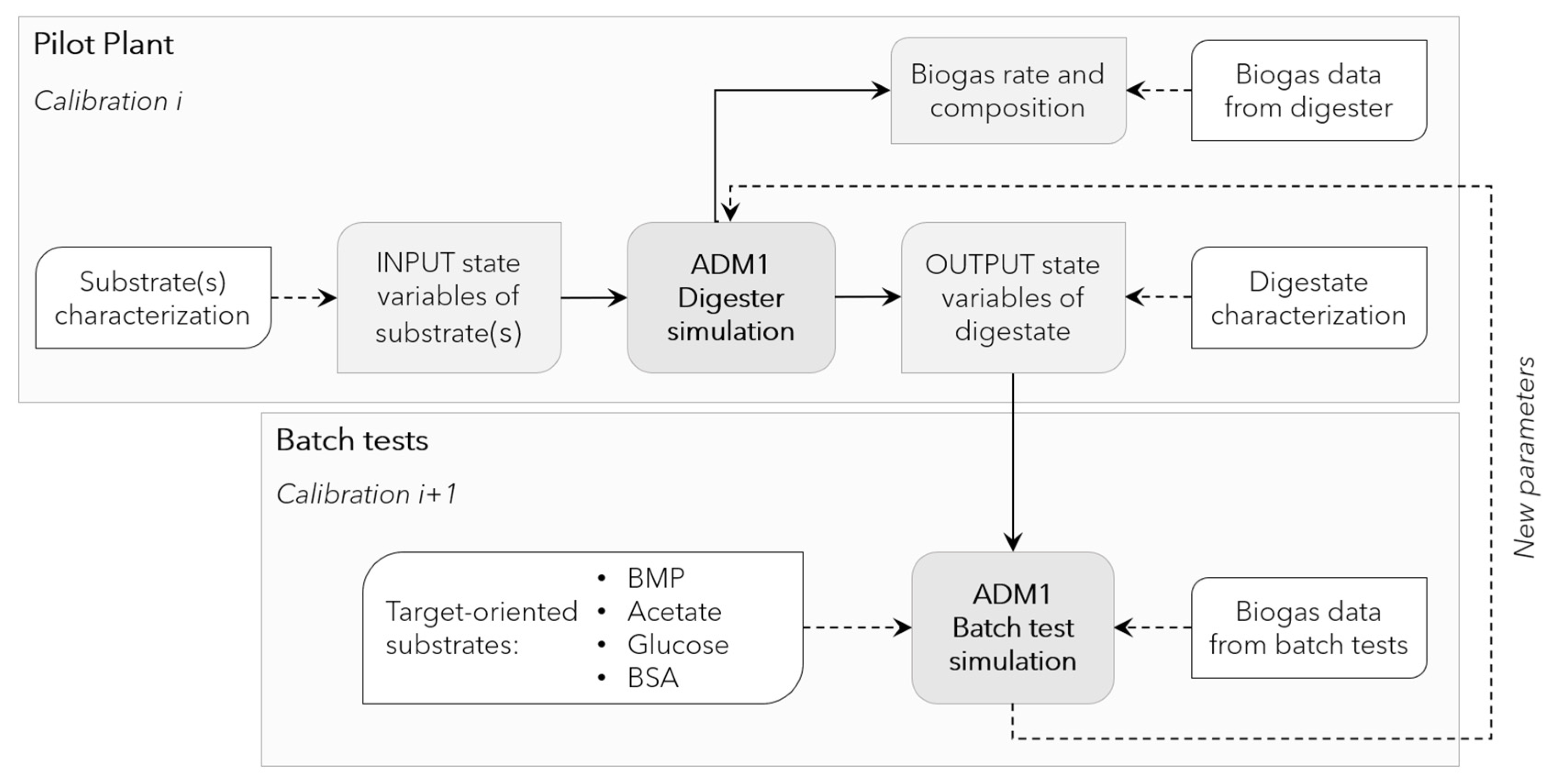
2.1. Pilot Plant
2.2. Monitoring Plan
2.3. Analytical Methods
2.4. Batch Experiments
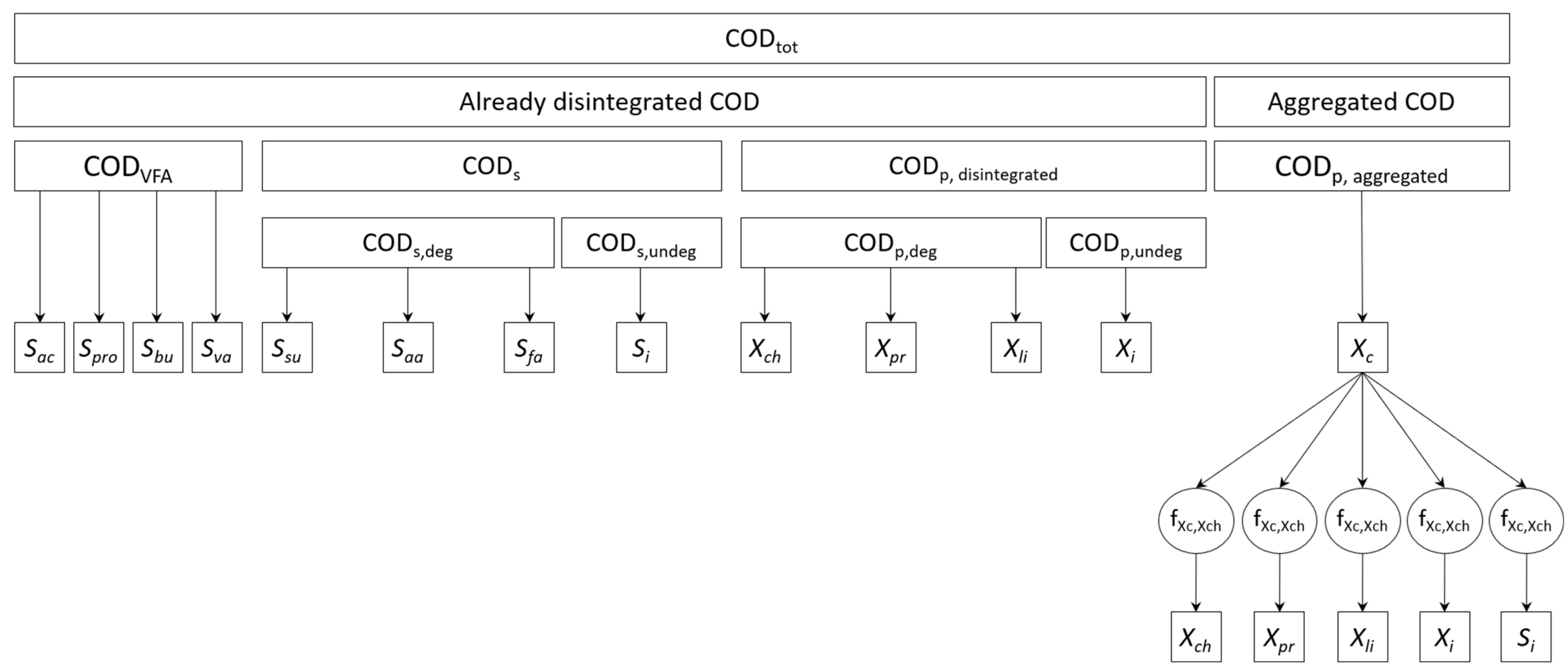
