Are Genetic Reference Libraries Sufficient for Environmental DNA Metabarcoding of Mekong River Basin Fish?
Abstract
:1. Introduction
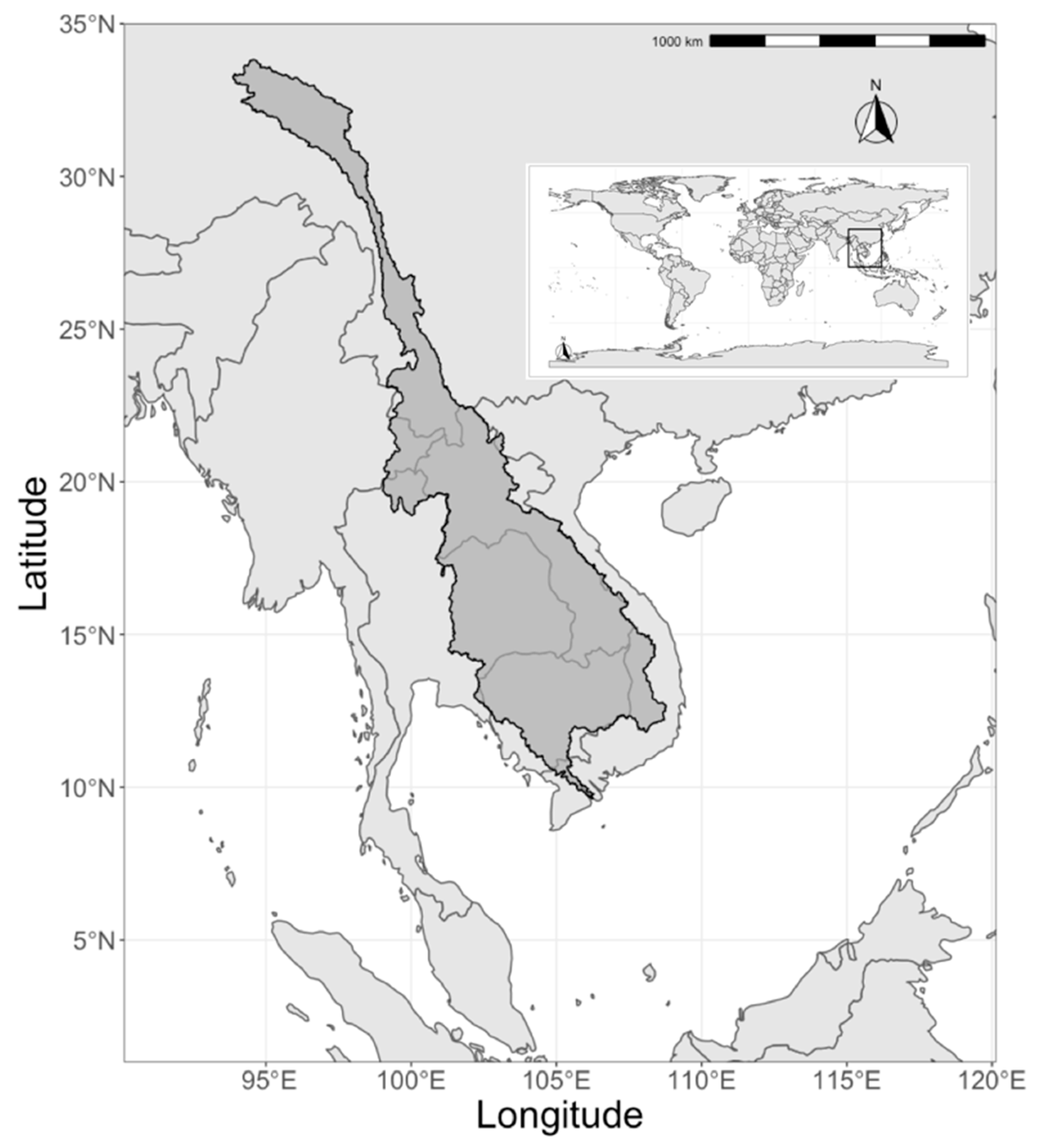
2. Materials and Methods
3. Results
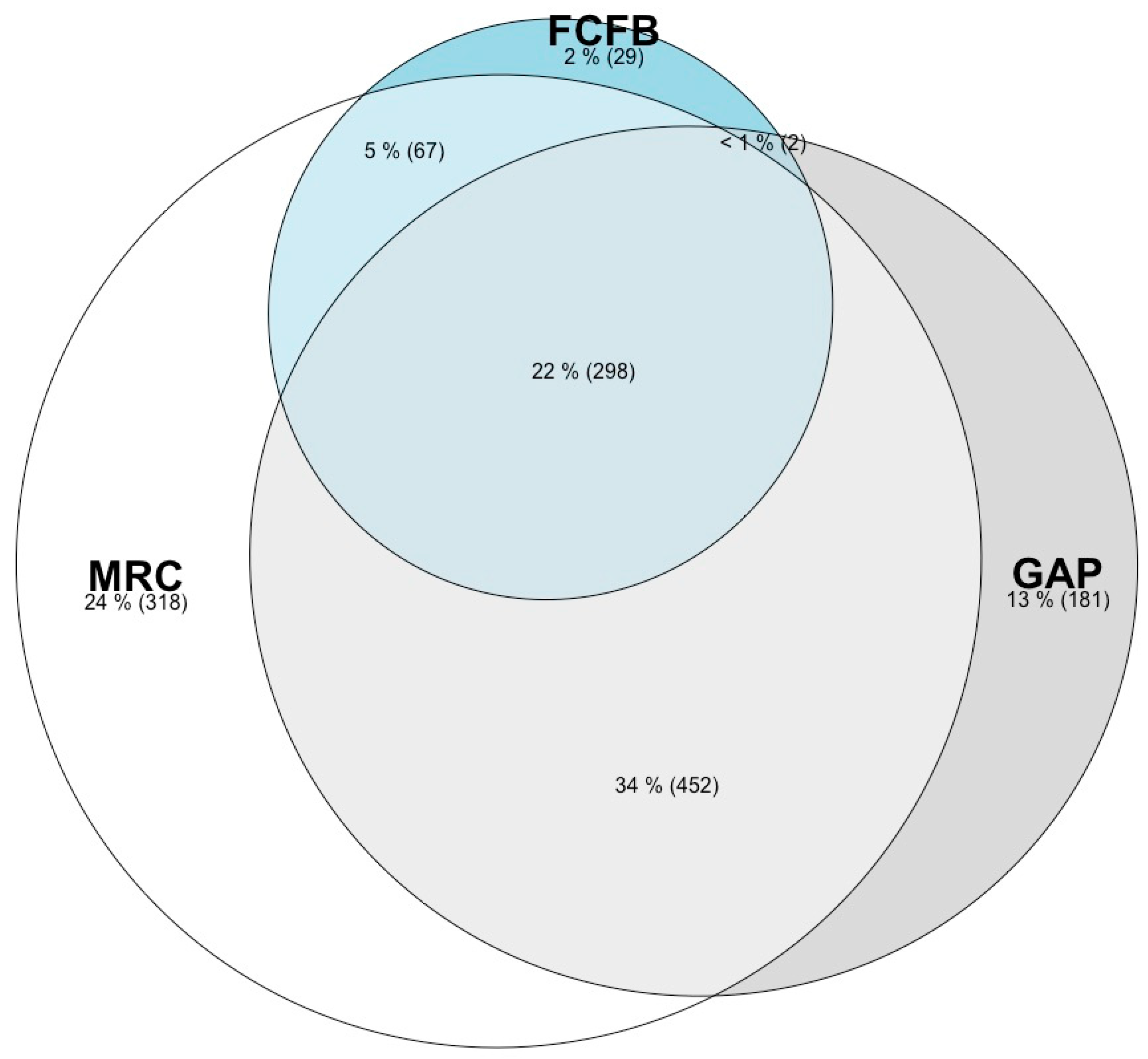
3.1. Species Lists
3.2. Single- and Multi-Primer Coverage
3.3. Species Specificity
3.4. IUCN Status
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomsen, P.F.; Kielgast, J.O.S.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Lacoursière-Roussel, A.; Deiner, K. Environmental DNA is not the tool by itself. J. Fish. Biol. 2021, 98, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L. Can we manage fisheries with the inherent uncertainty from eDNA? J. Fish. Biol. 2021, 98, 341–353. [Google Scholar] [CrossRef]
- McElroy, M.E.; Dressler, T.L.; Titcomb, G.C.; Wilson, E.A.; Deiner, K.; Dudley, T.L.; Jerde, C.L.; Eliason, E.J.; Evans, N.T.; Gaines, S.D.; et al. Calibrating environmental DNA metabarcoding to conventional surveys for measuring fish species richness. Front. Ecol. Evol. 2020, 8, 276. [Google Scholar] [CrossRef]
- Evans, N.T.; Olds, B.P.; Renshaw, M.A.; Turner, C.R.; Li, Y.; Jerde, C.L.; Lodge, D.M.; Mahon, A.R.; Pfrender, M.E.; Lamberti, G.A. Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol. Ecol. Res. 2016, 16, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKelvey, K.S.; Young, M.K.; Knotek, W.L.; Carim, K.J.; Wilcox, T.M.; Padgett-Stewart, T.M.; Schwartz, M.K. Sampling large geographic areas for rare species using environmental DNA: A study of bull trout Salvelinus confluentus occupancy in western Montana. J. Fish. Biol. 2016, 88, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Winfield, I.J.; Blackman, R.C.; Oliver, A. Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 2016, 25, 3101–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, M.; Tucker, A.; Chadderton, W.L.; Jerde, C.L.; Mahon, A.R. Active and passive environmental DNA surveillance of aquatic invasive species. Can. J. Fish. Aquat. Sci. 2016, 73, 76–83. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Das Mishu, M.; Charlop-Powers, Z. Improved environmental DNA reference library detects overlooked marine fishes in New Jersey, United States. Front. Mar. Sci. 2020, 7, 226. [Google Scholar] [CrossRef]
