Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Areas and Sampling Procedure
2.2. Laboratory Procedures
2.3. Bacterioplankton Analysis
2.3.1. DNA Extraction and PCR Amplification
2.3.2. DGGE
DNA Sequencing of Excised DGGE Bands
2.4. Statistical Analysis
3. Results
3.1. Environmental Variables and Nutrient Concentrations
3.2. Bacterioplankton Community
3.2.1. Bacterial Community of Water
3.2.2. Relations between Bacterial Communities and Environmental Parameters
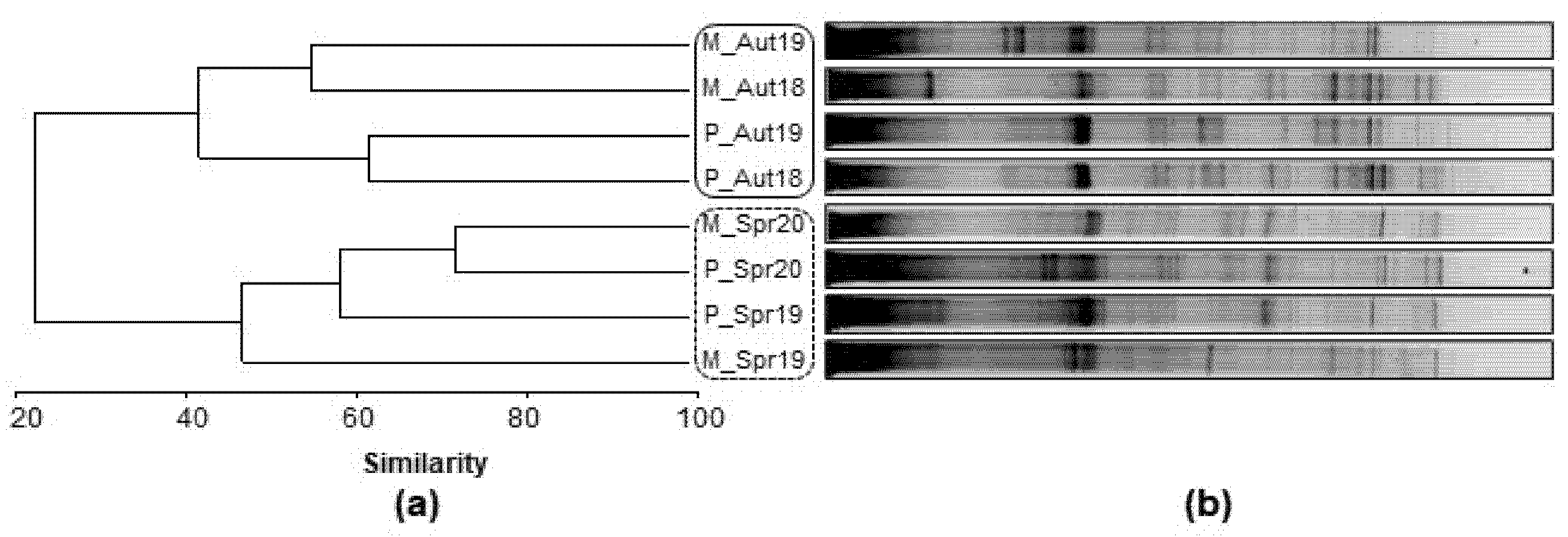
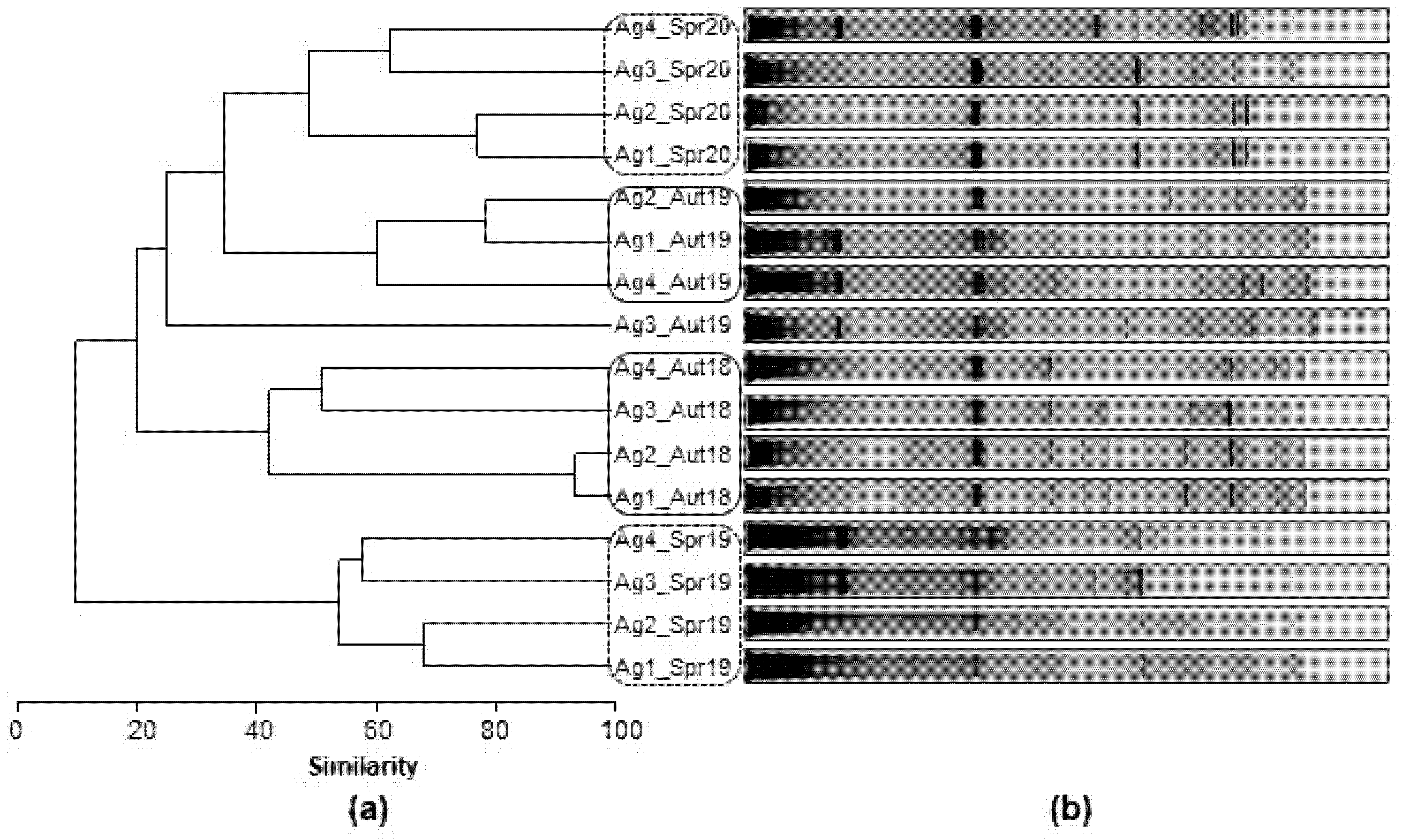
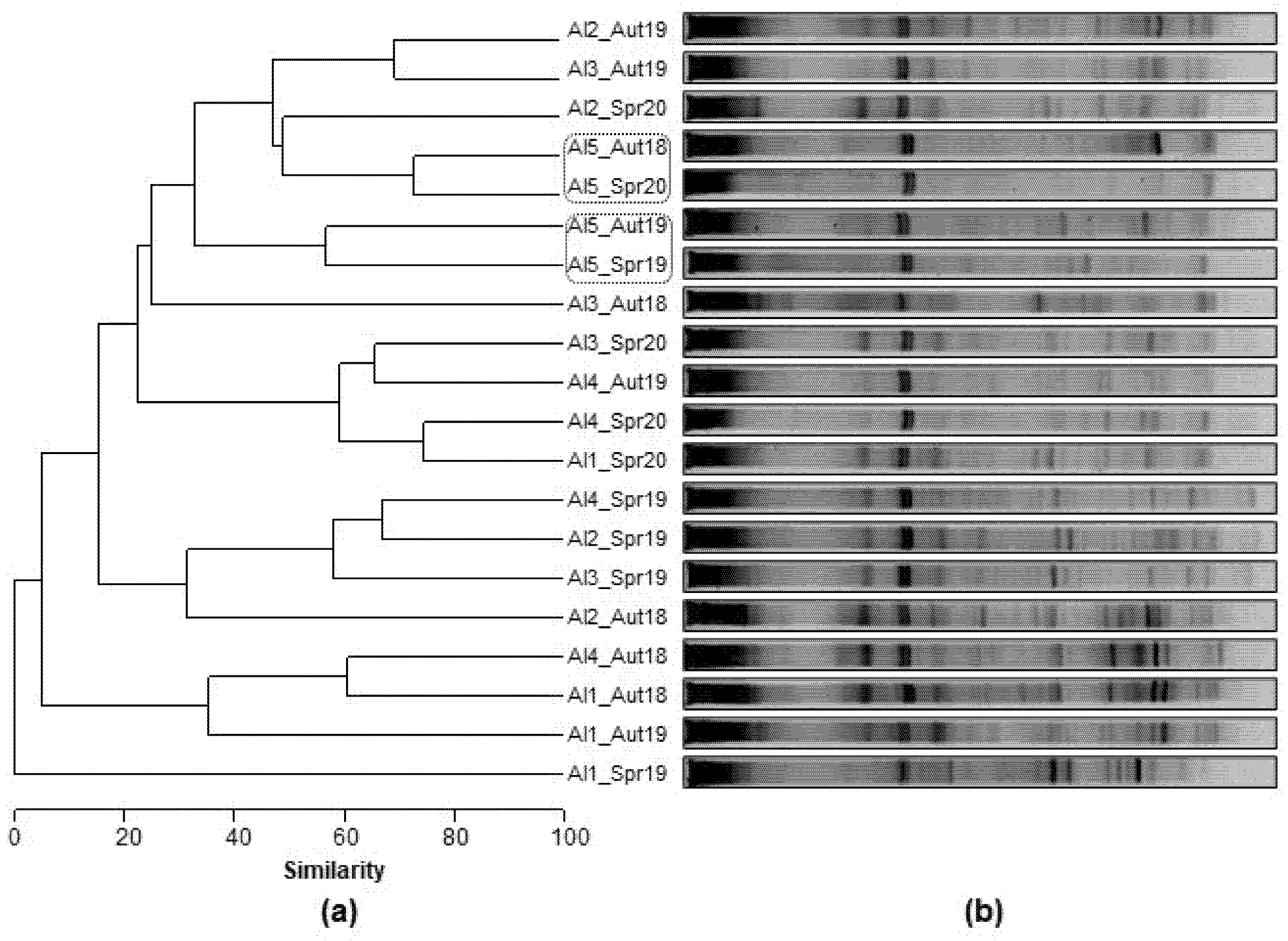
3.2.3. Spatial and Temporal Bacterial Dynamic
3.2.4. Microbial Community Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vasconcelos, V. Freshwater cyanobacteria and their toxins in Portugal. In Cyanotoxins–Occurrence, Causes, Consequences; Chorus, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 62–67. [Google Scholar]
- Liu, M.; Liu, L.; Chen, H.; Yu, Z.; Yang, J.R.; Xue, Y.; Huang, B.; Yang, J. Community Dynamics of Free-Living and Particle-Attached Bacteria following a Reservoir Microcystis Bloom. Sci. Total Environ. 2019, 660, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Farley, M. Encyclopedia of Lakes and Reservoirs; Bengtsson, L., Herschy, R.W., Fairbridge, R.W., Eds.; Encyclopedia of Earth Sciences Series; Springer: Dordrecht, The Netherlands, 2012; pp. 258–270. ISBN 978-1-4020-5616-1. [Google Scholar]
- European Community Commission (ECC). Commission Directive 2000/60/EC of the European Parliament and the Council establishing the framework for community action in the field of water policy. Off. J. Eur. Union 2000, 327, 1–73. [Google Scholar]
- Llirós, M.; Inceoğlu, Ö.; García-Armisen, T.; Anzil, A.; Leporcq, B.; Pigneur, L.-M.; Viroux, L.; Darchambeau, F.; Descy, J.-P.; Servais, P. Bacterial Community Composition in Three Freshwater Reservoirs of Different Alkalinity and Trophic Status. PLoS ONE 2014, 9, e116145. [Google Scholar] [CrossRef] [PubMed]
