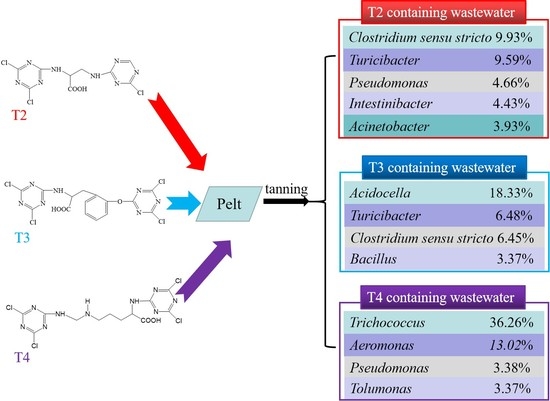
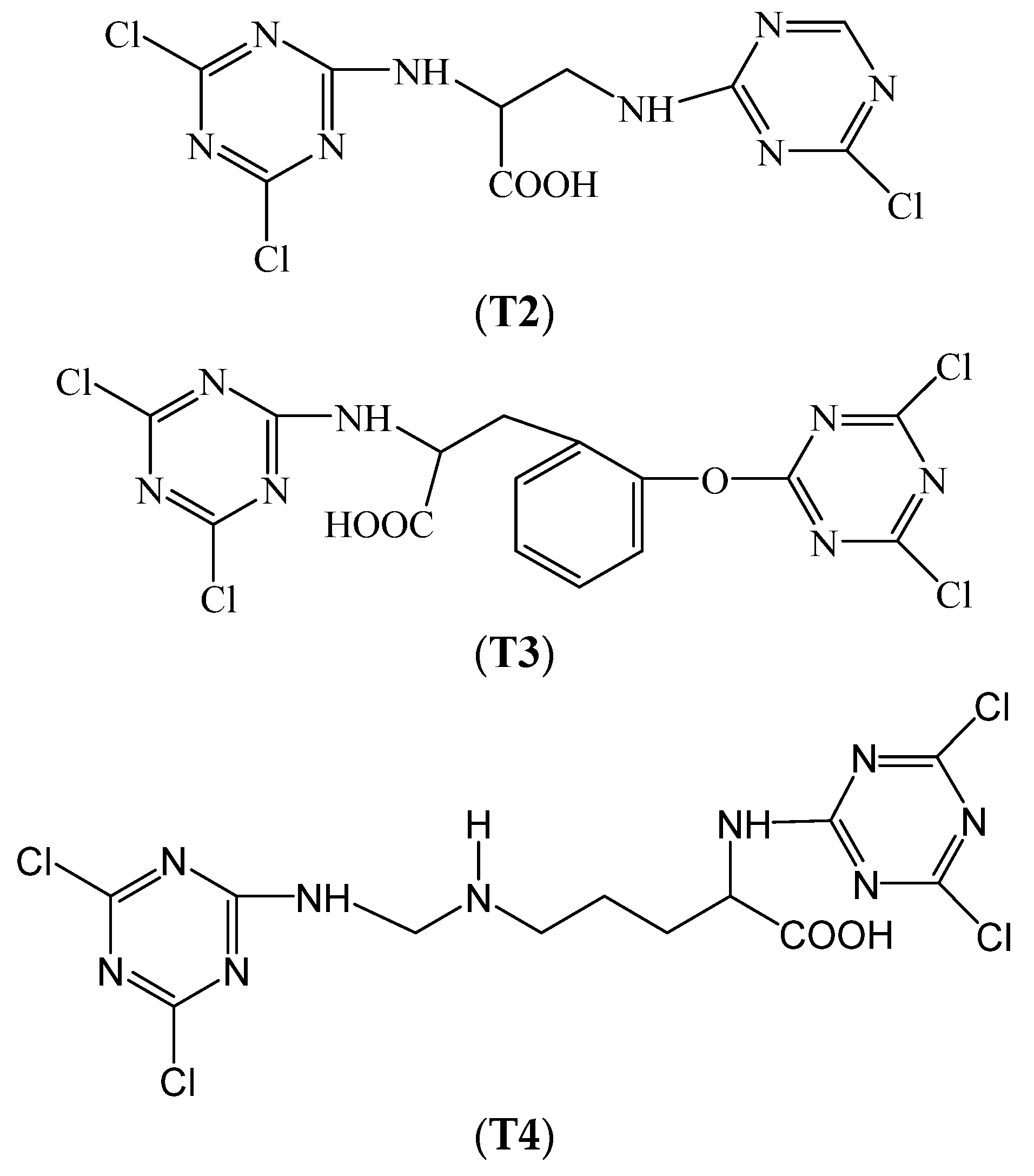
Impact of Non-Metallic Organic Tanning Agents with a Double-Triazine Structure on the Microbial Community Structure in Wastewater
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Analysis of Physicochemical Properties of Bistriazine Ring Derivative Wastewater
2.3. Extraction of DNA, Library Preparation, and Sequencing
2.3.1. Extraction of DNA
2.3.2. PCR Amplification
2.3.3. Illumina Miseq Sequencing
2.4. Analysis of Alpha and Beta Diversity
2.5. Data Processing
2.6. Canonical Correspondence Analysis (CCA) Method
3. Discussion and Results
3.1. Physicochemical Properties of Bistriazine Ring Derivative Wastewater after Degradation
3.2. Analysis of the Structure of the Bacterial Community in the Bistriazine Ring Derivative Wastewater
3.2.1. The Alpha Diversity Analysis of Bacteria
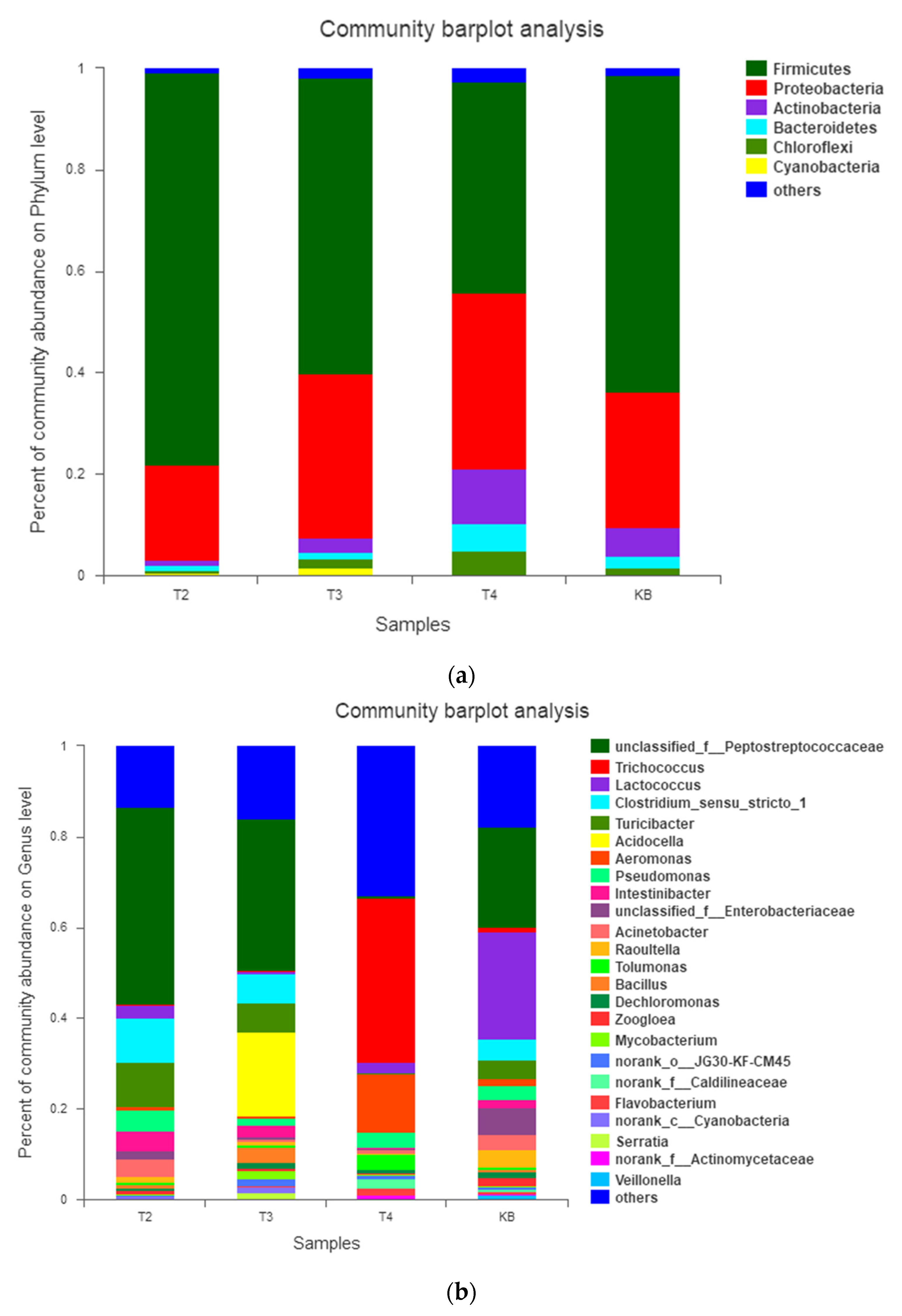
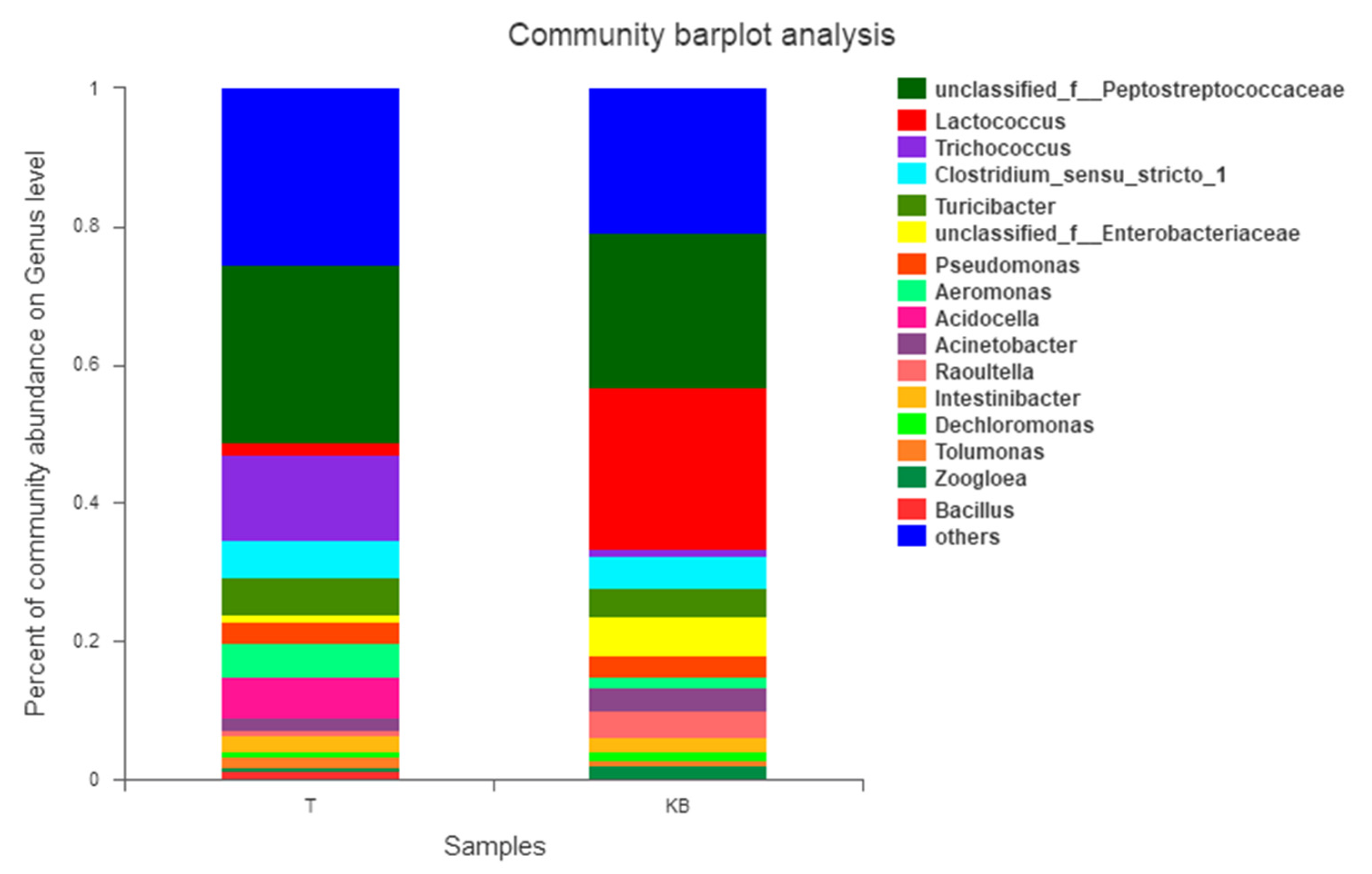
3.2.2. Effect of Derivatives Containing Different Bistriazine Rings on the Bacterial Microbial Community in the Wastewater
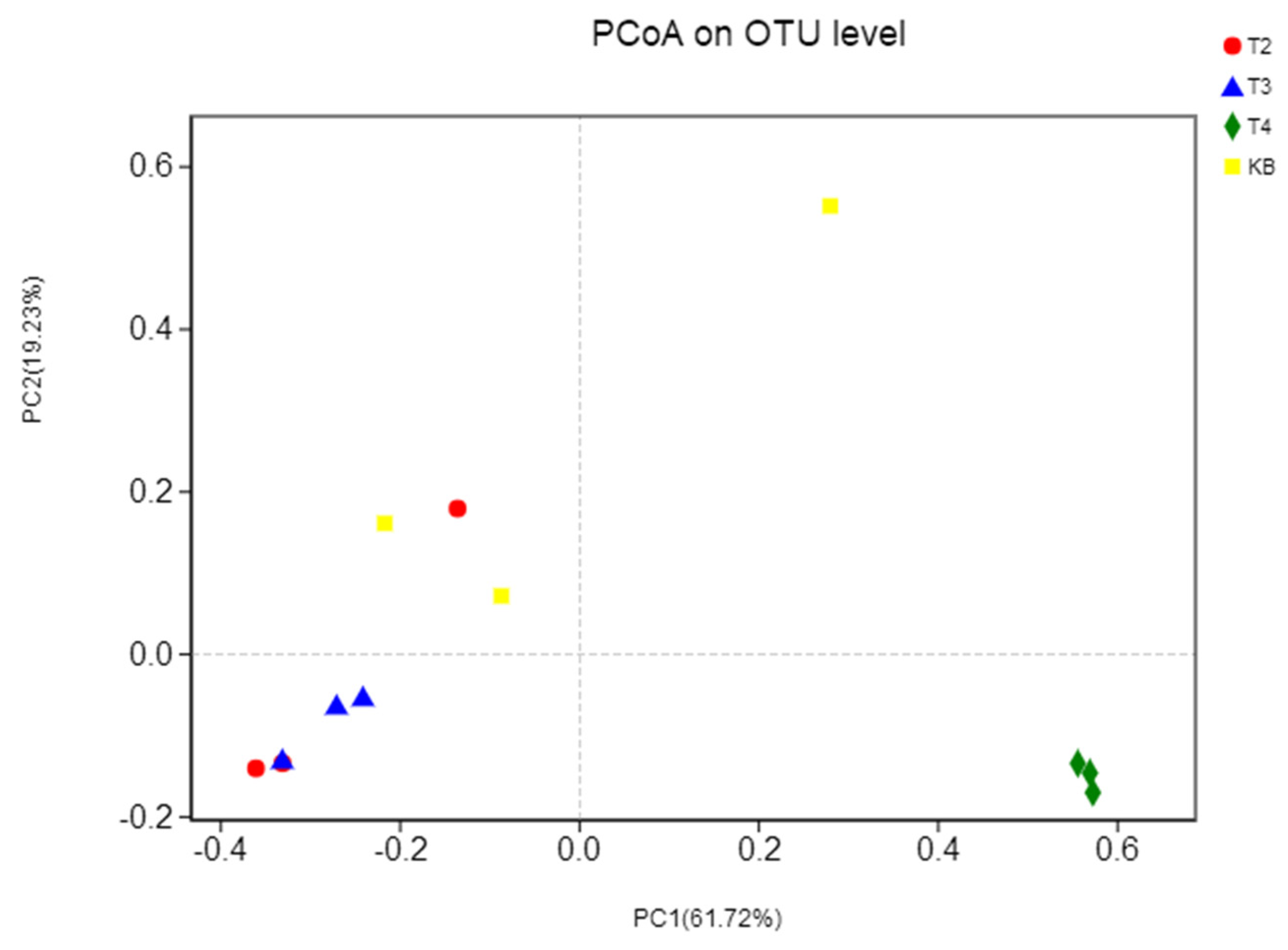
3.3. β-Diversity Analysis
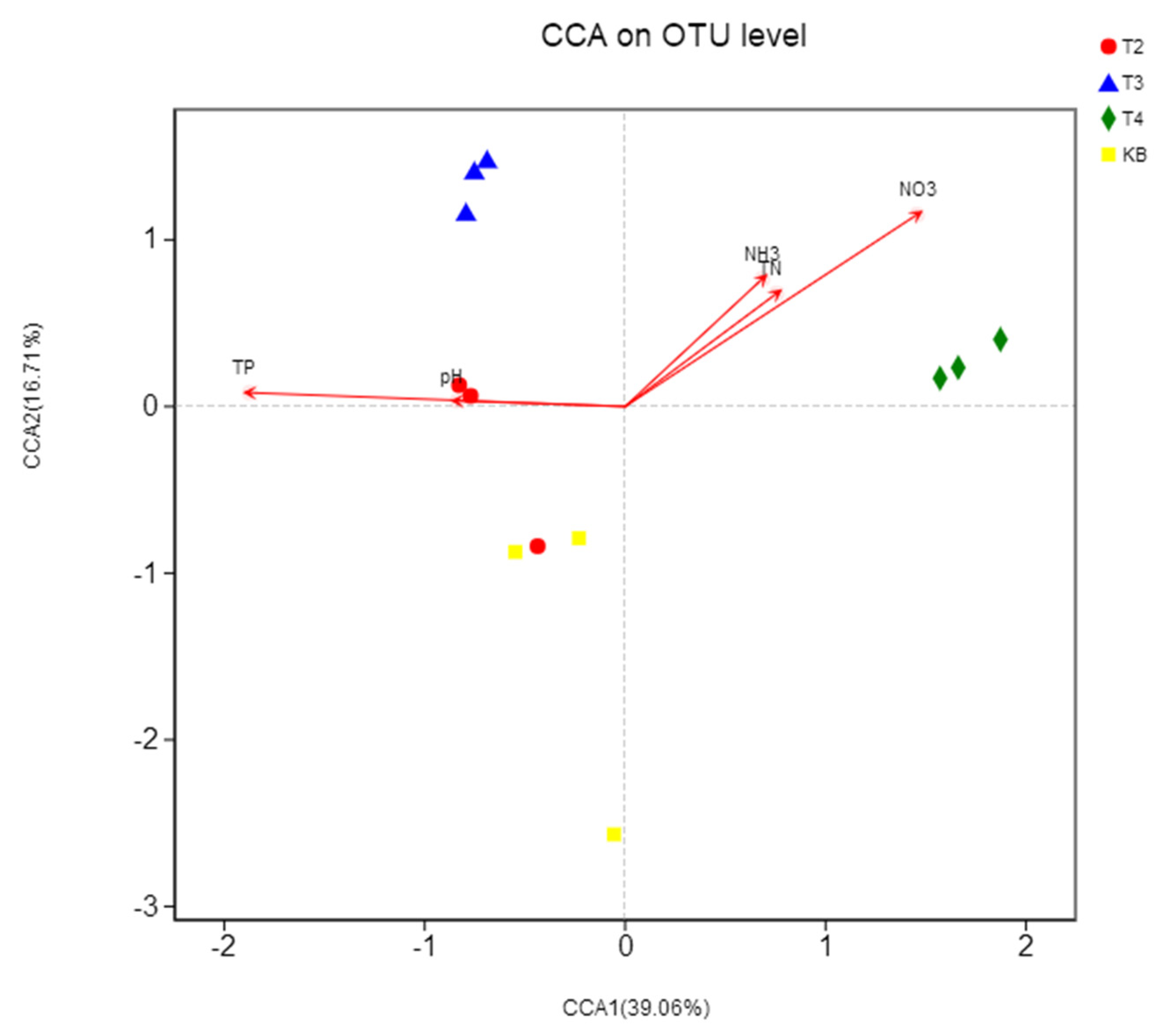
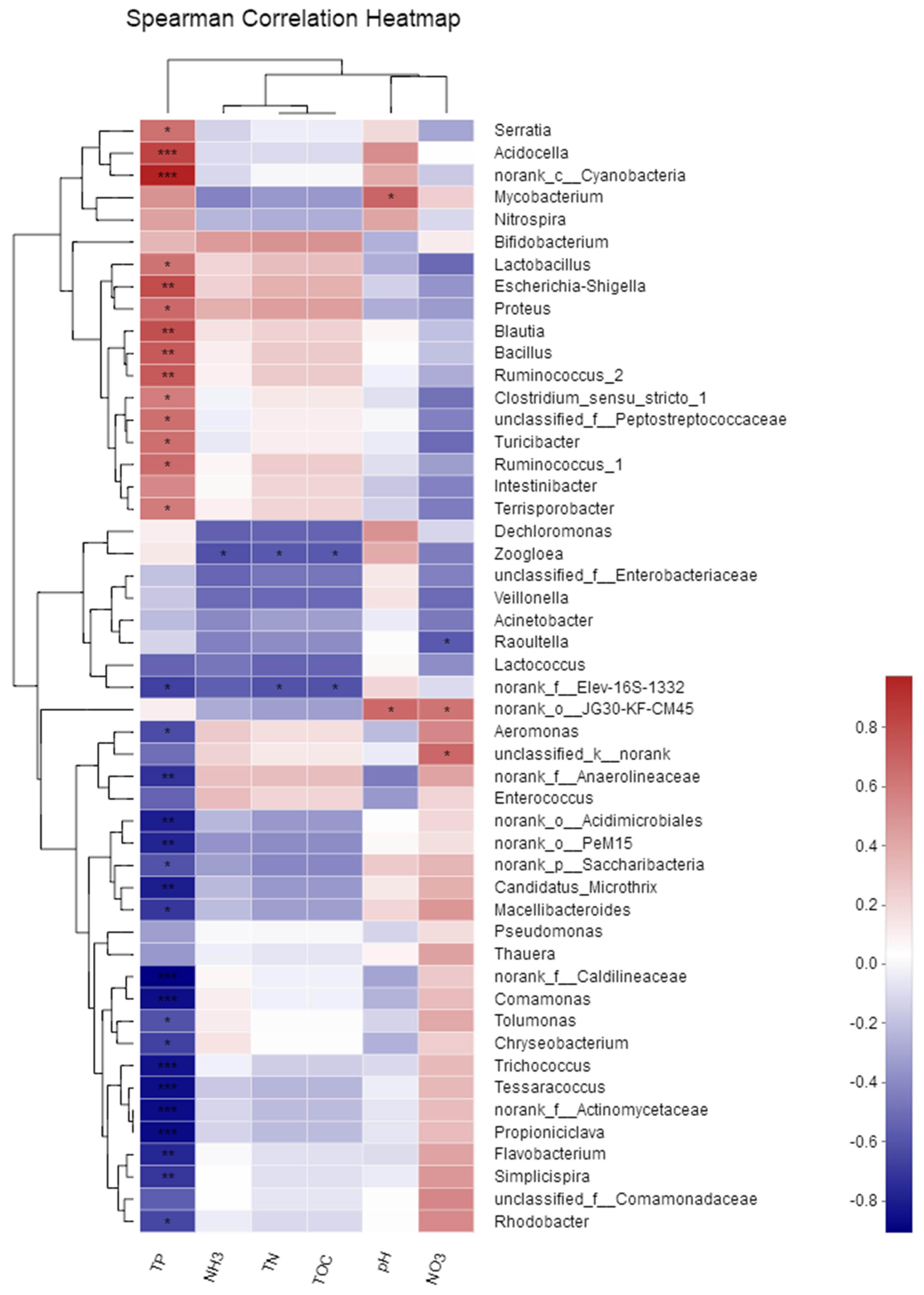
3.4. Correlation between Bacterial Community Composition and Physicochemical Properties of Bistriazine Ring Derivative Wastewater
3.5. Relationship between Bacterial Richness and Physicochemical Properties of Bistriazine Ring Derivative Wastewater
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.T.; Gao, W.W.; Dai, G.C.; Li, Z.J. Research progress of non-chromium tanning agents and chrome-free tanning technology based on silicon. China Leather 2019, 48, 21–27. [Google Scholar]
- Zhou, Y.L.; He, L.R.; Shi, J.B.; Lin, W. Research Progress of Non-metal Tanning Technology. China Leather 2011, 40, 47–51. [Google Scholar]