2.5. Input State Variables’ Determination
2.6. Model Implementation, Calibration, and Evaluation
3. Results
3.1. Substrates and Digestate Characterization
3.2. Model Calibration
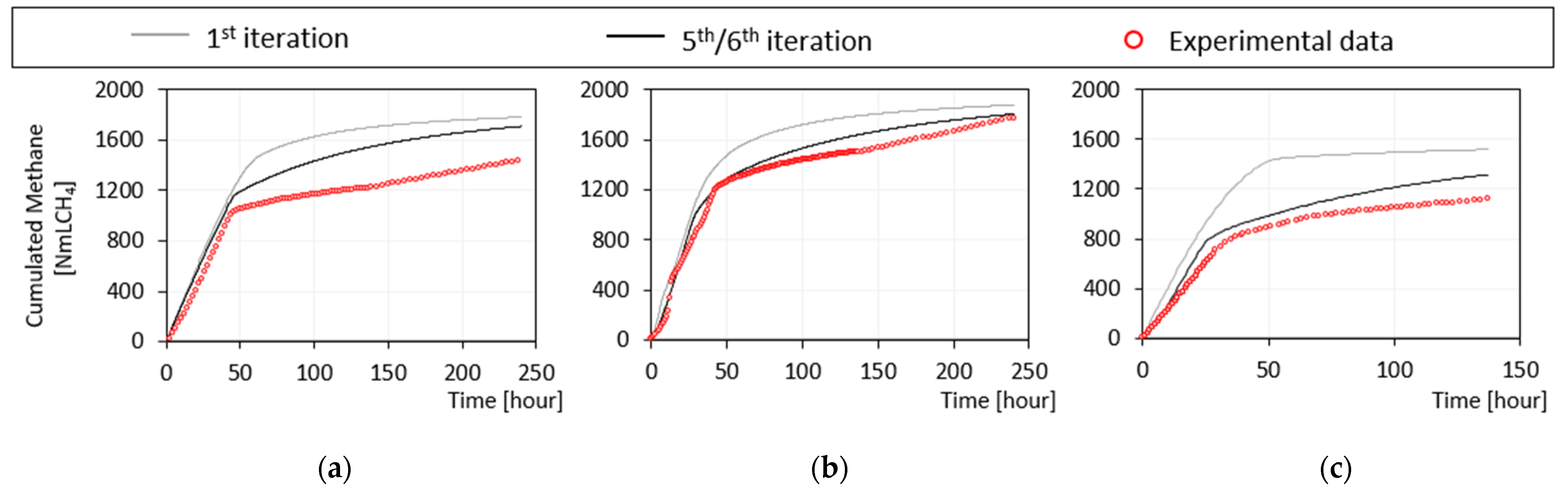
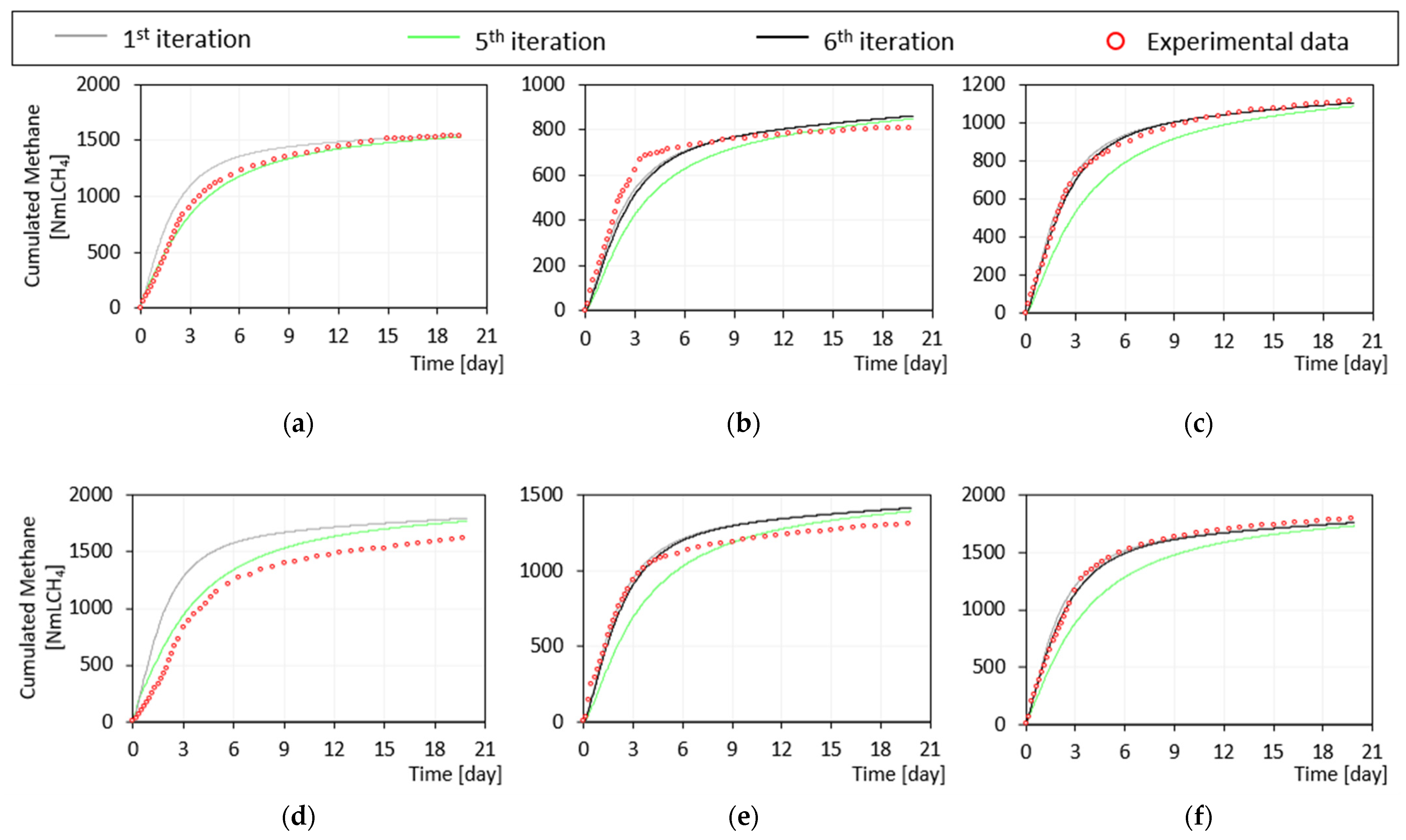
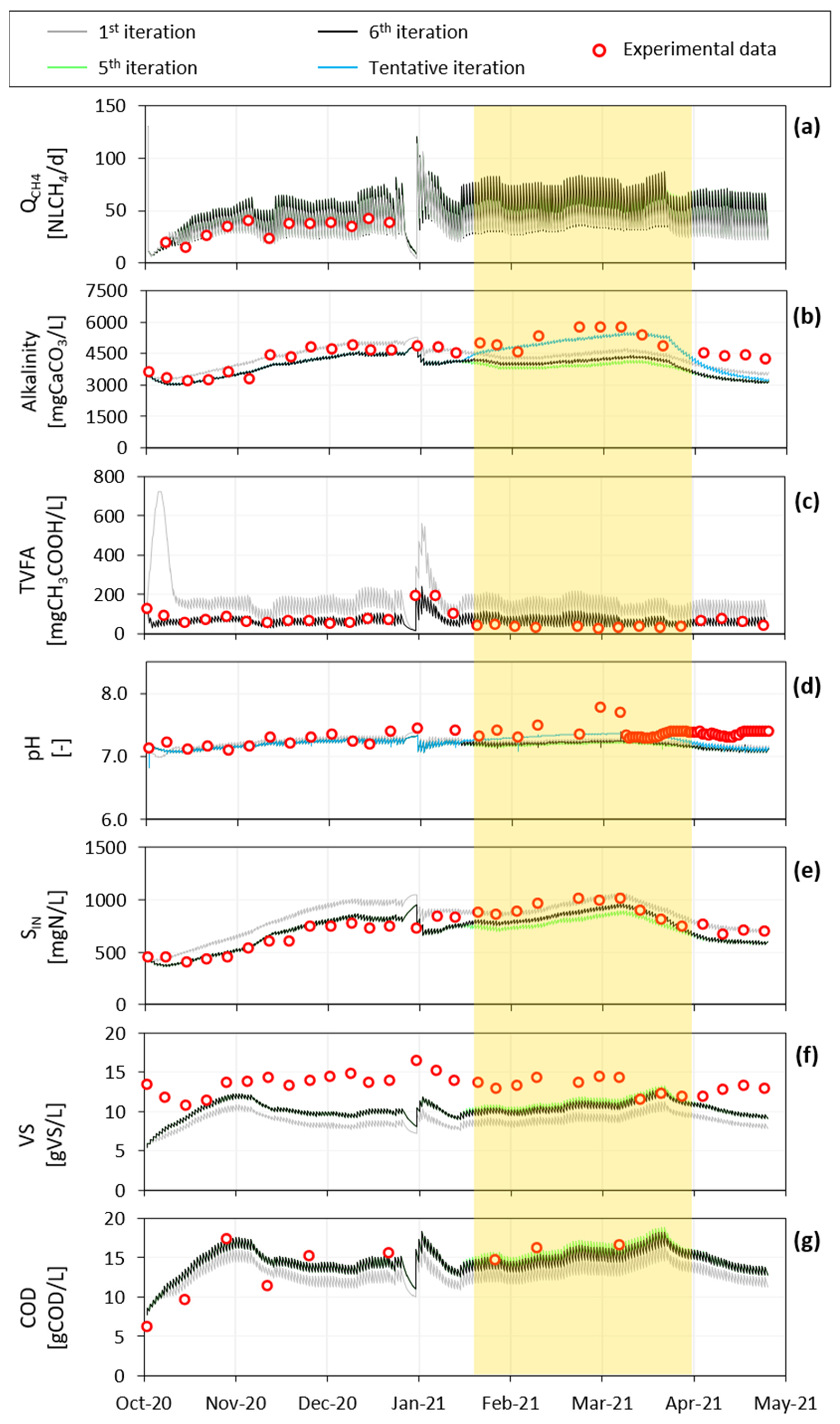
3.2.1. Iteration Steps from First to Fifth
3.2.2. Iteration Step from Fifth to Sixth
- Disintegration constants are different for waste sludge and yogurt, depending on the type of substrate and not only on biomass adaptation.
