Abstract
Aquatic macrophytes rhizosphere are biogeochemical cycling hotspots in freshwater ecosystems. However, little is known regarding the effect of aquatic macrophytes on bacterial community and interactions in the riparian zones. We investigated the bacterial community composition and network structures along a gradient of the riparian zone as follows: The supralittoral and eulittoral zones with Phragmites australis, the eulittoral and infralittoral zones without P. australi. The bacterial communities in the four zones differed significantly based on taxonomic dissimilarity, but the two zones with P. australis exhibited phylogenetic closeness of the bacterial communities. The characteristics of the bacterial networks, such as connectivity, modularity, and topological roles of OTUs, were totally different between the P. australis and non-P. australis zones. Some bacterial phyla enriched in the P. australis zones were found to be putative keystone taxa in the networks, which might be involved in the regulation of bacterial interactions and plant growth. Moreover, the hydrological regime and particle size were shown to be determinants of the bacterial community and network structures in the riparian zones. In summary, our results show that the role of P. australis and local factors are crucial for constructing bacterial community and interactions in the riparian zones of lakes.
1. Introduction
In the context of lake hydrological changes, the riparian zone, the interface between the terrestrial and aquatic domains, can provide a heterogeneous environment and diverse habitats for variety of flora and fauna. The riparian zones of lakes can also maintain the stability of lake ecosystems by receiving and transferring the majority of terrestrial allochthonous input [1]. Aquatic macrophytes are an important component in complex ecosystems of the riparian zones of lakes [2]. Meanwhile, the rhizosphere of aquatic plant has also become widely recognized biogeochemical hotspots because of interactions between roots and microorganisms [3,4], which effectively enhances the function of wetlands in purifying environmental contaminants [5]. The function of the riparian zones of lakes is inseparable from the diversity and structure of riparian zone-associated microorganisms. Aquatic macrophytes and local factors are crucial for the bacterial community distribution and assembly along the gradient of riparian zones. Although numerous studies have shown that the environment factors of riparian zones are crucial for microbial community composition and diversity [6,7,8], we lack an understanding of the effect of aquatic macrophytes on riparian bacterial community composition and interactions in the riparian zones of freshwater lakes.
Plant roots interact with surrounding soil/sediment, resulting in plant species-specific rhizosphere microorganisms, which has consequences for variation in microbial community composition and interactions [9]. Aquatic plant root exudates, such as organic acids, sugars, and other secondary metabolites, have been regarded as a driving forcing in determining microbial community composition and activity [10], especially promoting nitrogen cycling processes [11]. With the development of high-throughput sequencing technology, 16S rRNA gene sequencing has provided a deeper understanding of aquatic plant rhizosphere microbiota. Thus, more taxa of rhizosphere-associated microbiota have been shown to contribute to the processes of pollutant degradation and plant fitness in wetlands [4,12]. Some studies have also shown that the species and lineages of aquatic plants play a crucial role in the distribution and composition of microbial communities [4,13,14]. Moreover, interactions among members of a microbial community are an indispensable part the construction of a microbial community and impacts its functions [15]. Network analysis is an effective method to predict correlations between microbes and the dynamics of microbial interactions [16,17]. Recently, an increasing number of studies have investigated the influence of the rhizosphere on bacterial networks, with the results showing that the characteristics of rhizosphere microbial community networks are completely different from those of the surroundings, including properties such as connectivity, potential links, and complexity [18,19,20]. The putative key species of rhizosphere microbial networks have been recognized to be involved in plant growth and health [21,22].
Microbial communities are governed by abiotic (environment factors) and biotic (species and diversity of plant) factors in different ecosystems [9]. The characteristics of riparian microbial communities are typically considered to be highly sensitive to environment factors, such as nitrogen content, organic matter, and pH [8,23,24]. Dynamic littoral hydrology can also predominately affect riparian bacterial community assembly and carbon cycling [6,7,25]. In addition, aquatic macrophytes are adapted to a changing environment by being able to regulate their physiological characteristics. For example, the relatively high evapotranspiration rates of P. australis can help protect plants and affects nutrient uptake under flooding conditions [2,26]. In addition, aquatic macrophytes gradually modify the physiochemical properties of soil/sediment in a variety of ways, such as nutrient uptake by roots, plant residue decomposition, and root exudates [27,28,29]. These results suggest that interactions between aquatic plants and local factors are crucial to the distribution and interactions of bacterial communities in the riparian zones of lakes.