- Liu, D.; Qiang, X.H.; Hu, Y. Synthesis and Tanning Properties of N, N′-Bis-(4, 6-dichloro-[1,3,5]-triazin-2-yl)-2, 6-diaminohexanic Acid. Fine Chem. 2019, 36, 136–141, 148. [Google Scholar]
- Yu, L.D.; Qiang, X.H.; Ma, H.F.; Liu, D.; Chen, B.; Wang, X.K. Synthesis and Tanning Properties of p-(4,6-Dichloro-1,3,5-triazin-2-yloxy) benzenesulfonic Acid Sodium Salt. Fine Chem. 2019, 36, 1415–1421. [Google Scholar]
- Cupples, A.M. RDX degrading microbial communities and the prediction of microorganisms responsible for RDX bioremediation. Int. Biodeter. Biodegr. 2013, 85, 260–270. [Google Scholar] [CrossRef]
- Jiang, C.; Lu, Y.C.; Xu, J.Y.; Song, Y.; Song, Y.; Zhang, S.H.; Ma, L.Y.; Lu, F.F.; Wang, Y.K.; Yang, H. Activity, biomass and composition of microbial communities and their degradation pathways in exposed propazine soil. Ecotox. Environ. Saf. 2017, 145, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jin, R.; Liu, G.; Tian, T.; Zhou, J. Effects of hexavalent chromium on performance, extracellular polymeric substances and microbial community structure of anaerobic activated sludge in a sequencing batch reactor. J. Chem. Technol. Biotechnol. 2017, 92, 2719–2730. [Google Scholar] [CrossRef]
- Moga, I.C.; Ardelean, I.; Petrescu, G.; Crǎciun, N.; Popa, R. The potential of biofilms from moving bed bioreactors to increase the efficiency of textile industry wastewater treatment. Ind. Text. 2018, 69, 412–418. [Google Scholar]
- Stan-Lotter, H.; Fendrihan, S. Adaption of Microbial Life to Environmental Extremes: Novel Research Results and Application, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–342. [Google Scholar]
- Gao, P.P.; Mao, D.Q.; Luo, Y.; Wang, L.M.; Xu, B.J.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.C.; Wang, H.Y. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Xu, H.Z.; Pei, H.Y.; Jin, Y. High-throughput sequencing reveals microbial communities in drinking water treatment sludge from six geographically distributed plants, including potentially toxic cyanobacteria and pathogens. Sci. Total Environ. 2018, 634, 769–779. [Google Scholar] [CrossRef]