- Marques, V.; Milhau, T.; Albouy, C.; Dejean, T.; Manel, S.; Mouillot, D.; Juhel, J.B. GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Divers. Dist. 2020, 1–13. [Google Scholar] [CrossRef]
- Tedesco, P.A.; Beauchard, O.; Bigorne, R.; Blanchet, S.; Buisson, L.; Conti, L.; Oberdorff, T.; Cornu, J.-F.; Dias, M.S.; Grenouillet, G.; et al. A global database on freshwater fish species occurrence in drainage basins. Sci. Data 2017, 4, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, N.T.; Shirey, P.D.; Wieringa, J.G.; Mahon, A.R.; Lamberti, G.A. Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 2017, 42, 90–99. [Google Scholar] [CrossRef]
- Mekong River Commission. State of the Basin Report 2019; Mekong River Commission Secretariat: Vientiane, Lao, 2019; p. 272. [Google Scholar]
- Jordan, C.; Tiede, J.; Lojek, O.; Visscher, J.; Apel, H.; Nguyen, H.Q.; Quang, C.N.X.; Schlurmann, T. Sand mining in the Mekong Delta revisited-current scales of local sediment deficits. Sci. Rep. 2019, 9, 17823. [Google Scholar] [CrossRef] [PubMed]
- Anthony, E.; Brunier, G.; Besset, M.; Goichot, M.; Dussouillez, P.; Nguyen, V.L. Linking rapid erosion of the Mekong River delta to human activities. Sci. Rep. 2015, 5, 14745. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.X.; Li, S.; Kummu, M.; Padawangi, R.; Wang, J.J. Observed changes in the water flow at Chiang Saen in the lower Mekong: Impacts of Chinese dams? Quarter. Inter. 2014, 336, 145–157. [Google Scholar] [CrossRef]