P. australis is widely distributed in land–lake ecotones, and larger roots systems promote its effect over a wide range [2]. In particular, P. australis can inhibit the growth of other aquatic macrophytes [30], which is useful for investigating the effects of single aquatic macrophytes on bacterial communities along the gradient of the riparian zones of lakes. P. australis grow in different hydrology conditions along the riparian zone. Meanwhile, gradient environment conditions of riparian zone can be considered as a good model to evaluate the relative contribution of local factors and P. australis on bacterial community characteristics of riparian zone. In this study, 16S rRNA gene amplicon sequencing and co-occurrence network analysis [31] were performed to explore bacterial community and network structures in the riparian zones of a lake. We hypothesized that the P. australis rhizosphere selection exerts a strong effect on bacterial community and network structures. The objectives of this study were (i) to understand the P. australis rhizosphere bacterial community composition and core species of bacterial networks; and (ii) to elucidate the correlations among P. australis, local factors, and bacterial communities in the riparian zone of freshwater lake.
2. Materials and Methods
2.1. Site Description and Sampling
Lake Taihu, a shallow eutrophic lake in a region of the Yangtze Delta in China, has a mean depth of 1.9 m and a surface area of 2338 m2. The study area is located in a riparian zone in the southeastern portion of Lake Taihu (Figure 1), which is covered with lush P. australis. Given the environmental heterogeneity of regions within the riparian zone of Lake Taihu with respect to hydrological characteristics and the presence of P. australis, we divided the area into two parts along the gradient of riparian zone (30 m long), where one zone with P. australis is the rhizosphere zone (RZ) and another zone without aquatic macrophytes is the nonrhizosphere zone (NZ). In addition, the two zones were subdivided into four sites, and six replicated plots (1 × 1 m) were placed along each transect (Figure 1), as follows: The supralittoral zone with P. australis (RZa) in the nonflooded zone; the eulittoral zone with P. australis (RZb), and the eulittoral zone without any aquatic macrophytes (NZc), which are closely adjacent to each other and located at the middle areas of the riparian zones of the lake, where the zones are irregularly inundated and regulated by the hydrological characteristics of Lake Taihu; and the infralittoral zone without any aquatic macrophytes (NZd), which is located at the lowest region and is inundated. Twenty-four composite samples were collected in May 2016. At each plot, three parallel samples were randomly collected and evenly combined into a single composite sample. The RZ samples were collected by lightly shaking roots [32]. The NZ samples (5–10 cm, similar depth as rhizosphere samples) were collected using a polymer glass tube (6 cm in diameter and 60 cm in length). All the samples were stored on ice and transported to the laboratory. For each sample, one portion was stored at −80 °C for molecular analysis, while another portion was stored at 4 °C to assess the chemical properties.
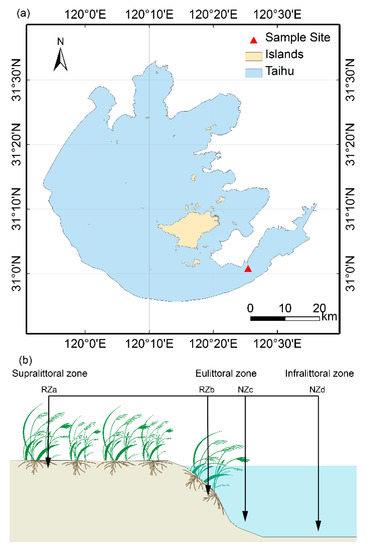
Figure 1.
(a) Location of sampling site in lake Taihu, China, (b) the distribution of the four sampling zones, including, supralittoral zone with P. australis (RZa), eulittoral zone with P. australis (RZb), eulittoral zone without P. australis (NZc), infralittoral zone without P. australis (NZd).
