Prevalence of Antibiotic Resistance Genes in Pharmaceutical Wastewaters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites and Sample Collection
2.2. Metagenomic DNA Extraction
2.3. Preparation of Oligonucleotide Primers Solutions
2.4. PCR Screening for Antibiotic Resistance Genes and Class 1 integrons
3. Results
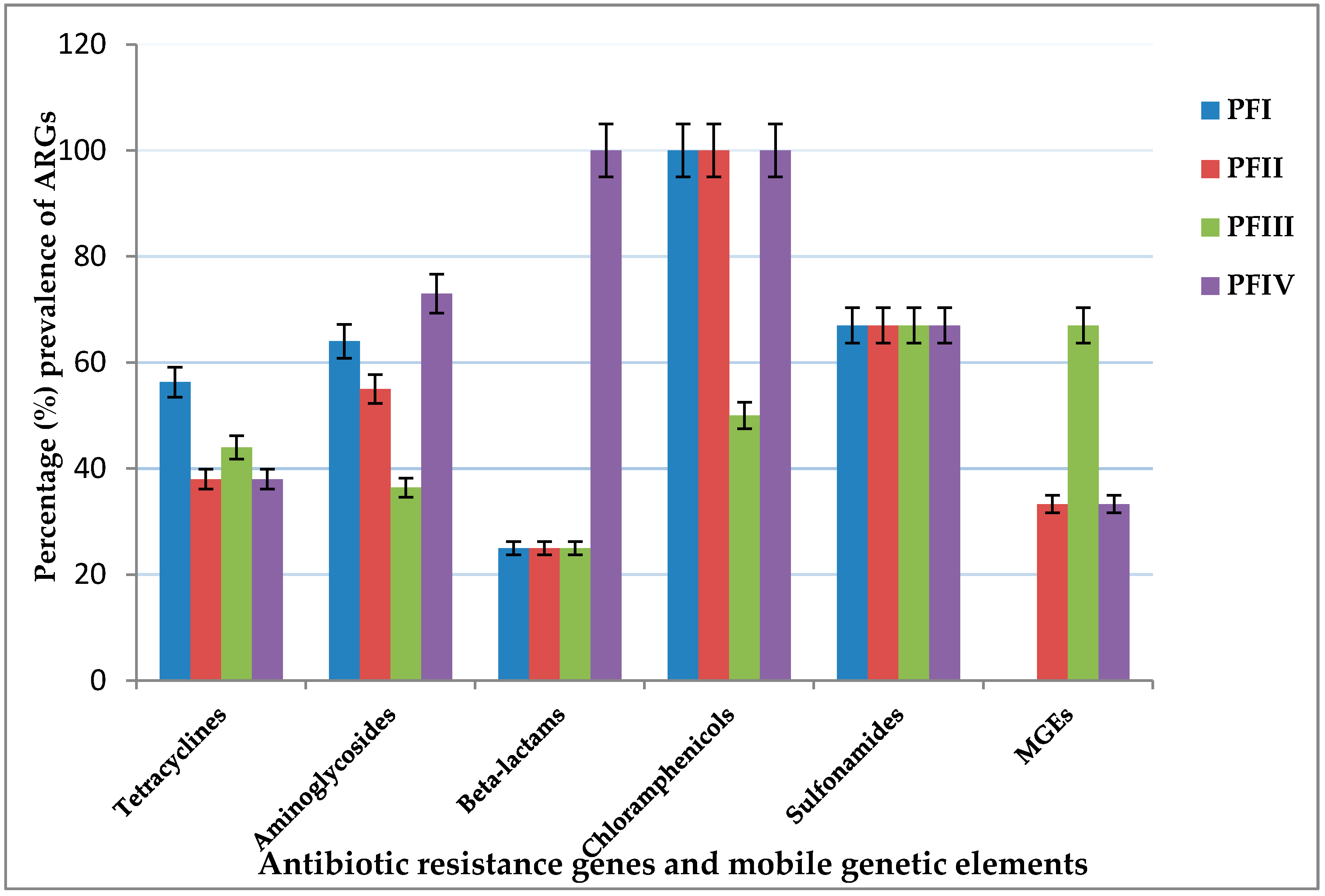
3.1. Antibiotic Resistance Gene Screening
3.1.1. Tetracycline Resistance Genes
3.1.2. Aminoglycoside Resistance Genes
3.1.3. β-Lactams and Penicillin Resistance Genes
3.1.4. Chloramphenicol and Sulfonamides Resistance Genes
3.1.5. Mobile Genetic Elements
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pruden, A. Balancing water sustainability and public health goals in the face of growing 650 concerns about antibiotic resistance. Environ. Sci. Technol. 2014, 48, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Obayiuwana, A.C.; Ogunjobi, A.; Yang, M.; Ibekwe, M. Characterization of bacterial communities and their antibiotic resistance profiles in wastewaters obtained from pharmaceutical facilities in Lagos and Ogun States, Nigeria. Int. J. Environ. Res. Public Health 2018, 15, 1365–1378. [Google Scholar] [CrossRef] [Green Version]
- Obayiuwana, A.C.; Ibekwe, A.M. Antibiotic resistance genes occurrence in wastewaters from selected pharmaceutical facilities in Nigeria. Water 2020, 15, 1897. [Google Scholar] [CrossRef]
- Thakali, O.; Brooks, J.P.; Shahin, S.; Sherchan, S.P.; Haramoto, E. Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA. Water 2020, 12, 1729. [Google Scholar] [CrossRef]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-resistance genes in waste water-review. Trends Microbiol. 2017, 26, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Moles, S.; Mosteo, R.; Gómez, J.; Szpunar, J.; Gozzo, S.; Castillo, J.R.; Ormad, M.P. Towards the Removal of Antibiotics Detected in Wastewaters in the POCTEFA Territory: Occurrence and TiO2 Photocatalytic Pilot-Scale Plant Performance. Water 2020, 12, 1453. [Google Scholar] [CrossRef]
- Li, L.; Guo, C.; Fan, S.; Lv, J.; Zhang, Y.; Xu, Y.; Xu, J. Dynamic transport of antibiotics and antibiotic resistance genes under different treatment processes in a typical pharmaceutical wastewater treatment plant. Environ. Sci. Pollut. Res. 2018, 25, 30191–30198. [Google Scholar] [CrossRef]
- Watkinson, A.J.; Micalizzi, G.B.; Graham, G.M.; Bates, J.B.; Costanzo, S.D. Antibiotic resistant Escherichia coli in wastewaters, surface waters, and Oysters from an urban riverine system. Appl. Environ. Microbiol. 2007, 73, 5667–5670. [Google Scholar] [CrossRef] [Green Version]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Munck, C.; Albertsen, M.; Telke, A.; Ellabaan, M.; Nielsen, P.H.; Sommer, M.O. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 2015, 6, 8452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aminov, R.I. Evolution in action: Dissemination of tet(x) into pathogenic microbiota. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Cui, X.; Xu, H.; Liu, W.; Tao, F.; Shao, T.; Pan, X.; Zheng, B. Whole genome sequencing of Extended-Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli isolated from a wastewater treatment plant in China. Front. Microbiol. 