- Some soluble proteins were already available from yogurt.
3.2.3. Yogurt Alkalinity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Nomenclature
| Cac, Cpro, Cbu, Cva | concentrations of acetic, propionic, butyric, and valeric acids [g∙L−1]; |
| γac, γpro, γbu, γva | stoichiometric conversion coefficients from moles of acetic, propionic, butyric, and valeric acids to COD [mol∙gCOD−1]; |
| MWac, MWpro, MWbu, MWva | molecular weights of acetic, propionic, butyric, and valeric acids [g∙mol−1]; |
| CHtot, PRtot | total concentration of carbohydrates and proteins [g∙L−1]; |
| LIp | particulate concentration of lipids [g∙L−1]; |
| LIs | soluble concentration of lipids [g∙L−1]; |
| CHs, PRs, LIs | soluble concentrations of carbohydrates, proteins, and lipids [g∙L−1]; |
| αch,COD, αpr,COD, αli,COD | conversion coefficients from carbohydrates, proteins, and lipids to COD [g∙g COD−1]; |
| fY | percentage of COD used for cell growth during BMP test [gCOD∙gCOD−1]; |
| CIN | concentrations of ammoniacal nitrogen [gN∙L−1]; |
| MWN | molecular weight of nitrogen [gN∙mol−1]; |
| Alk | concentration of alkalinity [M]; |
| Sac-, Spro-, Sbu-, Sva- | concentrations of dissociated species of acetic, propionic, butyric, and valeric acids [M]. |
Appendix A
| Variable Name | n. | |
|---|---|---|
| Soluble COD from VFAs | (1) | |
| Total COD | (2) | |
| Soluble COD Excluding VFAs | (3) | |
| Particulate COD | (4) | |
| Undegradable total COD | (5) | |
| Undegradable particulate COD | (6) | |
| Undegradable soluble COD | (7) | |
| Degradable total COD | (8) | |
| Degradable particulate COD | (9) | |
| Degradable soluble COD | (10) |
| Variable Name 1 | Equation | n. |
|---|---|---|
| Particulate inert | (1) | |
| Soluble inert | (2) | |
| Particulate carbohydrates | (3) | |
| Particulate proteins | (4) | |
| Particulate lipids | (5) | |
| Soluble sugars | (6) | |
| Soluble amino acids | (7) | |
| Soluble fatty acids | (8) | |
| Soluble total acetic acid | (9) | |
| Soluble total propionic acid | (10) | |
| Soluble total butyric acid | (11) | |
| Soluble total valeric acid | (12) | |
| Soluble inorganic nitrogen | (13) | |
| Soluble bicarbonate | (14) |
| Coefficient Name | n. | |
|---|---|---|
| Yield of particulate carbohydrates | (1) | |
| Yield of particulate proteins | (2) | |
| Yield of particulate lipids | (3) | |
| Yield of particulate inert | (4) | |
| Yield of soluble inert | (5) |
References
- Hanum, F.; Yuan, L.C.; Kamahara, H.; Aziz, H.A.; Atsuta, Y.; Yamada, T.; Daimon, H. Treatment of Sewage Sludge Using Anaerobic Digestion in Malaysia: Current State and Challenges. Front. Energy Res. 2019, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Mata-Alvarez, J.; Dosta, J.; Romero-Güiza, M.S.; Fonoll, X.; Peces, M.; Astals, S. A Critical Review on Anaerobic Co-Digestion Achievements between 2010 and 2013. Renew. Sustain. Energy Rev. 2014, 36, 412–427. [Google Scholar] [CrossRef]
- Montusiewicz, A.; Lebiocka, M. Co-Digestion of Intermediate Landfill Leachate and Sewage Sludge as a Method of Leachate Utilization. Bioresour. Technol. 2011, 102, 2563–2571. [Google Scholar] [CrossRef]
- Mata-Alvarez, J.; Dosta, J.; Macé, S.; Astals, S. Codigestion of Solid Wastes: A Review of Its Uses and Perspectives Including Modeling. Crit. Rev. Biotechnol. 2011, 31, 99–111. [Google Scholar] [CrossRef]
- Zhou, H.; Ying, Z.; Cao, Z.; Liu, Z.; Zhang, Z.; Liu, W. Feeding Control of Anaerobic Co-Digestion of Waste Activated Sludge and Corn Silage Performed by Rule-Based PID Control with ADM1. Waste Manag. 2020, 103, 22–31. [Google Scholar] [CrossRef]