- Schöler, A.; Jacquiod, S.; Vestergaard, G.; Schulz, S.; Schloter, M. Analysis of soil microbial communities based on amplicon sequencing of marker genes. Biol. Fert. Soils 2017, 53, 485–489. [Google Scholar] [CrossRef]
- Tao, K.; Liu, Y.X.; Ke, T.; Zhang, Y.R.; Xiao, L.; Li, S.X.; Wei, S.J.; Chen, L.Z.; Hu, T.S. Patterns of bacterial and archaeal communities in sediments in response to dam construction and sewage discharge in Lhasa River. Ecotox. Environ. Saf. 2019, 178, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.S. Water and Wastewater Monitoring and Analysis Methods, 4th ed.; China Environment Science Press: Beijing, China, 2002. [Google Scholar]
- Li, C.; Pan, G.; Wang, X.; Qiang, X.; Qiang, T. The effects of non-metallic organic tanning agents on the microbial community structure in wastewater. J. Clean. Prod. 2020, 279, 123553. [Google Scholar] [CrossRef]
- Wang, J.; Huang, M.; Wang, Q.; Sun, Y.; Zhao, Y.; Huang, Y. LDPE microplastics significantly alter the temporal turnover of soil microbial communities. Sci. Total. Environ. 2020, 726, 138682. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.L.; McGarrell, D.M.; Sun, Y.N.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ter Braak, C.J.F. The analysis of vegetation-environment relationships by canonical correspondence analysis. Vegetatio 1987, 69, 69–77. [Google Scholar] [CrossRef]
- Jogman, R.H.G.; ter Braak, C.J.F.; van Tongeren, O.F.R. (Eds.) Data Analysis in Community and Landscape Ecology; Pudoc: Wageningen, The Netherlands, 1995; p. 321. [Google Scholar]
- Ter Braak, C.J.F. Interpreting canonical correlation analysis through biplots of structural correlations and weights. Psychometrica 1990, 55, 519–531. [Google Scholar] [CrossRef]