2.2. Sample Chemical Properties
For all the samples, the sample water content (SWC) was measured as gravimetric weight loss after drying at 105 °C for 24 h and reweighing, while the organic matter content (OM) was estimated by the loss-on-ignition method after combustion at 550 °C in a muffle furnace for 4 h. The pH values of the samples were determined using a soil to water ratio of 1:5 (v/v) with a pH meter. Total carbon (TC) and total nitrogen (TN) were analyzed with an elemental analyzer (Euro Vector EA3000) and then used to calculate the TC/TN. The dissolved organic carbon (DOC) was measured for the sample extracts (5 g freeze-dried samples, 50 mL pure water) in a TOC analyzer. Particle size distributions (silt, clay, and sand) were determined using a laser particle size analyzer. To assess ammonium nitrogen (NH4+) and nitrate nitrogen (NO3−) contents, samples were extracted with 2 M KCl (1:10 sample: Extraction ratio) and the extract was analyzed using a continuous flowing analyzer.
2.3. S rRNA Gene Pyrosequencing and Bioinformatics Analysis
Total DNA was extracted from 0.5 g of dry soil/sediment using a FastDNA SPIN for Soil kit (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. The V4-V5 hypervariable regions of the bacterial 16S rRNA gene was amplified using specific primers with the barcodes (515F GTGCCAGCMGCCGCGGTAA and 907R CCGTCAATTCCTTTGAGTTT). All PCR-amplification reactions were in a 30 μL volume with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA), 0.2 μM forward and reverse primers, and approximately 10 ng of template DNA. The thermocycling conditions were as follows: 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s, with a final extension at 72 °C for 5 min. The PCR products were assessed by electrophoresis on a 2% agarose gel, and a mixture of PCR products was purified with a GeneJET Gel Extraction Kit (Thermo Scientific, Waltham, MA, USA). Sequencing libraries were generated using an Illumina TruSeq DNA PCR-Free Library Preparation kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations, and index codes were added. The library quality was assessed using a Qubit@ 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina HiSeq platform, and 250 bp paired-end reads were generated. All of the sequences were deposited in to the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP235199).
2.4. Network Construction and Analysis
Networks were constructed for the RZ and NZ bacterial communities based on the relative abundances of OTUs. A random matrix theory (RMT)-based network analysis was performed using the molecular ecological network analysis pipeline, and additional information regarding the structure and theories of the pipeline are described elsewhere [31]. Only OTUs detected in six samples of 12 replicates were selected to construct the networks. The primary procedures performed in this study were as follows. First, the OTU table, environmental factors data, and OTU taxonomy file were prepared as guided in the pipeline. Second, a similarity matrix was constructed based on the Spearman’s Rho between pairwise OTUs. A cutoff value (similarity threshold, St) for the similarity matrix was generated automatically following the default settings. When the correlation between the abundance of OTUs is larger than the St, a link is assigned between the pair of OTUs. Third, the “Global Network Properties”, “Individual Nodes’ Centrality”, and “Module Separation and Modularity” functions were performed to calculate some network properties (e.g., average path, average degree, and average clustering coefficient). A module is a group of nodes that is more highly connected to each other than to nodes outside the group. Modularity (M) is a value that measures how well a network is divided into a module (M > 0.4 as the threshold) [33]. The network hub analysis was based on the connectivity of each node, which was determined by the within-module connectivity (Zi) and among-module connectivity (Pi). The node topologies are divided into four categories: Network hubs (Zi > 2.5 and Pi > 0.62), module hubs (Zi > 2.5 and Pi < 0.62), connectors (Zi < 2.5 and Pi > 0.62), and peripherals (Zi < 2.5 and Pi < 0.62). Fourth, “Output for Cytoscape visualization” was run, and three files were generated for network graph visualization using Cytoscape. Fifth, “Randomize the network structure and calculate network properties” was performed to compare the properties of the empirical and random-generated networks. Sixth, “Gene/OTU significances (GS)” with environmental traits were calculated, and a Mantel test was performed to assess the correlations between the GS and network connectivity. Finally, “Module-EigenGene analyses” were performed to identify relationships between local factors and network modules.