- Ziv, G.; Baran, E.; Nam, S.; Rodríguez-Iturbe, I.; Levin, S.A. Trading-off fish biodiversity, food security, and hydropower in the Mekong River Basin. Proc. Nat. Acad. Sci. USA 2012, 109, 5609–5614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winemiller, K.O.; McIntyre, P.B.; Castello, L.; Fluet-Chouinard, E.; Giarrizzo, T.; Nam, S.; Sáenz, L.; Baird, I.G.; Darwall, W.; Lujan, N.K.; et al. Balancing hydropower and biodiversity in the Amazon, Congo, and Mekong. Science 2016, 351, 128–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.; Phommakone, S.; Vuthy, L.Y.; Samphawamana, T.; Son, N.; Khumsri, M.; Bun, N.P.; Sovanara, K.; Degen, P.; Starr, P. Lower Mekong fisheries estimated to be worth around $17 billion a year. Catch Cult. 2015, 21, 4–7. [Google Scholar]
- Belle, C.C.; Stoeckle, B.C.; Geist, J. Taxonomic and geographical representation of freshwater environmental DNA research in aquatic conservation. Aquat. Conserv. Mar. Fresh Ecosyst. 2019, 29, 1996–2009. [Google Scholar] [CrossRef]
- Bellemain, E.; Patricio, H.; Gray, T.; Ficetola, G.F.; Valentini, A.; Miaud, C.; Dejean, T. Trails of river monsters: Detecting critically endangered Mekong giant catfish Pangasianodon gigas using environmental DNA. Global Ecol. Conserv. 2016, 7, 148–156. [Google Scholar]
- Osathanunkul, M.; Minamoto, T. A molecular survey based on eDNA to assess the presence of a clown featherback (Chitala ornata) in a confined environment. Peer J. 2020, 8, e10338. [Google Scholar] [CrossRef] [PubMed]
- Osathanunkul, M.; Minamoto, T. eDNA-based detection of a vulnerable crocodile newt (Tylototriton uyenoi) to influence government policy and raise public awareness. Divers. Distrib. 2021, 1–8. [Google Scholar] [CrossRef]
- Gillet, B.; Cottet, M.; Destanque, T.; Kue, K.; Descloux, S.; Chanudet, V.; Hughes, S. Direct fishing and eDNA metabarcoding for biomonitoring during a 3-year survey significantly improves number of fish detected around a South East Asian reservoir. PLoS ONE 2018, 13, e0208592. [Google Scholar] [CrossRef] [Green Version]
- McInnes, J.C. The Development and Application of DNA Metabarcoding to Non-Invasively Assess Seabird Diets, Using Albatrosses as a Model. Ph.D. Thesis, University of Tasmania, Hobart, Tasmania, 2017. [Google Scholar]