- Conley, D.J.; Paerl, H.W.; Howarth, R.W. Controlling Eutrophication: Nitrogen and Phosphorus. Science 2009, 323, 1014–1015. [Google Scholar] [CrossRef]
- Hu, W.F.; Lo, W.; Chua, H.; Sin, S.N.; Yu, P.H. Nutrient release and sediment oxygen demand in a eutrophic land-locked embayment in Hong Kong. Environ. Int. 2001, 26, 369–375. [Google Scholar] [CrossRef]
- Chen, H.H.; Wan, J.J.; Chen, K.F.; Luo, G.; Fan, J.J.; Clark, J.; Zhang, S.C. Biogas production from Hydrothermal Liquefaction Waste Water (HTLWW): Focusing on the microbial communities as revealed by high-throughput sequencing of full-length 16S rRNA genes. Water Res. 2016, 106, 98–107. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhang, X.X.; Huang, K.L.; Miao, Y.; Shi, P.; Liu, B.; Long, C.; Li, A.M. Metagenomic Profiling of Antibiotic Resistance Genes and Mobile Genetic Elements in a Tannery Wastewater Treatment Plant. PLoS ONE 2013, 8, e76079. [Google Scholar] [CrossRef]
- Liu, B.-Y.; Huan, H.-L.; Gu, H.-R.; Xu, N.-X.; Shen, Q.; Ding, C.-L. Dynamics of a microbial community during ensiling and upon aerobic exposure in lactic acid bacteria inoculation-treated and untreated barley silages. Bioresour. Technol. 2019, 273, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Xu, X.-Y.; Zhu, L. Activated sludge bacterial communities of typical wastewater treatment plants: Distinct genera identification and metabolic potential differential analysis. AMB Express 2018, 8, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-H.; Tian, R.; Chen, Z.-C.; Guo, J.; Jia, Y. Effects of NaCl salinity on wastewater pollutants removal and microorganism in A2/O technology process. Trans. Chin. Soc. Agric. Eng. 2018, 34, 231–237. [Google Scholar]