2.5. Statistical Analysis
One-way ANOVA with Tukey’s post-hoc tests were used to assess the effects of the riparian zones on environmental characteristics, alpha diversity indices, and the relative abundances of phyla (significance level of 0.05). If the distribution of dates were non-normal, the Kruskal–Wallis ANOVA was used. OTUs with relative abundances of >0.2% were selected to study the differences in dominant OTUs in four sampling sites and were visualized using the “ggplot2” package. The characterization of microbial features differentiated along riparian sampling sites at different taxonomic levels was performed using the linear discriminant analysis (LDA) effect size (LEfSe) method for biomarker discovery, and we used an LDA > 3.0 to indicate significant differences [34]. We used the principal coordinate analysis (PCoA) to study the shifts in bacterial community structure based on Bray–Curtis/Weighted UniFrac distances permutational multivariate analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (ADONIS) to test whether the bacterial communities in the four sites were significantly different. Redundancy analysis (RDA) was performed using R to explore the linkages between bacterial community structure and local factors (i.e., NH4+, NO3−, pH, OM, SWC, DOC, and silt). To elucidate the relationships between the local factors distance and bacterial community dissimilarities (Bray–Curtis/Weighted UniFrac distances), Mantle tests were performed in R. The analyses described above were performed in R.
3. Results
3.1. Local Factors in Sampling Zones
The results revealed that the NO3− and DOC concentrations of sediments exhibited significant differences between the RZ and NZ in the following order: NO3− (RZ > NZ) and DOC (RZ < NZ) (Table S1). The NH4+ concentrations in the NZ samples ranged from 21.87 to 63.36 mg·kg−1, which was much higher than those observed in the RZ samples (5.85 to 5.89 mg·kg−1) (p < 0.05) (Table S1). However, the SWC was significantly different among four zones and increased in the order of RZa, RZb, NZd, and NZc (p < 0.05) (Table S1). For the pH values, the lowest concentrations were observed in the RZb samples (5.28 ± 0.30). Particle size, including clay, silt, and sand differed among the four zones (p < 0.05). From the supralittoral to infralittoral regions, the clay content increased, whereas the silt content notably decreased.
3.2. Bacterial Community Composition and Diversity in the Riparian Zone of Lake Taihu
The obtained OTUs were affiliated with 16 phyla and subphyla (relative abundance >1%) (Figure S1). The abundances of Alphaproteobacteria, Acidobcteria, and Gemmatimonadetes were significantly higher in the RZa and RZb samples compared to the NZc and NZd samples, whereas Chloroflexi and Firmicutes were enriched in the NZc and NZd samples. Bacterroidetes was dominant in the NZc samples compared with the other zones. The richness and diversity of the bacterial communities did not exhibit remarkable differences among the four zones based on the Chao, Ace, Shannon, and phylogenetic diversity (PD) indices (p > 0.05) (Table S2). Furthermore, the 30 most abundant bacterial OTUs were visualized by hierarchical clustering analysis (Figure 2). The results revealed that the top 30 OTUs in the RZa and RZb samples clearly grouped together, whereas the top 30 OTUs in the NZc and NZd samples exhibited obvious differences, suggesting that the P. australis rhizosphere had similar abundant bacterial OTUs in different riparian zones.

Figure 2.
Heat map depicting the relative abundance of dominant OTUs (average relative abundance >0.2%) in the different sampling sites. The relative abundance dates were normalization and clusters were generated using Euclidean distance in R. Abbreviations: P. australis zone (RZ); non-P. australis zone (NZ); supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd).
To further explore the significance of discriminatively specific bacterial taxa, the LEfSe analysis was performed from the phylum to genus level among the four sampling zones using the default logarithmic (LDA) value of 3 (Figure 3). The clade graph showed that 11 groups of bacteria were enriched in the RZa samples, namely, Gemmatimonadetes (from the phylum to genus levels), Rhizobiales (Pseudolabrys), and Caulobacterales (Phenylobacterium). The discriminative clade of RZb was affiliated with Alphaproteobacteria, Acidobacteria_Gp1, and Myxococcales. We observed that the RZa and RZb samples had similar biomarkers, such as those affiliated with Alphaproteobacteria. In contrast, Bacteroidetes (Cryomorphaceae) and Clostridium_sensu_stricto were biomarkers of the NZc samples. In the NZd samples, Chloroflexi (from the phylum to family levels), Actinobacteria, and Clostridium_XI appeared to be the primary discriminant clades.