- Mekong River Commission. Updated MRC database estimates 1,148 fish species in Mekong Basin. Catch Cult. 2019, 25, 20–21. [Google Scholar]
- Olds, B.P.; Jerde, C.L.; Renshaw, M.A.; Li, Y.; Evans, N.T.; Turner, C.R.; Deiner, K.; Mahon, A.R.; Brueseke, M.A.; Lamberti, G.A.; et al. Estimating species richness using environmental DNA. Ecol. Evol. 2016, 6, 4214–4226. [Google Scholar] [CrossRef] [Green Version]
- Van Zalinge, N.; Degen, P.; Pongsri, C.; Nuov, S.; Jensen, J.G.; Nguyen, V.H.; Choulamany, X. The Mekong River System; FAO Regional Office for Asia and the Pacific; RAP Publication: Bangkok, Thailand, 2004. [Google Scholar]
- Holtgrieve, G.W.; Arias, M.E.; Irvine, K.N.; Lamberts, D.; Ward, E.J.; Kummu, M.; Koponen, J.; Sarkkula, J.; Richey, J.E. Patterns of ecosystem metabolism in the Tonle Sap Lake, Cambodia with links to capture fisheries. PLoS ONE 2013, 8, e71395. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.E.; Cochrane, T.A.; Elliott, V. Modelling future changes of habitat and fauna in the Tonle Sap wetland of the Mekong. Environ. Conserv. 2014, 41, 165–175. [Google Scholar] [CrossRef] [Green Version]
- World Wildlife Fund. First Contact in the Greater Mekong–New Species Discoveries; World Wildlife Fund: Hanoi, Vietnam, 2009; p. 39. [Google Scholar]
- Valbo-Jorgensen, J.; Visser, T. The MRC Mekong Fish Database: An information Base on Fish of a Major International River Basin. In MRC Conference Series; MRC: Phnom Penh, Cambodia, 2003. [Google Scholar]
- So, N.; Utsugi, K.; Shibukawa, K.; Thach, P.; Chhuoy, S.; Kim, S.; Chin, D.; Nen, P.; Chheng, P. Fishes of Cambodian Freshwater Bodies; Inland Fisheries Research and Development Institute, Fisheries Administration: Phnom Penh, Cambodia, 2018; p. 197. [Google Scholar]
- Froese, R.; Pauly, D. (Eds.) FishBase 2000: Concepts Designs and Data Sources; CLARM: Los Banos, Philippines, 2000; Volume 1594. [Google Scholar]