- Zhang, L.-H.; Zhang, J.-C.; Zeng, G.-M.; Dong, H.-R.; Chen, Y.-N.; Huang, C.; Zhu, Y.; Xu, R.; Cheng, Y.-J.; Hou, K.-J.; et al. Multivariate relationships between microbial communities and environmental variables during co-composting of sewage sludge and agricultural waste in the presence of PVP-AgNPs. Bioresour. Technol. 2018, 261, 10–18. [Google Scholar] [CrossRef]
- Yao, Y.; Zhu, J.-R.; Tang, M.; Chen, G.; Huang, J.; Chen, T.; Liao, W. Application of aerobic granular sludge technology on treatment of villages and towns sewage. Res. Environ. Sci. 2018, 31, 379–388. [Google Scholar]
- Jayamani, I. Assessing the Biodegradation of Toluene, Ethylbenzene and RDX and the Identification of the Microorganisms Involved Using Stable Isotope Probing and High Throughput Amplicon Sequencing. Ph.D. Thesis, Michigan State University, East Lansing, MI, USA, 2015. [Google Scholar]
- Livermore, J.A. Microbial Ecology of a Managed Aquifer near the Iowa Army Ammunition Plant (Middletown, IA). Ph.D. Thesis, The University of Lowa, Iowa City, IA, USA, 2011. [Google Scholar]
- Jayakumar, G.C.; Kumar, G.; Tesema, A.F.; Thi NB, D.; Kobayashi, T.; Xu, K. Bioremediation for tanning industry: A future perspective for zero emission. In Management of Hazardous Wastes; IntechOpen: London, UK, 2016. [Google Scholar]
- Kapahi, M.; Sachdeva, S. Bioremediation Options for Heavy Metal Pollution. J. Health Pollut. 2019, 9, 191203. [Google Scholar] [CrossRef] [Green Version]
- Isazadeh, S.; Jauffur, S.; Frigon, D. Bacterial community assembly in activated sludge: Mapping beta diversity across environmental variables. Microbiol. Open 2016, 5, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, T.; Tsushima, I.; Tsumori, J. Comparative analyses of microbial structures and gene copy numbers in the anaerobic digestion of various types of sewage sludge. Bioresour. Technol. 2018, 253, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Lu, D.; Bai, Y.; Hu, W.; Li, M. Influence of effluent from sewage treatment plants on the biofilm of receiving rivers. Coal Chem. Ind. 2018, 41, 145–148. [Google Scholar]