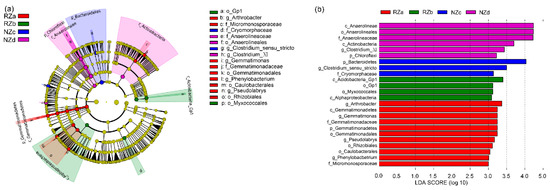
Figure 3.
(a) Visualizes the output of the linear discriminant analysis effect size (LEfSe)algorithm, which identifies taxonomically consistent among four sites as following: Supralittoral zone with P. australis (RZa), eulittoral zone with P. australis (RZb), eulittoral zone without P. australis (NZc), and infralittoral zone without P. australis (NZd). (b) Indicator bacterial groups within the four zones with linear discriminant analysis (LDA) scores higher than 3.
Principal coordinate analysis of the taxonomic (Bray–Curtis distance matrix) and phylogenetic (Weighted UniFrac distance matrix) dissimilarity were performed to investigate patterns of separations among the bacterial communities in the four zones. We observed that RZ and NZ samples separated across the first principal coordinate explained 31.23% and 49.35% of the variability in the bacterial communities with respect to taxonomic- and phylogenetic-based dissimilarity, respectively (Figure 4). Moreover, the ANOSIM and ADONIS results demonstrated that the structures of the bacterial communities among the four zones were significantly different based on taxonomic dissimilarity (p < 0.05), whereas the structure of the bacterial communities in the RZa and RZb samples were similar based on phylogenetic dissimilarity (p > 0.05) (Table S3), indicating that P. australis roots may enrich phylogenetic closeness bacterial communities in different riparian zones.
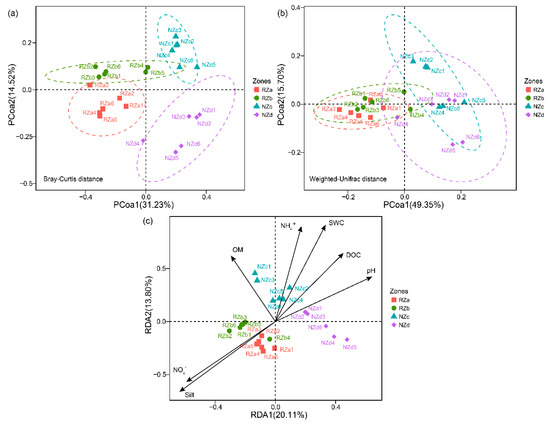
Figure 4.
Principal coordinate analysis (PCoA) plots derived from the taxonomy-based (Bray–Curtis dissimilarity matrix) (a) and phylogeny-based (Weighted UniFrac dissimilarity matrix) (b) dissimilarities of the bacterial communities. Color symbols correspond to different sampling sites. (c) Redundancy analysis (RDA) diagram illustrating the relationship between the bacterial community structures and local factors. Abbreviations: Supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd); ammonium nitrogen (NH4+); nitrate nitrogen (NO3−); sample water content (SWC); organic matter content (OM); dissolved organic carbon (DOC).
3.3. Relationships between Bacterial Communities and Local Factors in the Riparian Zone of Lake Taihu
To investigate the associations of bacterial community dissimilarity with local factors in the riparian zone of Lake Taihu, Mantel tests were performed (Table 1). The results showed that the taxonomic and phylogenetic dissimilarity in the NZ samples was significantly correlated with local factors (p < 0.05). The local factors also exhibited a significant effect on the RZ-associated bacterial community taxonomic dissimilarity (r = 0.37, p < 0.05), whereas they were uncorrelated with the RZ-associated bacterial community phylogenetic dissimilarity (r = 0.13, p > 0.05) (Table 1). These results suggested that the selection of P. australis roots strongly influenced the bacterial community at the phylogenetic level. In addition, Mantel tests showed that the NZ-associated bacterial community dissimilarity was strongly correlated with NH4+, DOC, OM, N%, and C%. We also observed that the SWC and particle size was associated with the RZ- and NZ-associated bacterial community dissimilarity (Table 1). Furthermore, RDA were performed to assess associations between local factors and bacterial community structure (Figure 4c). The RDA results showed that two axes (RDA1 and RDA2) explained 33.91% of the bacterial community variance. The results of Monte Carlo permutations indicated that the local factors NH4+, NO3−, SWC, DOC, OM, pH, and silt were significantly correlated with the bacterial community structures (p < 0.05) (Table S4).