2019, 10, 1797. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Zhang, T. Occurrence, abundance, and diversity of tetracycline resistance genes in 15 sewage treatment plants across china and other global locations. Environ. Sci. Technol. 2011, 45, 2598–2604. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Yang, M.; Tian, Z.; Ren, L.; Zhang, S. Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environ. Sci. Technol. 2012, 46, 7551–7557. [Google Scholar] [CrossRef]
- Walsh, F.; Duffy, B. The culturable soil antibiotic resistome: A community of multi-drug resistant bacteria. PLoS ONE 2014, e65567. [Google Scholar] [CrossRef] [Green Version]
- Schmiede, R.; Edwards, R. Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012, 7, 73–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.P.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a New bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, gkw1004. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Chen, H.; Bond, P.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 7, 002. [Google Scholar] [CrossRef]
- Nesme, J.; Cécillon, S.; Delmont, T.O.; Monier, J.M.; Vogel, T.M.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Yang, Y.; Ma, L.; Ju, F.; Guo, F.; Tiedje, J.M.; Zhang, T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015, 9, 2490–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef] [PubMed]
- Sandegren, L. Selection of antibiotic resistance at very low antibiotic concentrations. Upsala J. Med. Sci. 2014, 119, 103–107. [Google Scholar] [CrossRef]
- Lundström, S.V.; Östman, M.; Bengtsson-Palme, J.; Rutgersson, C.; Thoudal, M.; Sircar, T.; Blanck, H.; Eriksson, K.M.; Tysklind, M.; Flach, C.; et al. Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci. Total Environ. 2016, 553, 587–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Zhao, L.; Dong, Y.H. Oxidative degradation kinetics and products of chlortetracycline by manganese dioxide. J. Hazard. Mater. 2011, 193, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, Y.; Lee, C.C.; Liu, L.; Huang, Y. Hydrophilic interaction chromatography separation mechanisms of tetracyclines on amino-bonded silica column. J. Sep. Sci. 2011, 34, 1508–1516. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechnol. 2002, 20, 261–283. [Google Scholar] [CrossRef]
- Moore, I.F.; Hughes, D.W.; Wright, G.D. Tigecycline is modified by the flavin-dependent monooxygenase tet(x). Biochemistry. 2005, 44, 11829–11835. [Google Scholar] [CrossRef]
- Shakil, S.; Khan, R.; Zarrilli, R.; Khan, A.U. Aminoglycosides versus bacteria—A description of the action, resistance mechanism, and nosocomial battle ground. J. Biomed. Sci. 2008, 15, 5–14. [Google Scholar] [CrossRef]
- Heuer, H.; Krögerrecklenfort, E.; Wellington, E.M.H.; Egan, S.; van Elsas, J.D.; van Overbeek, L.; Collard, J.M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol. Ecol. 2002, 42, 289–302. [Google Scholar] [CrossRef]