- Shin, S.G.; Koo, T.; Lee, J.; Han, G.; Cho, K.; Kim, W.; Hwang, S. Correlations between bacterial populations and process parameters in four full-scale anaerobic digesters treating sewage sludge. Bioresour. Technol. 2016, 214, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.Y.; Li, Y.; Wu, J.S. Diversity and distribution of bacteria in a multistage surface flow constructed wetland to treat swine wastewater in sediments. Appl. Microbiol. Biot. 2018, 102, 10755–10765. [Google Scholar] [CrossRef]
- Jin, N.B.; Shou, Z.Q.; Yuan, H.P.; Lou, Z.Y.; Zhu, N.W. Effects of ferric nitrate additions under different pH conditions on autothermal thermophilic aerobic digestion for sewage sludge. RSC Adv. 2015, 5, 90127–90134. [Google Scholar] [CrossRef]
- Huang, W.; Liu, L.Y.; Wu, M.H.; Chen, L.H.; Lü, X.; Ye, M.F.; Lin, D.Y.; Song, Y.K. Microbial community structure and dynamics in swine wastewater treatment system. J. Environ. Sci. 2019, 39, 839–848. [Google Scholar]
- Yang, H.; Zhang, G.Z.; Yang, X.N.; Wu, F.P.; Zhao, W.; Zhang, H.W.; Zhang, X. Microbial Community Structure and Diversity in Cellar Water by 16S rRNA High-throughput Sequencing. Environ. Sci. 2017, 38, 1704–1716. [Google Scholar]
- Zhang, H.; Sun, Z.L.; Liu, B.; Xuan, Y.M.; Jiang, M.; Pan, Y.S.; Zhang, Y.M.; Gong, Y.P.; Lu, X.P.; Yu, D.S.; et al. Dynamic changes of microbial communities in Litopenaeus vannamei cultures and the effects of environmental factors. Aquaculture 2016, 455, 97–108. [Google Scholar] [CrossRef]
- Shi, M.Y.; Wen, G.C.; Liu, H.F.; Jian, G.D.; Chen, Y.Q. Influence of initial pH on bioleaching of river sediments to achieve deep dehydration. Environ. Sci. Pollut. R 2019, 26, 17183–17194. [Google Scholar] [CrossRef]