- Šimek, K.; Horňák, K.; Jezbera, J.; Nedoma, J.; Znachor, P.; Hejzlar, J.; Sed’a, J. Spatio-Temporal Patterns of Bacterioplankton Production and Community Composition Related to Phytoplankton Composition and Protistan Bacterivory in a Dam Reservoir. Aquat. Microb. Ecol. 2008, 51, 249–262. [Google Scholar] [CrossRef][Green Version]
- Berdjeb, L.; Ghiglione, J.F.; Domaizon, I.; Jacquet, S. A 2-Year Assessment of the Main Environmental Factors Driving the Free-Living Bacterial Community Structure in Lake Bourget (France). Microb. Ecol. 2011, 61, 941–954. [Google Scholar] [CrossRef]
- Cole, J.J.; Findlay, S.; Pace, M.L. Bacterial Production in Fresh and Saltwater Ecosystems: A Cross-System Overview. Mar. Ecol. Prog. Ser. 1988, 43, 1–10. [Google Scholar] [CrossRef]
- Lindström, E.S. Bacterioplankton Community Composition in Five Lakes Differing in Trophic Status and Humic Content. Microb. Ecol. 2000, 40, 104–113. [Google Scholar] [CrossRef]
- Höfle, M.G.; Haas, H.; Dominik, K. Seasonal Dynamics of Bacterioplankton Community Structure in a Eutrophic Lake as Determined by 5S RRNA Analysis. Appl. Environ. Microbiol. 1999, 65, 3164–3174. [Google Scholar] [CrossRef]
- Lindström, E.S.; Kamst-Van Agterveld, M.P.; Zwart, G. Distribution of Typical Freshwater Bacterial Groups Is Associated with PH, Temperature, and Lake Water Retention Time. Appl. Environ. Microbiol. 2005, 71, 8201–8206. [Google Scholar] [CrossRef]
- Donner, G.; Schwarz, K.; Hoppe, H.G.; Muyzer, G. Profiling the Succession of Bacterial Populations in Pelagic Chemoclines. Arch. Hydrobiol. Spec. Issues Advanc. Limnol. 1996, 48, 7–14. [Google Scholar]
- Van der Gucht, K.; Sabbe, K.; De Meester, L.; Vloemans, N.; Zwart, G.; Gillis, M.; Vyverman, W. Contrasting Bacterioplankton Community Composition and Seasonal Dynamics in Two Neighbouring Hypertrophic Freshwater Lakes. Environ. Microbiol. 2001, 3, 680–690. [Google Scholar] [CrossRef]
- Yannarell, A.C.; Kent, A.D.; Lauster, G.H.; Kratz, T.K.; Triplett, E.W. Temporal Patterns in Bacterial Communities in Three Temperate Lakes of Different Trophic Status. Microb. Ecol. 2003, 46, 391–405. [Google Scholar] [CrossRef]