- Tennstedt, T.; Szczepanowski, R.; Braun, S.; Pühler, A.; Schlüter, A. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 2003, 45, 239–252. [Google Scholar] [CrossRef]
- Tennstedt, T.; Szczepanowski, R.; Krahn, I.; Pühler, A.; Schlüter, A. Sequence of the 68,869 bp IncP-1a plasmid pTB11 from a wastewater treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 2005, 53, 218–238. [Google Scholar] [CrossRef]
- Livermore, D.M. Are all beta-lactams created equal? Scand. J. Infect. Dis. Suppl. 1996, 101, 33–43. [Google Scholar] [PubMed]
- Craig, W.A. Basic pharmacodynamics of antibacterials with clinical applications to the use of β-lactams, glycopeptides, and linezolid. Infect. Dis. Clin. N. Am. 2003, 17, 479–501. [Google Scholar] [CrossRef]
- Benito-Peña, E.; Partal-Rodera, A.I.; León-González, M.E.; Moreno-Bondi, M.C. Evaluation of mixed mode solid phase extraction cartridges for the preconcentration of beta-lactam antibiotics in wastewater using liquid chromatography with UV-DAD detection. Anal. Chim. Acta 2006, 556, 415–422. [Google Scholar] [CrossRef]
- Brown, K.D.; Kulis, J.; Thomson, B.; Chapman, T.H.; Mawhinney, D.B. Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 2006, 366, 772–783. [Google Scholar] [CrossRef]
- Bailón-Pérez, M.I.; García-Campaña, A.M.; Cruces-Blanco, C.; del Olmo Iruela, M. Trace determination of β-lactam antibiotics in environmental aqueous samples using off-line and on-line preconcentration in capillary electrophoresis. J. Chromatogr. 2008, 1185, 273–280. [Google Scholar] [CrossRef]
- Poirel, L.; Marque, S.; Heritier, C.; Segonds, C.; Chabanon, G.; Nordmann, P. OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 202–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caucci, S.; Karkman, A.; Cacace, D.; Rybicki, M.; Timpel, P.; Voolaid, V.; Gurke, R.; Virta, M.; Berendonk, T.U. Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol. Ecol. 2016, 92, fiw060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karkman, A.; Johnson, T.A.; Lyra, C.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hultman, J.; Tamminen, M.; Pärnänen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [Green Version]
- Heuer, H.; Szczepanowski, R.; Schneiker, S.; Phler, A.; Top, E.M.; Schlüter, A. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1b group without any accessory genes. Microbiology 2004, 150, 3591–3599. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.Y.; Ren, J.; Song, L.S.; Sun, S.; An, L.G. Dominant chloramphenicol-resistant bacteria and resistance genes in coastal marine waters of Jiaozhou Bay, China. World J. Microb. Biot. 2008, 24, 209–217. [Google Scholar] [CrossRef]
- Then, R.L. Mechanisms of resistance to trimethoprim, the sulfonamides, and trimethoprim-sulfamethoxazole. Rev. Infect. Dis. 1982, 4, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, X.; Jiao, S.; Zhang, J.; Ye, B.; Gao, S. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu province, Southeastern China. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Wilson, C.A.; Novak, J.T.; Riffat, R.; Aynur, S.; Murthy, S.; Pruden, A. Effect of various sludge digestion conditions on sulfonamide, macrolide, and tetracycline resistance genes and class I integrons. Environ. Sci. Technol. 2011, 45, 7855–7861. [Google Scholar] [CrossRef]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef] [PubMed]