- Carvalho, J.R.S.; Amaral, F.M.; Florencio, L.; Kato, M.T.; Delforno, T.P.; Gavazza, S. Microaerated UASB reactor treating textile wastewater: The core microbiome and removal of azo dye Direct Black 22. Chemosphere 2020, 242, 125127. [Google Scholar] [CrossRef]
- García-Ruíz, M.J.; Castellano-Hinojosa, A.; Armato, C.; González-Martínez, A.; González-López, J.; Osorio, F. Biogas production and microbial community structure in a stable-stage of a two-stage anaerobic digester. Aiche J. 2020, 66, e16807. [Google Scholar] [CrossRef]
- Luo, Y.S.; Yao, J.Q.; Wang, X.Y.; Zheng, M.Y.; Guo, D.Y.; Chen, Y.G. Efficient municipal wastewater treatment by oxidation ditch process at low temperature: Bacterial community structure in activated sludge. Sci. Total Environ. 2020, 703, 135031. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Rahaman, M.H.; Chen, X.; Xiao, H.W.; Liao, K.S.; Li, X.T.; Duan, C.R.; Zhang, B.Y.; Tao, G.L.; John, Y. New nitrogen removal pathways in a full-scale hybrid constructed wetland proposed from high-throughput sequencing and isotopic tracing results. Ecol. Eng. 2016, 97, 434–443. [Google Scholar] [CrossRef]
- Lin, X.; Liu, F.; Hu, X.M. Influence of Acid and Alkali Modified Fly Ash on Handling Ammonia Nitrogen Wastewater by SBR Reactor. J. Northeast. Univ. (Nat. Sci.) 2018, 39, 1783–1787. [Google Scholar]
- Eleni, V.; Petros, G. Effects of chromium on activated sludge and on the performance of wastewater treatment plants: A review. Water Res. 2012, 46, 549–570. [Google Scholar]
- Miao, Y.; Liao, R.-H.; Zhang, X.-X.; Wang, Y.; Wang, Z.; Shi, P.; Liu, B.; Li, A.-M. Metagenomic insights into Cr(VI) effect on microbial communities and functional genes of an expanded granular sludge bed reactor treating high-nitrate wastewater. Water Res. 2015, 76, 43–52. [Google Scholar] [CrossRef]
| Sample | pH | (Chemical Oxygen Demand) COD(mg/L) |
|---|---|---|
| T2 | 1.74 | 3.43 × 104 |
| T3 | 6.27 | 3.69 × 104 |
| T4 | 2.12 | 2.99 × 104 |
| Index | T2 | T3 | T4 | KB |
|---|---|---|---|---|