- Bylemans, J.; Gleeson, D.M.; Hardy, C.M.; Furlan, E. Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray–Darling Basin (Australia). Ecol. Evol. 2018, 8, 8697–8712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiBattista, J.D.; Coker, D.J.; Sinclair-Taylor, T.H.; Stat, M.; Berumen, M.L.; Bunce, M. Assessing the utility of eDNA as a tool to survey reef-fish communities in the Red Sea. Coral Reefs 2017, 36, 1245–1252. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Dewaard, J.R.; Hebert, P.D. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef] [Green Version]
- Kitano, T.; Umetsu, K.; Tian, W.; Osawa, M. Two universal primer sets for species identification among vertebrates. Inter. J. Legal Med. 2007, 121, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Nat. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miya, M.; Nishida, M. Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion. Mol. Phylogenetics Evol. 2000, 17, 437–455. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Iwasaki, W.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. Roy. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [Green Version]
- Palumbi, S.R. What can molecular genetics contribute to marine biogeography? An urchin’s tale. J. Exp. Mar. Biol. Ecol. 1996, 203, 75–92. [Google Scholar] [CrossRef]
- Shaw, J.L.; Clarke, L.J.; Wedderburn, S.D.; Barnes, T.C.; Weyrich, L.S.; Cooper, A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol. Conserv. 2016, 197, 131–138. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Møller, P.R.; Rasmussen, M.; Willerslev, E. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 2012, 7, e41732. [Google Scholar]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Dejean, T.; Bellemain, E.; Besnard, A.; Coissac, E.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Phil. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Larsson, J. R Package ‘Eulerr’: Area-Proportional Euler and Venn Diagrams with Ellipses, version 6.1.0; Available online: https://cran.r-project.org/web/packages/eulerr/index.html (accessed on 18 June 2021).