- Šimek, K.; Pernthaler, J.; Weinbauer, M.G.; Hornák, K.; Dolan, J.R.; Nedoma, J.; Mašín, M.; Amann, R. Changes in Bacterial Community Composition and Dynamics and Viral Mortality Rates Associated with Enhanced Flagellate Grazing in a Mesoeutrophic Reservoir. Appl. Environ. Microbiol. 2001, 67, 2723–2733. [Google Scholar] [CrossRef]
- de Figueiredo, D.R.; Pereira, M.J.; Moura, A.; Silva, L.; Bárrios, S.; Fonseca, F.; Henriques, I.; Correia, A. Bacterial Community Composition over a Dry Winter in Meso- and Eutrophic Portuguese Water Bodies. FEMS Microb. Ecol. 2007, 59, 638–650. [Google Scholar] [CrossRef] [PubMed]
- Bordalo, A.A.; Teixeira, R.; Wiebe, W.J. A Water Quality Index Applied to an International Shared River Basin: The Case of the Douro River. Environ. Manag. 2006, 38, 910–920. [Google Scholar] [CrossRef]
- Rodrigues, S.; Pinto, I.; Martins, F.; Formigo, N.; Antunes, S.C. Can Biochemical Endpoints Improve the Sensitivity of the Biomonitoring Strategy Using Bioassays with Standard Species, for Water Quality Evaluation? Ecotoxicol. Environ. Saf. 2021, 215, 112151. [Google Scholar] [CrossRef]
- Resolução do Conselho de Ministros n.o 186. Plano de Ordenamento da Albufeira da Aguieira; 1.a Série-N.o 246; Diário Da República: Lisbon, Portugal, 2007. [Google Scholar]
- Pádua, J.; Bernardo, J.M.; Alves, M.H. Exercício de Intercalibração Em Massas de Água Fortemente Modificadas–Albufeiras, No Âmbito Da Directiva Quadro Da Água. In Proceedings of the 9th Congresso da Água, Cascais, Portugal, 2–4 April 2005; pp. 1–14. [Google Scholar]
- Pérez, J.R.; Loureiro, S.; Menezes, S.; Palma, P.; Fernandes, R.M.; Barbosa, I.R.; Soares, A.M.V.M. Assessment of Water Quality in the Alqueva Reservoir (Portugal) Using Bioassays. Environ. Sci. Pollut. Res. 2010, 17, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Empresa de Desenvolvimento e Infra-estruturas do Alqueva (EDIA) Massas de Água de Alqueva. 2017. Available online: https://www.edia.pt/wp-content/uploads/2019/05/PublicacaoMassasAgua_07032018.pdf (accessed on 15 May 2020).