| TN/mg·L−1 | 0.171 ± 0.000 a | 0.109 ± 0.001 b | 0.168 ± 0.002 a | 0.066 ± 0.001 c |
| TP/mg·L−1 | 0.019 ± 0.000 a | 0.020 ± 0.001 a | 0.011 ± 0.000 b | 0.018 ± 0.000 a |
| NH3–N/mg·L−1 | 52.400 ± 0.444 a | 45.420 ± 0.414 b | 51.650 ± 0.628 a | 39.680 ± 0.100 c |
| NO3–N/mg·L−1 | 0.237 ± 0.007 a | 0.083 ± 0.003 c | 0.157 ± 0.003 b | 0.003 ± 0.003 d |
| pH | 1.810 ± 0.006 d | 6.020 ± 0.009 a | 2.250 ± 0.006 c | 5.860 ± 0.010 b |
| TOC/mg·L−1 | 721.000 ± 0.577 a | 358.000 ± 1.732 c | 640.000 ± 1.155 b | 203.000 ± 1.732 d |
| Sample | Shannon | Simpson | Chao | Coverage | Seq_Num | OTUs |
|---|---|---|---|---|---|---|
| T2 | 2.757 | 0.233 | 826.161 | 0.995 | 41,495 | 692 |
| T3 | 3.208 | 0.153 | 1119.642 | 0.992 | 43,229 | 878 |
| T4 | 3.835 | 0.153 | 1102.665 | 0.994 | 58,522 | 923 |
| KB | 3.580 | 0.121 | 1154.254 | 0.993 | 53,440 | 840 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Ma, H.; Hassan, A.; Li, C.; Qiang, X. Impact of Non-Metallic Organic Tanning Agents with a Double-Triazine Structure on the Microbial Community Structure in Wastewater. Water 2021, 13, 2438. https://doi.org/10.3390/w13172438
Xu Z, Ma H, Hassan A, Li C, Qiang X. Impact of Non-Metallic Organic Tanning Agents with a Double-Triazine Structure on the Microbial Community Structure in Wastewater. Water. 2021; 13(17):2438. https://doi.org/10.3390/w13172438
Chicago/Turabian StyleXu, Zhifen, Hongrui Ma, Areeb Hassan, Chengtao Li, and Xihuai Qiang. 2021. "Impact of Non-Metallic Organic Tanning Agents with a Double-Triazine Structure on the Microbial Community Structure in Wastewater" Water 13, no. 17: 2438. https://doi.org/10.3390/w13172438
APA StyleXu, Z., Ma, H., Hassan, A., Li, C., & Qiang, X. (2021). Impact of Non-Metallic Organic Tanning Agents with a Double-Triazine Structure on the Microbial Community Structure in Wastewater. Water, 13(17), 2438. https://doi.org/10.3390/w13172438