Table 1.
Mantel tests for the relationship between bacterial community dissimilarities and local factors. Bacterial community dissimilarity was calculated based on Bray–Curtis distances (BC) and on Weighted UniFrac distances (WU).
3.4. Co-Occurrence Bacterial Networks in the Riparian Zone of Lake Taihu
To explore the bacterial interactions in the RZ and NZ samples, we constructed bacterial co-occurrence networks using the random matrix theory (RMT) [31] (Figure 5). As shown in Table 2, the similarity threshold (St) values were 0.86 and 0.89 for the RZ and NZ network topologies, respectively. The modularity of each co-occurrence network was higher than 0.4, suggesting that the two networks were modular [33]. Moreover, the average path (GD), average degree (avgK), and average clustering coefficient (avgCC) values of the two networks were significantly higher than the values of random networks, indicating the occurrence of small-word behavior [35] (p < 0.05). Overall, the co-occurrence networks were markedly different between the RZ and NZ samples, revealing that the RZ network had more nodes but fewer links than the NZ network. Furthermore, the NZ network had a significantly higher avgK value (5.646) than the RZ network (2.95) (p < 0.05), and the avgCC value of the NZ network (0.311) was also notably higher than that observed for the RZ network (0.161) (p < 0.05), suggesting that the NZ-associated bacterial community had a greater potential for interactions.

Figure 5.
An overview of bacterial networks in P. australis zone (a) and non-P. australis zone (b). Node color represents bacterial phylum of the OUT. Red edges indicate positive relationships and blue edges indicate negative relationships. Modules lager than five nodes are labeled with a number. Abbreviations: P. australis zone (RZ); non-P. australis zone (NZ).

Table 2.
Topological properties of bacterial networks under RZ (P. australis zone) and NZ (non-P. australis zone), and their corresponding random networks.
The analysis of modular topological roles is important to identify keystone species that play distinct topological roles in a network. The nodes of the network module were classified into four categories based on their within-module connectivity (Zi) and among-module connectivity (Pi) values (Figure 6). Most of the nodes in each network were peripherals that had most of their links inside their modules. There were one module hub and four connectors in the RZ network, whereas the NZ network had three module hubs and two connectors. The module hub and connectors of the RZ network were affiliated with the genus Sphingomonas (Alphaproteobacteria), the order Gp3 (Acidobacteria), the class Betaproteobacteria (Proteobacteria), and the order Acidimicrobiales (Actinobacteria), whereas the module hubs and connectors of the NZ network belonged to the family Anaerolineaceae (Chloroflexi), the genus Ignavibacterium (Ignavibacteriae), and the genus Pelosinus (Firmicutes), suggesting that these taxa were crucial in the construction of co-occurrence networks (detailed taxonomic information is provided in Table S5). Furthermore, the connector of the RZ network appeared at the peripheral of the NZ network (OTU_166), whereas the module hub of the NZ network occurred at the peripheral of the RZ network (OTU_115) (Table S5), suggesting a role shift of the nodes between the two networks.
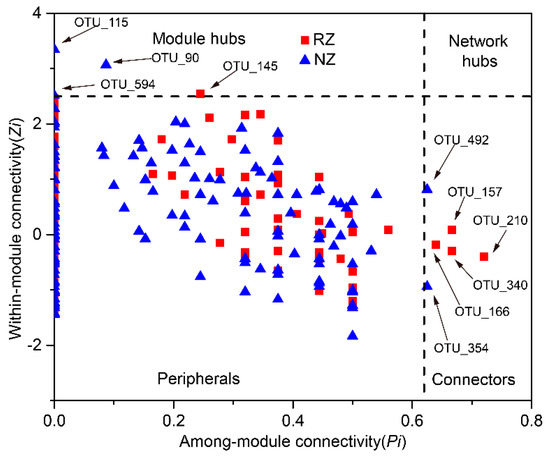
Figure 6.