| Gene Function/Mechanism of Drug Resistance | Resistance Genes | Pharmaceutical Facility | Total No. of Facilities with Positive Genes | |||
|---|---|---|---|---|---|---|
| PFI | PFII | PFIII | PFIV | |||
| Efflux Ribosomal Protection Enzymatic modification | tet(A) | + | + | + | − | 3 |
| tet(B) | + | − | + | + | 3 | |
| tet(C) | + | − | + | + | 3 | |
| tet(D) | + | + | + | − | 3 | |
| tet(E) | − | − | − | + | 1 | |
| tet(G) | + | + | + | + | 4 | |
| tet(30) | − | − | − | + | 1 | |
| tet(L) | + | − | − | − | 1 | |
| tet(M) | − | + | + | − | 2 | |
| tet(Q) | + | − | + | − | 2 | |
| tet(BP) | + | + | − | − | 2 | |
| tet(X) | + | + | − | + | 3 | |
| Acetyl transferases Nucleotidyl-transferases | aac(3)-II | + | + | − | + | 3 |
| aac(3)-IV | + | + | − | + | 3 | |
| aacA4 | − | − | − | + | 3 | |
| aadA | + | + | + | + | 4 | |
| aadB | + | − | + | + | 3 | |
| Phosphor-transferases | aphA1 | + | + | − | + | 3 |
| strA | + | + | + | + | 4 | |
| strB | + | + | + | + | 4 | |
| β-Lactamase | blaTEM | − | − | − | + | 1 |
| blaOXA | + | + | + | + | 4 | |
| blaCTX-M | − | − | − | + | 1 | |
| blaIMP | − | − | − | + | 1 | |
| Chloramphenicol acetyltransferase | catA1 | + | + | + | + | 4 |
| cmlA | + | + | − | + | 3 | |
| Dihydropteroate synthase | sulI | + | + | + | + | 4 |
| sulII | + | + | + | + | 4 | |
| Integrase | Intl1 | − | + | + | + | 3 |
| Intl2 | − | − | + | − | 1 | |
| * Pharmaceutical Facilities | PFI | PFII | PFIII | PFIV |
|---|---|---|---|---|
| Sources | WWHT | WWHT | WWHT | WWDP |
| Tetracycline Resistance Genes | tet(A), tet(B), tet(C), tet(D), tet(G), tet(L), tet(Q), tet(X), tet(BP) | tet(A), tet(D), tet(G), tet(M), tet(X) tet(BP) | tet(A), tet(B), tet(C), tet(D), tet(G), tet(M), tet(Q) | tet(B), tet(C), tet(E), tet(G), tet(X), tet(30) |
| Aminoglycoside Resistance Genes | aadA, aadB, aac(3)-II, aac(3)-IV, aphA1, strA, strB | aadA, aac(3)-II, aac(3)-IV, aphA1, strA, strB | aadA, aadB, strA, strB | aadA, aadB, aac(3)-II, aac(3)-IV, aphA1, aacA4, strA, strB |
| β-Lactams Resistance Genes | blaOXA | blaOXA | blaTEM, blaOXA, blaCTX-M, blaIMP | blaOXA |
| Chloramphenicol Resistance Genes | catA1, cmlA | catA1, cmlA | catA1 | catA1, cmlA |
| Sulphonamide Resistance Genes | sulI, sulII | sulI, sulII | sulI, sulII | sulI, sulII |
| Mobile Genetic Elements | - | intl1 | intl1, intl2, | intl1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obayiuwana, A.; Ogunjobi, A.; Ibekwe, A. Prevalence of Antibiotic Resistance Genes in Pharmaceutical Wastewaters. Water 2021, 13, 1731. https://doi.org/10.3390/w13131731
Obayiuwana A, Ogunjobi A, Ibekwe A. Prevalence of Antibiotic Resistance Genes in Pharmaceutical Wastewaters. Water. 2021; 13(13):1731. https://doi.org/10.3390/w13131731
Chicago/Turabian StyleObayiuwana, Amarachukwu, Adeniyi Ogunjobi, and Abasiofiok Ibekwe. 2021. "Prevalence of Antibiotic Resistance Genes in Pharmaceutical Wastewaters" Water 13, no. 13: 1731. https://doi.org/10.3390/w13131731