- Astals, S.; Batstone, D.J.; Mata-Alvarez, J.; Jensen, P.D. Identification of Synergistic Impacts during Anaerobic Co-Digestion of Organic Wastes. Bioresour. Technol. 2014, 169, 421–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisgativa, H.; Zennaro, B.; Charnier, C.; Richard, C.; Accarion, G.; Béline, F. Comprehensive Determination of Input State Variables Dataset Required for Anaerobic Digestion Modelling (ADM1) Based on Characterisation of Organic Substrates. Data Brief 2020, 29, 105212. [Google Scholar] [CrossRef] [PubMed]
- Batstone, D.J.; Keller, J.; Angelidaki, I.; Kalyuzhnyi, S.V.; Pavlostathis, S.G.; Rozzi, A.; Sanders, W.T.M.; Siegrist, H.; Vavilin, V.A. The IWA Anaerobic Digestion Model No 1 (ADM1). Water Sci. Technol. 2002, 45, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Girault, R.; Bridoux, G.; Nauleau, F.; Poullain, C.; Buffet, J.; Steyer, J.-P.; Sadowski, A.G.; Béline, F. A Waste Characterisation Procedure for ADM1 Implementation Based on Degradation Kinetics. Water Res. 2012, 46, 4099–4110. [Google Scholar] [CrossRef]
- Rosen, C.; Jeppsson, U. Aspects on ADM1 Implementation within the BSM2 Framework; Department of Industrial Electrical Engineering and Automation, Lund Institute of Technology: Lund, Sweden, 2005. [Google Scholar]
- Li, P.; Pei, Z.; Liu, D.; Shi, F.; Wang, S.; Li, W.; Sun, Y.; Liu, J.; Gao, Y.; Yu, Q. Application of Anaerobic Digestion Model No. 1 for Modeling Anaerobic Digestion of Vegetable Crop Residues: Fractionation of Crystalline Cellulose. J. Clean. Prod. 2021, 285, 124865. [Google Scholar] [CrossRef]
- Razaviarani, V.; Buchanan, I.D. Calibration of the Anaerobic Digestion Model No. 1 (ADM1) for Steady-State Anaerobic Co-Digestion of Municipal Wastewater Sludge with Restaurant Grease Trap Waste. Chem. Eng. J. 2015, 266, 91–99. [Google Scholar] [CrossRef]
- Baquerizo, G.; Fiat, J.; Buffiere, P.; Girault, R.; Gillot, S. Modelling the Dynamic Long-Term Performance of a Full-Scale Digester Treating Sludge from an Urban WRRF Using an Extended Version of ADM1. Chem. Eng. J. 2021, 423, 128870. [Google Scholar] [CrossRef]
- Girault, R.; Rousseau, P.; Steyer, J.P.; Bernet, N.; Béline, F. Combination of Batch Experiments with Continuous Reactor Data for ADM1 Calibration: Application to Anaerobic Digestion of Pig Slurry. Water Sci. Technol. 2011, 63, 2575–2582. [Google Scholar] [CrossRef] [PubMed]
- Batstone, D.J.; Tait, S.; Starrenburg, D. Estimation of Hydrolysis Parameters in Full-Scale Anerobic Digesters. Biotechnol. Bioeng. 2009, 102, 1513–1520. [Google Scholar] [CrossRef]
- Xie, S.; Hai, F.I.; Zhan, X.; Guo, W.; Ngo, H.H.; Price, W.E.; Nghiem, L.D. Anaerobic Co-Digestion: A Critical Review of Mathematical Modelling for Performance Optimization. Bioresour. Technol. 2016, 222, 498–512. [Google Scholar] [CrossRef]
- Souza, T.S.O.; Carvajal, A.; Donoso-Bravo, A.; Peña, M.; Fdz-Polanco, F. ADM1 Calibration Using BMP Tests for Modeling the Effect of Autohydrolysis Pretreatment on the Performance of Continuous Sludge Digesters. Water Res. 2013, 47, 3244–3254. [Google Scholar] [CrossRef] [PubMed]
- Mottet, A.; Ramirez, I.; Carrère, H.; Déléris, S.; Vedrenne, F.; Jimenez, J.; Steyer, J.P. New Fractionation for a Better Bioaccessibility Description of Particulate Organic Matter in a Modified ADM1 Model. Chem. Eng. J. 2013, 228, 871–881. [Google Scholar] [CrossRef]