- Presidência do Conselho de Ministros. Resolução Do Conselho de Ministros n.o 94/2006: Regulamento Do Plano de Ordenamento Das Albufeiras Do Alqueva e Pedrógão; Presidência do Conselho de Ministros: Lisboa, Portugal, 2006. [Google Scholar]
- NF EN ISO 10304-1. Water Quality—Determination of Dissolved Anions by Liquid Chromatography of Ions—Part 1: Determination of Bromide, Chloride, Fluoride, Nitrate, Nitrite, Phosphate and Sulfate; ISO: Geneva, Switzerland, 2007. [Google Scholar]
- Brower, J.E.; Zar, J.H.; von Ende, C.N. Field and Laboratory Methods for General Ecology, 4th ed.; WCB McGraw-Hill: Boston, MA, USA, 1997. [Google Scholar]
- APHA. Standard Methods for the Examination of Water and Wastewater, 17th ed.; APHA: Washington, DC, USA, 1989; ISBN 978-0875532356. [Google Scholar]
- NF EN ISO 25663. Water Quality—Determination of Kjeldahl Nitrogen—Method after Mineralization with Selenium; ISO: Geneva, Switzerland, 1984. [Google Scholar]
- NF EN ISO 17294-2. Water Quality—Application of Inductively Coupled Plasma Mass Spectrometry (ICP-MS)—Part 2: Determination of Selected Elements Including Uranium Isotopes; ISO: Geneva, Switzerland, 2016; p. 31. [Google Scholar]
- NF EN ISO 15923-1. Water Quality—Determination of Selected Parameters by Discrete Analysis Systems—Part 1: Ammonium, Nitrate, Nitrite, Chloride, Orthophosphate, Sulfate and Silicate with Photometric Detection; ISO: Geneva, Switzerland, 2013; p. 25. [Google Scholar]
- Agência Portuguesa do Ambiente. Plano de Gestão de Região Hidrográfica—Parte 2—Caracterização e Diagnóstico—Anexos—Região Hidrográfica Do Vouga, Mondego E Lis (Rh4); Agência Portuguesa do Ambiente: Lisboa, Portugal, 2016. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Cabecinha, E.; Cortes, R.; Alexandre Cabral, J.; Ferreira, T.; Lourenço, M.; Pardal, M. Multi-Scale Approach Using Phytoplankton as a First Step towards the Definition of the Ecological Status of Reservoirs. Ecol. Indic. 2009, 9, 240–255. [Google Scholar] [CrossRef]
- Instituto da Água. Modelação Matemática Da Qualidade Da Água Em Albufeiras Com Planos de Ordenamento-Lll-Albufeira Da Aguieira; Ministério da Agricultura, Mar, Ambiente e Ordenamento do Território; Instituto da Água, I.P.: Lisboa, Portugal, 2011. [Google Scholar]
- Vasconcelos, V.; Morais, J.; Vale, M. Microcystins and Cyanobacteria Trends in a 14 Year Monitoring of a Temperate Eutrophic Reservoir (Aguieira, Portugal). J. Environ. Monit. 2011, 13, 668–672. [Google Scholar] [CrossRef]
- Silva, H.; Morais, M.; Rosado, J.; Serafim, A.; Pedro, A.; Sarmento, P.; Fialho, A. South Portugal Reservoirs–Status and Major Concerns. In Proceedings of the 12th International Specialized Conference on Watershed and River Basin Management, Recife, Brazil, 14–16 September 2011. [Google Scholar]
- Pinto, I.; Rodrigues, S.; Lage, O.M.; Antunes, S.C. Assessment of Water Quality in Aguieira Reservoir: Ecotoxicological Tools in Addition to the Water Framework Directive. Ecotoxicol Environ. Saf. 2021, 208, 111583. [Google Scholar] [CrossRef]
- Agência Portuguesa do Ambiente. Plano de Ordenamento Da Albufeira Da Aguieira; Agência Portuguesa do Ambiente: Lisboa, Portugal, 2005. [Google Scholar]
- SNIRH. Sistema Nacional de Informação de Recursos Hídricos SNIRH: Sistema Nacional de Informação de Recursos Hídricos. Available online: https://snirh.apambiente.pt/index.php?idMain= (accessed on 24 September 2019).
- Luo, X.; Xiang, X.; Huang, G.; Song, X.; Wang, P.; Fu, K. Bacterial Abundance and Physicochemical Characteristics of Water and Sediment Associated with Hydroelectric Dam on the Lancang River China. Int. J. Environ. Res. Public Health 2019, 16, 2031. [Google Scholar] [CrossRef]