- Adamson, E.A.; Britz, R.; Lieng, S. Channa auroflammea, a new species of snakehead fish of the Marulius group from the Mekong River in Laos and Cambodia (Teleostei: Channidae). Zootaxa 2019, 4571, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Vu, A.V.; Baumgartner, L.J.; Mallen-Cooper, M.; Howitt, J.A.; Robinson, W.A.; So, N.; Cowx, I.G. Diadromy in a large tropical river, the Mekong: More common than assumed, with greater implications for management. J. Ecohydraulics 2020, 1–13. [Google Scholar] [CrossRef]
- Doble, C.J.; Hipperson, H.; Salzburger, W.; Horsburgh, G.J.; Mwita, C.; Murrell, D.J.; Day, J.J. Testing the performance of environmental DNA metabarcoding for surveying highly diverse tropical fish communities: A case study from Lake Tanganyika. Environ. DNA 2020, 2, 24–41. [Google Scholar] [CrossRef]
- Jerde, C.L.; Wilson, E.A.; Dressler, T.L. Measuring global fish species richness with eDNA metabarcoding. Mol. Ecol. Res. 2019, 19, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef]
- Darling, J.A.; Jerde, C.L.; Sepulveda, A.J. What do you mean by false positive? Environ. DNA 2021. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Collins, R.A.; Bakker, J.; Wangensteen, O.S.; Soto, A.Z.; Corrigan, L.; Sims, D.W.; Genner, M.J.; Mariani, S. Non-specific amplification compromises environmental DNA metabarcoding with COI. Meth. Ecol. Evol. 2019, 10, 1985–2001. [Google Scholar] [CrossRef]
- Deagle, B.E.; Jarman, S.N.; Coissac, E.; Pompanon, F.; Taberlet, P. DNA metabarcoding and the cytochrome c oxidase subunit I marker: Not a perfect match. Biol. Lett. 2014, 10, 20140562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficetola, G.F.; Coissac, E.; Zundel, S.; Riaz, T.; Shehzad, W.; Bessière, J.; Taberlet, P.; Pompanon, F. An in silico approach for the evaluation of DNA barcodes. BMC Genom. 2010, 11, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, M.V.; Hester, J.; Shalkhauser, A.; Chan, E.R.; Logue, K.; Small, S.T.; Serre, D. In silico assessment of primers for eDNA studies using PrimerTree and application to characterize the biodiversity surrounding the Cuyahoga River. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, W.; Zhou, Y.; Zhang, Y.; Lu, Y.; Wang, X.; Zhao, D.; Yang, Y.; Zhang, C. MFEprimer-2.0: A fast thermodynamics-based program for checking PCR primer specificity. Nucleic Acids Res. 2012, 40, W205–W208. [Google Scholar] [CrossRef] [PubMed]
- Siva, C.; Kumar, R.; Sharma, L.; Laskar, M.A.; Sumer, S.; Barat, A.; Sahoo, P.K. The complete mitochondrial genome of a stream loach (Schistura reticulofasciata) and its phylogeny. Conserv. Gen. Res. 2018, 10, 829–832. [Google Scholar] [CrossRef]
- Pentinsaari, M.; Ratnasingham, S.; Miller, S.E.; Hebert, P.D. BOLD and GenBank revisited–Do identification errors arise in the lab or in the sequence libraries? PLoS ONE 2020, 15, e0231814. [Google Scholar] [CrossRef]
- Leray, M.; Knowlton, N.; Ho, S.L.; Nguyen, B.N.; Machida, R.J. GenBank is a reliable resource for 21st century biodiversity research. Proc. Nat. Acad. Sci. USA 2019, 116, 22651–22656. [Google Scholar] [CrossRef] [Green Version]

| Step | Primer Pair(s) | Species with Sequences | Percent of Species with Sequences (n = 782) | Percent of Total Species in UNION (n = 1345) |
|---|---|---|---|---|
| Step 1 | 16S Shaw | 643 | 82.1% | 47.8% |
| Step 2 | 16S Shaw CytB Thomsen cb | 723 | 92.5% | 53.8% |
| Step 3 | 16S Shaw CytB Thomsen cb CytB Miya | 745 | 95.3% | 55.4% |
| Step 4 | 16S Shaw CytB Thomsen cb CytB Miya 12S Bylemans | 759 | 97.1% | 56.4% |