- Arnell, M.; Astals, S.; Åmand, L.; Batstone, D.J.; Jensen, P.D.; Jeppsson, U. Modelling Anaerobic Co-Digestion in Benchmark Simulation Model No. 2: Parameter Estimation, Substrate Characterisation and Plant-Wide Integration. Water Res. 2016, 98, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Poggio, D.; Walker, M.; Nimmo, W.; Ma, L.; Pourkashanian, M. Modelling the Anaerobic Digestion of Solid Organic Waste—Substrate Characterisation Method for ADM1 Using a Combined Biochemical and Kinetic Parameter Estimation Approach. Waste Manag. 2016, 53, 40–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Gen, S.; Sousbie, P.; Rangaraj, G.; Lema, J.M.; Rodríguez, J.; Steyer, J.-P.; Torrijos, M. Kinetic Modelling of Anaerobic Hydrolysis of Solid Wastes, Including Disintegration Processes. Waste Manag. 2015, 35, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Lübken, M.; Kosse, P.; Koch, K.; Gehring, T.; Wichern, M. Influent Fractionation for Modeling Continuous Anaerobic Digestion Processes. In Biogas Science and Technology; Guebitz, G.M., Bauer, A., Bochmann, G., Gronauer, A., Weiss, S., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer International Publishing: Cham, Switzerland, 2015; Volume 151, pp. 137–169. ISBN 978-3-319-21992-9. [Google Scholar]
- Kleerebezem, R.; Van Loosdrecht, M.C.M. Waste Characterization for Implementation in ADM1. Water Sci. Technol. 2006, 54, 167–174. [Google Scholar] [CrossRef]
- Astals, S.; Esteban-Gutiérrez, M.; Fernández-Arévalo, T.; Aymerich, E.; García-Heras, J.L.; Mata-Alvarez, J. Anaerobic Digestion of Seven Different Sewage Sludges: A Biodegradability and Modelling Study. Water Res. 2013, 47, 6033–6043. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Lübken, M.; Gehring, T.; Wichern, M.; Horn, H. Biogas from Grass Silage—Measurements and Modeling with ADM1. Bioresour. Technol. 2010, 101, 8158–8165. [Google Scholar] [CrossRef]
- Wichern, M.; Gehring, T.; Fischer, K.; Andrade, D.; Lübken, M.; Koch, K.; Gronauer, A.; Horn, H. Monofermentation of Grass Silage under Mesophilic Conditions: Measurements and Mathematical Modeling with ADM 1. Bioresour. Technol. 2009, 100, 1675–1681. [Google Scholar] [CrossRef]
- Zaher, U.; Grau, P.; Benedetti, L.; Ayesa, E.; Vanrolleghem, P. Transformers for Interfacing Anaerobic Digestion Models to Pre- and Post-Treatment Processes in a Plant-Wide Modelling Context. Environ. Model. Softw. 2007, 22, 40–58. [Google Scholar] [CrossRef]
- Zaher, U.; Buffiere, P.; Steyer, J.-P.; Chen, S. A Procedure to Estimate Proximate Analysis of Mixed Organic Wastes. Water Environ. Res. 2009, 81, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Standard Methods for the Examination of Water and Wastewater, 23th ed.; American Public Health Association (APHA): Washington, DC, USA, 2017.
- Jimenez, J.; Vedrenne, F.; Denis, C.; Mottet, A.; Déléris, S.; Steyer, J.-P.; Cacho Rivero, J.A. A Statistical Comparison of Protein and Carbohydrate Characterisation Methodology Applied on Sewage Sludge Samples. Water Res. 2013, 47, 1751–1762. [Google Scholar] [CrossRef] [PubMed]
- Hassam, S.; Ficara, E.; Leva, A.; Harmand, J. A Generic and Systematic Procedure to Derive a Simplified Model from the Anaerobic Digestion Model No. 1 (ADM1). Biochem. Eng. J. 2015, 99, 193–203. [Google Scholar] [CrossRef]
- VDI 4630.Pdf. Available online: https://infostore.saiglobal.com/en-us/Standards/VDI-4630-2016-1115305_SAIG_VDI_VDI_2590568/ (accessed on 29 October 2021).