- Tamaki, H.; Sekiguchi, Y.; Hanada, S.; Nakamura, K.; Nomura, N.; Matsumura, M.; Kamagata, Y. Comparative Analysis of Bacterial Diversity in Freshwater Sediment of a Shallow Eutrophic Lake by Molecular and Improved Cultivation-Based Techniques. Appl. Environ. Microbiol. 2005, 71, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A Guide to the Natural History of Freshwater Lake Bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed]
- Eiler, A.; Bertilsson, S. Composition of Freshwater Bacterial Communities Associated with Cyanobacterial Blooms in Four Swedish Lakes. Environ. Microbiol. 2004, 6, 1228–1243. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.L.; Hahn, M.W. High Predictability of the Seasonal Dynamics of a Species-like Polynucleobacter Population in a Freshwater Lake. Environ. Microbiol. 2006, 8, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Lymperopoulou, D.S.; Kormas, K.A.; Karagouni, A.D. Variability of Prokaryotic Community Structure in a Drinking Water Reservoir (Marathonas, Greece). Microbes Environ. 2011, 27, 1–8. [Google Scholar] [CrossRef]
- Kang, I.; Kim, S.; Islam, M.R.; Cho, J.C. The First Complete Genome Sequences of the AcI Lineage, the Most Abundant Freshwater Actinobacteria, Obtained by Whole-Genome-Amplification of Dilution-to-Extinction Cultures. Sci. Rep. 2017, 7, 42252. [Google Scholar] [CrossRef]
- Neuenschwander, S.M.; Ghai, R.; Pernthaler, J.; Salcher, M.M. Microdiversification in Genome-Streamlined Ubiquitous Freshwater Actinobacteria. ISME J. 2018, 12, 185–198. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Helmus, M.R.; McMahon, K.D. Phylogenetic Ecology of the Freshwater Actinobacteria AcI Lineage. Appl. Environ. Microbiol. 2007, 73, 7169–7176. [Google Scholar] [CrossRef]
- Burkert, U.; Warnecke, F.; Babenzien, D.; Zwirnmann, E.; Pernthaler, J. Members of a Readily Enriched β-Proteobacterial Clade Are Common in Surface Waters of a Humic Lake. Appl. Environ. Microbiol. 2003, 69, 6550–6559. [Google Scholar] [CrossRef] [PubMed]
- van der Gucht, K.; Vandekerckhove, T.; Vloemans, N.; Cousin, S.; Muylaert, K.; Sabbe, K.; Gillis, M.; Declerk, S.; de Meester, L.; Vyverman, W. Characterization of Bacterial Communities in Four Freshwater Lakes Differing in Nutrient Load and Food Web Structure. FEMS Microbiol. Ecol. 2005, 53, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Henson, M.W.; Lanclos, V.C.; Brant, F.C.; Thrash, J. Cameron Cultivation and Genomics of the First Freshwater SAR11 (LD12) Isolate. ISME J. 2018, 12, 1846–1860. [Google Scholar] [CrossRef] [PubMed]
- Tabraiz, S.; Shamurad, B.; Petropoulos, E.; Quintela-Baluja, M.; Charlton, A.; Dolfing, J.; Sallis, P.J. Mitigation of Membrane Biofouling in Membrane Bioreactor Treating Sewage by Novel Quorum Quenching Strain of Acinetobacter Originating from a Full-Scale Membrane Bioreactor. Bioresour. Technol. 2021, 334, 125242. [Google Scholar] [CrossRef] [PubMed]
- Agência Portuguesa do Ambiente. Plano de Gestão de Região Hidrográfica, Parte 5-Objetivos, Anexo II.1, Região Hidrográfica Do Douro (RH3); Agência Portuguesa do Ambiente: Lisboa, Portugal, 2016. [Google Scholar]
- Agência Portuguesa do Ambiente. Plano de Gestão de Região Hidrográfica, Parte 5-Objetivos, Anexo II.2, Região Hidrográfica Do Douro (RH3); Agência Portuguesa do Ambiente: Lisboa, Portugal, 2016. [Google Scholar]