| Step 5 | 16S Shaw CytB Thomsen cb CytB Miya 12S Bylemans 12S Kelly | 766 | 98.0% | 56.9% |
| Step 6 | 16S Shaw CytB Thomsen cb CytB Miya 12S Bylemans 12S Kelly CytB Thomsen 2deg | 772 | 98.7% | 57.4% |
| Species Identity | Sequence Presence in Shaw 16S (Yes/No) | Within Sequence Similarity (Shaw 16S) | Between Species with >0.05 Genetic Similarity (Shaw 16S) | Number of Primer Pairs with Sequences (23 Max) |
|---|---|---|---|---|
| Channa spp. | ||||
| C. gachua | Yes | 6% | None | 18 |
| C. lucius | Yes | 4% | None | 19 |
| C. marulioides | Yes | N.E. | C. auroflammea C. marulius | 5 |
| C. marulius | Yes | 1% | C. auroflammea C. marulioides | 20 |
| C. melanoptera | No | N.E. | N.E. | 0 |
| C. melasoma | Yes | 1% | None | 5 |
| C. micropeltes | Yes | 1% | None | 17 |
| C. orientalis | Yes | N.E. | None | 5 |
| C. striata | Yes | 3% | None | 19 |
| C. auroflammea1 | Yes | N.E. | C. marulioides C. marulius | N.E. |
| Henicorhynchus spp. | ||||
| H. caudimaculatus | No | N.E. | N.E. | 1 |
| H. entmema | Yes | N.E. | N.E. | 15 |
| H. lineatus | Yes | 7% | H. entmema H. ornatipinnis H. siamensis | 18 |
| H. ornatipinnis | Yes | N.E. | H. entmema H. ornatipinnis H. siamensis | 4 |
| H. siamensis | Yes | 0% | H. entmema H. ornatipinnis H. siamensis | 16 |
| Pangasius spp. and Pangasianodon spp. | ||||
| Pangasius bocourti | Yes | 1% | P. polyuranodon P. macronema P. hypophthalmus | 12 |
| P. conchophilus | Yes | 0% | P. macronema P. hypophthalmus | 10 |
| P. djambal | Yes | 0% | P. macronema P. hypophthalmus | 3 |
| P. elongatus | Yes | N.E. | P. hypophthalmus | 4 |
| P. krempfi | Yes | 0% | P. macronema P. hypophthalmus | 13 |
| P. kunyit | No | N.E. | N.E. | 0 |
| P. larnaudii | Yes | 0% | P. polyuranodon P. macronema P. hypophthalmus | 19 |
| P. macronema | Yes | 0% | Most Pangasius sp. with sequences | 14 |
| P. mekongensis | No | N.E. | N.E. | 0 |
| P. nasutus | Yes | 1% | P. hypophthalmus | 10 |
| P. pangasius | Yes | 0% | P. macronema P. hypophthalmus | 20 |
| P. polyuranodon | Yes | N.E. | P. larnaudii P. bocourti P. hypophthalmus | 10 |
| P. sanitwongsei | Yes | 1% | P. macronema P. hypophthalmus | 13 |
| Panagasianodon gigas | Yes | 0% | P. macronema | 19 |
| P. hypophthalmus | Yes | 16% | All Pangasius sp. with sequences | 20 |
| Schistura spp. | ||||
| S. fasciolata | Yes | 5% | N.E. | 16 |
| S. kaysonei | Yes | N.E. | N.E. | 16 |
| +73 Schistura spp. | No | N.E. | N.E. | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jerde, C.L.; Mahon, A.R.; Campbell, T.; McElroy, M.E.; Pin, K.; Childress, J.N.; Armstrong, M.N.; Zehnpfennig, J.R.; Kelson, S.J.; Koning, A.A.; et al. Are Genetic Reference Libraries Sufficient for Environmental DNA Metabarcoding of Mekong River Basin Fish? Water 2021, 13, 1767. https://doi.org/10.3390/w13131767
Jerde CL, Mahon AR, Campbell T, McElroy ME, Pin K, Childress JN, Armstrong MN, Zehnpfennig JR, Kelson SJ, Koning AA, et al. Are Genetic Reference Libraries Sufficient for Environmental DNA Metabarcoding of Mekong River Basin Fish? Water. 2021; 13(13):1767. https://doi.org/10.3390/w13131767
Chicago/Turabian StyleJerde, Christopher L., Andrew R. Mahon, Teresa Campbell, Mary E. McElroy, Kakada Pin, Jasmine N. Childress, Madeline N. Armstrong, Jessica R. Zehnpfennig, Suzanne J. Kelson, Aaron A. Koning, and et al. 2021. "Are Genetic Reference Libraries Sufficient for Environmental DNA Metabarcoding of Mekong River Basin Fish?" Water 13, no. 13: 1767. https://doi.org/10.3390/w13131767