- Siegrist, H.; Vogt, D.; Garcia-Heras, J.L.; Gujer, W. Mathematical Model for Meso- and Thermophilic Anaerobic Sewage Sludge Digestion. Environ. Sci. Technol. 2002, 36, 1113–1123. [Google Scholar] [CrossRef]
- Decostere, B.; De Craene, J.; Van Hoey, S.; Vervaeren, H.; Nopens, I.; Van Hulle, S.W.H. Validation of a Microalgal Growth Model Accounting with Inorganic Carbon and Nutrient Kinetics for Wastewater Treatment. Chem. Eng. J. 2016, 285, 189–197. [Google Scholar] [CrossRef]
- Hauduc, H.; Neumann, M.B.; Muschalla, D.; Gamerith, V.; Gillot, S.; Vanrolleghem, P.A. Efficiency Criteria for Environmental Model Quality Assessment: A Review and Its Application to Wastewater Treatment. Environ. Model. Softw. 2015, 68, 196–204. [Google Scholar] [CrossRef]
- Ras, M.; Girbal-Neuhauser, E.; Paul, E.; Spérandio, M.; Lefebvre, D. Protein Extraction from Activated Sludge: An Analytical Approach. Water Res. 2008, 42, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Raunkjær, K.; Hvitved-Jacobsen, T.; Nielsen, P.H. Measurement of Pools of Protein, Carbohydrate and Lipid in Domestic Wastewater. Water Res. 1994, 28, 251–262. [Google Scholar] [CrossRef]
- Sosulski, F.W.; Imafidon, G.I. Amino Acid Composition and Nitrogen-to-Protein Conversion Factors for Animal and Plant Foods. J. Agric. Food Chem. 1990, 38, 1351–1356. [Google Scholar] [CrossRef]
- Huang, M.; Li, Y.; Gu, G. Chemical Composition of Organic Matters in Domestic Wastewater. Desalination 2010, 262, 36–42. [Google Scholar] [CrossRef]
- Zaher, U.; Li, R.; Jeppsson, U.; Steyer, J.-P.; Chen, S. GISCOD: General Integrated Solid Waste Co-Digestion Model. Water Res. 2009, 43, 2717–2727. [Google Scholar] [CrossRef]
- Jimenez, J.; Charnier, C.; Kouas, M.; Latrille, E.; Torrijos, M.; Harmand, J.; Patureau, D.; Spérandio, M.; Morgenroth, E.; Béline, F.; et al. Modelling Hydrolysis: Simultaneous versus Sequential Biodegradation of the Hydrolysable Fractions. Waste Manag. 2020, 101, 150–160. [Google Scholar] [CrossRef]
- Shi, E.; Li, J.; Zhang, M. Application of IWA Anaerobic Digestion Model No. 1 to Simulate Butyric Acid, Propionic Acid, Mixed Acid, and Ethanol Type Fermentative Systems Using a Variable Acidogenic Stoichiometric Approach. Water Res. 2019, 161, 242–250. [Google Scholar] [CrossRef]

| Iteration | Parameter (Unit) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kdis (d−1) | khyd,ch (d−1) | khyd,pr (d−1) | khyd,li (d−1) | km,su (d−1) | km,aa (d−1) | km,ac (d−1) | KS,su (kgCOD∙m−3) | KS,aa (kgCOD∙m−3) | KS,ac (kgCOD∙m−3) | KS,pro (kgCOD∙m−3) | |
| 1st | 1.2 | 0.75 | 0.6 | 0.3 | 30 | 50 | 8 | 0.5 | 0.3 | 0.15 | 0.3 |
| 5th (6th) | 1.5 | 0.3 | 0.2 (0.5) | 0.5 | 8 | 8 | 8 | 0.3 | 0.2 | 0.01 | 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catenacci, A.; Grana, M.; Malpei, F.; Ficara, E. Optimizing ADM1 Calibration and Input Characterization for Effective Co-Digestion Modelling. Water 2021, 13, 3100. https://doi.org/10.3390/w13213100
Catenacci A, Grana M, Malpei F, Ficara E. Optimizing ADM1 Calibration and Input Characterization for Effective Co-Digestion Modelling. Water. 2021; 13(21):3100. https://doi.org/10.3390/w13213100
Chicago/Turabian StyleCatenacci, Arianna, Matteo Grana, Francesca Malpei, and Elena Ficara. 2021. "Optimizing ADM1 Calibration and Input Characterization for Effective Co-Digestion Modelling" Water 13, no. 21: 3100. https://doi.org/10.3390/w13213100
APA StyleCatenacci, A., Grana, M., Malpei, F., & Ficara, E. (2021). Optimizing ADM1 Calibration and Input Characterization for Effective Co-Digestion Modelling. Water, 13(21), 3100. https://doi.org/10.3390/w13213100