- Huang, W.H.; Asce, S.M.; Kao, C.M.; Asce, F. Bioremediation of Petroleum-Hydrocarbon Contaminated Groundwater under Sulfate-Reducing Conditions: Effectiveness and Mechanism Study. J. Environ. Eng. 2015. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Liang, S.H.; Asce, M.; Wang, S.Y.; Chen, S.H.; Kao, C.M.; Asce, F. Application of Emulsified Substrate Biobarrier to Remediate TCE-Contaminated Groundwater: Pilot-Scale Study. J. Hazard. Toxic Radioact. Waste 2014, 18. [Google Scholar] [CrossRef]
- Ministério do Equipamento, do Planeamento e da Administração do Território. Decreto-Lei No 344-A/98—Regulamento Da via Navegável Do Douro; Diário da República: Lisboa, Portugal, 1998; pp. 5964-(2)–5964-(32). [Google Scholar]
- Gurung, T.B.; Urabe, J. Temporal and Vertical Difference in Factors Limiting Growth Rate of Heterotrophic Bacteria in Lake Biwa. Microb. Ecol. 1999. [Google Scholar] [CrossRef]
- Bird, D.F.; Kalff, J. Empirical Relationships between Bacterial Abundance and Chlorophyll Concentration in Fresh and Marine Waters. Can. J. Fish. Aquat. Sci. 1984, 41, 1015–1023. [Google Scholar] [CrossRef]
- Agência Portuguesa do Ambiente. Plano de Gestão de Região Hidrográfica, Parte 5-Objetivos, Anexo II.1, Região Hidrográfica Do Vouga, Mongedo e Lis (RH4); Agência Portuguesa do Ambiente: Lisboa, Portugal, 2016. [Google Scholar]
- Daims, H.; Wagner, M. Nitrospira. Trends Microbiol. 2018, 26, 462–463. [Google Scholar] [CrossRef]
- Ushiki, N.; Fujitani, H.; Shimada, Y.; Morohoshi, T.; Sekiguchi, Y.; Tsuneda, S. Genomic Analysis of Two Phylogenetically Distinct Nitrospira Species Reveals Their Genomic Plasticity and Functional Diversity. Front. Microbiol. 2018, 8, 2637. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Dunlap, P.V.; Clark, D.P. Brock Biology of Microorganisms, 12th ed.; Benjamin Cummings: San Francisco, CA, USA, 2008; ISBN 978-0132324601. [Google Scholar]
- Graça, M.A.; Pardal, M.Â.; Marques, J.C.; Vasconcelos, V. Toxic cyanobacteria in the Mondego basin reservoirs: An overiew. In Aquatic ecology of the Mondego River Basin Global Importance of Local Experience; Imprensa da Universidade de Coimbra: Coimbra, Portugal, 2002; pp. 105–114. [Google Scholar]
- Sotero-Santos, R.B.; Silva, C.R.D.S.E.; Verani, N.F.; Nonaka, K.O.; Rocha, O. Toxicity of a Cyanobacteria Bloom in Barra Bonita Reservoir (Middle Tietê River, São Paulo, Brazil). Ecotoxicol. Environ. Saf. 2006, 64, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Gannon, V.P.J.; Duke, G.D.; Thomas, J.E.; VanLeeuwen, J.; Byrne, J.; Johnson, D.; Kienzle, S.W.; Little, J.; Graham, T.; Selinger, B. Use of In-Stream Reservoirs to Reduce Bacterial Contamination of Rural Watersheds. Sci. Total Environ. 2005, 348, 19–31. [Google Scholar] [CrossRef] [PubMed]
- He, L.M.; Lu, J.; Shi, W. Variability of Fecal Indicator Bacteria in Flowing and Ponded Waters in Southern California: Implications for Bacterial TMDL Development and Implementation. Water Res. 2007, 41, 3132–3140. [Google Scholar] [CrossRef] [PubMed]
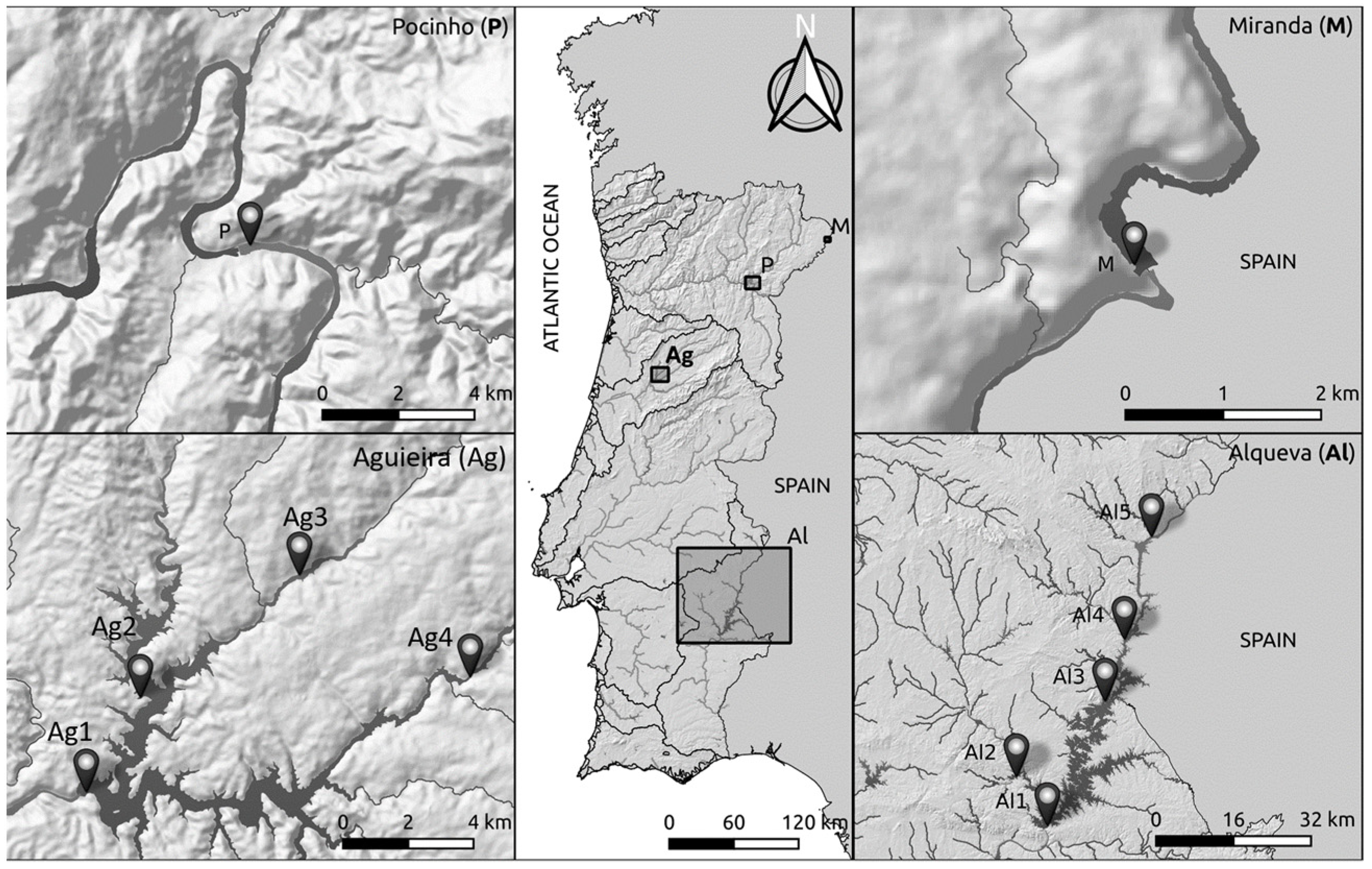

| pH | Cond (μS/cm) | Temp (°C) | O2 (mg/L) | O2 (%) | Turb (m−1) | TSS (mg/L) | NO3− (mg/L) | Ntotal (mg/L) | Ptotal (mg/L) | NH4+ (mg/L) | Chl a (µg/L) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Environmental Quality Standards (EQS) (1) | 6–9 | ≥5 | 60–120 (north *) 60–140 (south **) | ≤25 | ≤0.05 (north *) ≤0.07 (south **) | ≤7.90 (north) ≤9.50 (south and main course) | ||||||||
| Miranda * (main course) | Aut18 | M | 8.1 | 403 | 9.4 | 9.8 | 89.4 | 0.025 | 8.08 | 6.9 | <0.6 | 0.10 | <0.05 | 4.65 |
| Spr19 | M | 8.8 | 447 | 15.6 | 14.1 | 149.7 | 0.069 | 12.75 | 6.4 | <0.5 | 0.03 | <0.05 | 43.45 | |
| Aut19 | M | 7.9 | 341 | 18.3 | 4.4 | 50.0 | 0.016 | 8.82 | 2.3 | <0.5 | 0.01 | 0.06 | 2.34 | |
| Spr20 | M | 8.6 | 438 | 19.0 | 11.0 | 124.1 | 0.038 | 15.65 | 7.4 | <0.5 | 0.13 | <0.05 | 29.63 | |
| Pocinho * (main course) | Aut18 | P | 7.9 | 330 | 12.2 | 9.1 | 84.4 | 0.014 | 8.42 | 3.9 | <0.6 | 0.08 | <0.05 | 0.45 |
| Spr19 | P | 8.8 | 316 | 16.5 | 14.0 | 144.0 | 0.016 | 8.75 | <0.5 | <0.5 | 0.03 | <0.05 | 18.29 | |
| Aut19 | P | 8.0 | 306 | 19.2 | 8.2 | 90.3 | 0.000 | 7.38 | 2.3 | <0.5 | 0.04 | <0.05 | 6.74 | |
| Spr20 | P | 9.2 | 268 | 22.8 | 15.9 | 185.0 | 0.054 | 10.12 | 3.5 | 0.7 | 0.09 | 0.05 | 38.73 | |
| Aguieira (north) | Aut18 | Ag1 | 8.4 | 86 | 24.5 | 8.8 | 106.3 | 0.018 | 8.24 | 1.3 | <0.5 | <0.01 | <0.05 | 5.43 |
| Ag2 | 7.6 | 97 | 23.2 | 8.0 | 94.4 | 0.036 | 10.09 | <0.5 | <0.5 | 0.03 | <0.05 | 10.10 | ||
| Ag3 | 8.1 | 143 | 21.0 | 9.9 | 112.0 | 0.115 | 312.50 | <0.5 | 6.3 | 0.22 | <0.05 | 1082.23 | ||
| Ag4 | 7.4 | 87 | 24.5 | 7.4 | 89.0 | 0.038 | 10.75 | 2.4 | <0.5 | 0.03 | <0.05 | 3.44 | ||
| Spr19 | Ag1 | 9.2 | 83 | 14.4 | 11.9 | 119.4 | 0.072 | 13.45 | 2.8 | <0.5 | 0.01 | 0.07 | 26.32 | |
| Ag2 | 9.0 | 89 | 15.0 | 12.4 | 124.9 | 0.069 | 19.05 | 3.3 | <0.5 | 0.01 | <0.05 | 30.62 | ||
| Ag3 | 8.3 | 112 | 15.2 | 11.3 | 112.1 | 0.087 | 11.08 | 4.0 | <0.5 | 0.09 | 0.06 | 10.17 | ||
| Ag4 | 9.2 | 78 | 15.5 | 12.2 | 125.2 | 0.074 | 17.50 | 1.2 | 0.7 | 0.02 | 0.09 | 27.90 | ||
| Aut19 | Ag1 | 6.8 | 91 | 17.7 | 4.5 | 47.1 | 0.023 | 7.82 | 1.5 | <0.5 | <0.01 | <0.05 | 1.61 | |
| Ag2 | 6.7 | 92 | 17.9 | 5.3 | 55.9 | 0.035 | 10.75 | 1.2 | <0.5 | <0.01 | <0.05 | 2.59 | ||
| Ag3 | 6.7 | 101 | 16.3 | 9.0 | 91.8 | 0.046 | 12.58 | 2.3 | 2.2 | 0.09 | <0.05 | 10.96 | ||
| Ag4 | 6.8 | 88 | 17.3 | 6.9 | 72.3 | 0.016 | 5.55 | 1.0 | 0.6 | <0.01 | <0.05 | 5.30 | ||
| Spr20 | Ag1 | 9.6 | 74 | 21.9 | 12.9 | 150.2 | 0.008 | 15.19 | 2.7 | <0.5 | 0.02 | 0.07 | 26.16 | |
| Ag2 | 9.7 | 85 | 20.5 | 14.2 | 160.1 | 0.008 | 17.54 | 2.2 | 0.7 | 0.03 | 0.07 | 42.08 | ||
| Ag3 | 9.0 | 90 | 20.7 | 12.4 | 141.0 | 0.015 | 16.62 | 3.3 | <0.5 | 0.08 | 0.18 | 19.03 | ||
| Ag4 | 9.4 | 73 | 22.4 | 13.3 | 156.5 | 0.008 | 11.23 | 0.6 | <0.5 | 0.03 | 0.10 | 31.75 | ||
| Alqueva ** (main course) | Aut18 | Al1 | 7.9 | 501 | 16.6 | 6.6 | 68.8 | 0.002 | 4.28 | 4.5 | <1.0 | 0.06 | <0.05 | 0.98 |
| Al2 | 8.0 | 491 | 17.2 | 7.4 | 78.0 | 0.000 | 3.83 | 0.5 | <1.0 | 0.03 | <0.05 | 3.60 | ||
| Al3 | 8.1 | 515 | 16.8 | 7.0 | 73.2 | 0.023 | 4.72 | <0.5 | <1.1 | 0.07 | 0.17 | 1.81 | ||
| Al4 | 8.0 | 541 | 17.6 | 6.6 | 69.6 | 0.014 | 3.61 | 0.8 | <1.0 | 0.08 | 0.20 | 2.18 | ||
| Al5 | 8.4 | 692 | 16.6 | 11.6 | 120.9 | 0.044 | 15.42 | 5.6 | 1.3 | 0.16 | <0.05 | 31.26 | ||
| Spr19 | Al1 | 8.5 | 517 | 23.0 | 9.6 | 114.4 | 0.009 | 7.75 | <0.5 | 0.6 | 0.01 | 0.05 | 2.28 | |
| Al2 | 8.7 | 515 | 23.7 | 9.4 | 112.8 | 0.012 | 7.42 | <0.5 | 0.6 | <0.01 | <0.05 | 0.94 | ||
| Al3 | 8.8 | 538 | 23.1 | 10.0 | 118.2 | 0.002 | 7.42 | 0.6 | 0.6 | 0.01 | 0.11 | 2.49 | ||
| Al4 | 8.5 | 570 | 23.8 | 12.7 | 152.6 | 0.016 | 12.75 | 0.7 | 0.7 | 0.01 | 0.08 | 7.83 | ||
| Al5 | 9.1 | 714 | 23.0 | 16.9 | 199.5 | 0.074 | 20.42 | 0.9 | 2.1 | 0.09 | 0.58 | 56.65 | ||
| Aut19 | Al1 | 8.2 | 525 | 16.6 | 8.1 | 83.7 | 0.023 | 10.02 | <0.5 | <0.5 | 0.07 | <0.05 | 3.53 | |
| Al2 | 8.3 | 521 | 16.9 | 8.0 | 83.3 | 0.021 | 24.72 | <0.5 | 0.5 | 0.05 | <0.05 | 8.28 | ||
| Al3 | 8.3 | 545 | 16.9 | 8.5 | 88.8 | 0.018 | 7.88 | <0.5 | 0.7 | 0.04 | 0.19 | 2.75 | ||
| Al4 | 8.3 | 578 | 16.9 | 7.5 | 77.6 | 0.021 | 8.78 | 0.5 | 1.0 | 0.05 | 0.28 | 2.81 | ||
| Al5 | 8.4 | 769 | 14.6 | 11.0 | 108.6 | 0.092 | 29.97 | 1.7 | 1.6 | 0.07 | 0.81 | 40.10 | ||
| Spr20 | Al1 | 8.8 | 540 | 32.0 | 8.3 | 114.0 | 0.008 | 6.25 | <0.5 | <0.5 | 0.04 | <0.05 | 2.20 | |
| Al2 | 8.9 | 506 | 33.3 | 8.5 | 119.1 | 0.003 | 7.08 | <0.5 | <0.5 | 0.05 | <0.05 | 2.28 | ||
| Al3 | 9.0 | 558 | 31.7 | 8.4 | 115.2 | 0.008 | 6.32 | 0.8 | 0.6 | 0.03 | <0.05 | 3.68 | ||
| Al4 | 9.2 | 509 | 32.0 | 10.0 | 133.6 | 0.038 | 10.02 | <0.5 | 0.8 | 0.06 | <0.05 | 20.78 | ||
| Al5 | 8.6 | 588 | 32.0 | 7.0 | 97.0 | 0.082 | 28.50 | <0.5 | 1.0 | 0.18 | 0.16 | 45.42 | ||
| Total Abundance (OTUs) | Diversity Shannon-Wiener Index a | Richness Margalef Index b | |||
|---|---|---|---|---|---|
| Miranda | Aut18 | M | 18 | 2.86 | 1.04 |
| Spr19 | M | 15 | 2.67 | 0.87 | |
| Aut19 | M | 20 | 2.95 | 1.16 | |
| Spr20 | M | 17 | 2.81 | 0.99 | |
| Pocinho | Aut18 | P | 20 | 2.96 | 1.15 |
| Spr19 | P | 14 | 2.57 | 0.80 | |
| Aut19 | P | 17 | 2.78 | 0.98 | |
| Spr20 | P | 20 | 2.97 | 1.16 | |
| Aguieira | Aut18 | Ag1 | 27 | 3.26 | 1.56 |
| Ag2 | 27 | 3.26 | 1.56 | ||
| Ag3 | 14 | 2.59 | 0.81 | ||
| Ag4 | 16 | 2.74 | 0.92 | ||
| Spr19 | Ag1 | 13 | 2.55 | 0.75 | |
| Ag2 | 14 | 2.61 | 0.81 | ||
| Ag3 | 11 | 2.36 | 0.63 | ||
| Ag4 | 17 | 2.75 | 0.99 | ||
| Aut19 | Ag1 | 17 | 2.81 | 0.99 | |
| Ag2 | 16 | 2.76 | 0.93 | ||
| Ag3 | 16 | 2.76 | 0.92 | ||
| Ag4 | 17 | 2.80 | 0.98 | ||
| Spr20 | Ag1 | 16 | 2.72 | 0.92 | |
| Ag2 | 17 | 2.79 | 0.99 | ||
| Ag3 | 19 | 2.90 | 1.09 | ||
| Ag4 | 19 | 2.90 | 1.09 | ||
| Alqueva | Aut18 | Al1 | 17 | 2.74 | 1.03 |
| Al2 | 9 | 1.93 | 0.55 | ||
| Al3 | 7 | 1.92 | 0.41 | ||
| Al4 | 11 | 2.36 | 0.66 | ||
| Al5 | 5 | 1.37 | 0.28 | ||
| Spr19 | Al1 | 9 | 1.93 | 0.55 | |
| Al2 | 14 | 2.59 | 0.86 | ||
| Al3 | 6 | 1.73 | 0.35 | ||
| Al4 | 6 | 1.75 | 0.34 | ||
| Al5 | 5 | 1.35 | 0.29 | ||
| Aut19 | Al1 | 11 | 2.26 | 0.67 | |
| Al2 | 11 | 2.17 | 0.68 | ||
| Al3 | 8 | 2.04 | 0.48 | ||
| Al4 | 8 | 1.92 | 0.49 | ||
| Al5 | 7 | 1.91 | 0.41 | ||
| Spr20 | Al1 | 8 | 2.03 | 0.48 | |
| Al2 | 11 | 2.15 | 0.68 | ||
| Al3 | 5 | 1.57 | 0.29 | ||
| Al4 | 5 | 1.55 | 0.28 | ||
| Al5 | 4 | 1.31 | 0.21 |
| Aguieira | Alqueva | |||||
|---|---|---|---|---|---|---|
| Total Abundance (OTUs) | Diversity Shannon-Wiener Index | Richness Margalef Index | Total Abundance (OTUs) | Diversity Shannon-Wiener Index | Richness Margalef Index | |
| pH | - | - | - | −0.49 | −0.51 | −0.48 |
| Cond | - | - | - | −0.48 | −0.50 | −0.48 |
| Temp | 0.66 | 0.66 | 0.65 | - | - | - |
| O2 | - | - | - | −0.44 | −0.51 | −0.45 |
| Turb | −0.50 | −0.59 | −0.50 | −0.50 | −0.54 | −0.51 |
| NO3− | −0.52 | −0.54 | −0.52 | - | - | - |
| Ntotal | - | - | - | −0.56 | −0.63 | −0.56 |
| Ptotal | - | - | - | - | −0.46 | - |
| Chl a | - | - | - | −0.55 | −0.66 | −0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, I.; Calisto, R.; Serra, C.R.; Lage, O.M.; Antunes, S.C. Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment. Water 2021, 13, 2836. https://doi.org/10.3390/w13202836
Pinto I, Calisto R, Serra CR, Lage OM, Antunes SC. Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment. Water. 2021; 13(20):2836. https://doi.org/10.3390/w13202836
Chicago/Turabian StylePinto, Ivo, Rita Calisto, Cláudia R. Serra, Olga M. Lage, and Sara C. Antunes. 2021. "Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment" Water 13, no. 20: 2836. https://doi.org/10.3390/w13202836
APA StylePinto, I., Calisto, R., Serra, C. R., Lage, O. M., & Antunes, S. C. (2021). Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment. Water, 13(20), 2836. https://doi.org/10.3390/w13202836