Zi-Pi plots showing distribution of OTUs based on their topological roles in the P. australis zone network (red square) and non-P. australis zone network (blue triangle). Threshold values of Zi and Pi for categorizing OTUs were 2.5 and 0.62. More information on the module hubs and connectors are shown in Table S5. Abbreviations: P. australi zone (RZ); non-P. australis zone (NZ).
3.5. Relationships between Network Connectivity and Local Factors in the Riparian Zone of Lake Taihu
Overall, the Mantel test results revealed that the connectivity of the RZ network was uncorrelated with local factors, whereas that of the NZ network was significantly correlated with local factors (Table S6). Partial Mantel test results showed that pH was significantly correlated with the RZ network, whereas the variables NH4+ and DOC affected the NZ network. Notably, the silt and sand contents were both correlated with the two networks. Furthermore, network module eigengene analysis was performed to analyze the correlations between modules and local factors (Table S7). For the RZ and NZ networks, the eigengenes of the primary modules could explain 54–77% and 63–83%, respectively. pH was only correlated with the modules of RZ, whereas NH4+, NO3−, TC, OM, and DOC were only correlated with the NZ modules. Moreover, the variables TN, SWC, clay, silt, and sand affected some modules in both the RZ and NZ networks.
4. Discussion
4.1. Roots Enrich and Filter the Bacterial Community Composition in the Riparian Zone of Lake Taihu
Similar to the studies on the P. australis rhizosphere [12,14,36], Alphaproteobacteria and Acidobcteria were substantially enriched in the P. australis zones. Alphaproteobacteria exhibit fast growth by utilizing a broad range of root-derived carbon substrates [37], with members of this phylum being widely distributed in different types of plant rhizosphere soils, such as rice [38], lettuce [39], rabbiteye blueberry [20], and wheat [19]. Acidobacteria play an important role in the carbon cycle by performing functions such as cellulose and lignin degradation [40], and Acidobacteria has also been shown to be a primary phylum in rhizosphere soil [41,42]. Moreover, Rhizobiales and Gemmatimonadetes were shown to be biomarkers of the P. australis zones (Figure 3), members of which are associated with the metabolism of nitrogen and phosphorus [43,44], suggesting that the P. australis rhizosphere microbiota may be involved in promoting plant nutritional requirements. In contrast, Chloroflexi and Firmicutes were primarily dominated in non-P. australis zones (Figure 3 and Figure S1), consistent with the results of previous studies on anaerobic sediments, such as in Lake Taihu [45] and freshwater wetlands [19].
4.2. Co-Occurrence Network Structures in the Riparian Zone of Lake Taihu
Network analysis is crucial to understanding the community dynamics of bacterial interactions and niches [15,16,46], providing a new dimension to our understanding of plant rhizosphere bacterial community ecology [18]. For the RZ and NZ networks, positive links accounted for 78.8% and 87.7% of the total links, respectively, suggesting that the nodes (i.e., OTUs) of the two networks represent cooperative interactions (interdependency) or a response to a common local factor [16,18]. Similar to the results of a study on wheat [19], the bacterial network of the P. australis zones exhibited lower connectivity than the surroundings. In contrast, rhizosphere bacterial networks have shown to be of greater complexity and connectivity compared with nonrhizosphere bacterial networks as plants grow, where the flowering stage of plants is particularly more complex than the vegetative stage [18]. These contradictory results indicate that plant species, growth stages, and habitats can affect the characteristics of bacterial networks. Thus, we need to pay greater attention to the bacterial networks of aquatic macrophyte rhizospheres at different growth stages. In addition, our results showed that bacterial network of the P. australis zones had more nodes and higher modularity than the zones without P. australis. Higher modularity was shown beneficial for increasing the stability of microbial networks and the resistance profiles of microbial communities during environment changes [47].
With respect to topology, numerous studies have focused on putative keystone species that play distinct roles in co-occurrence networks [16,18]. We showed that the module hubs and connectors of the networks were completely different between the P. australis and non-P. australis zones, suggesting that P. australis roots can affect ecological functions and structuring bacterial networks in the riparian zone of Lake Taihu. Acidimicrobiales (Actinobacteria) and Gp3 (Acidobacteria) were module connectors in the bacterial network of the P. australis zones. Similarly, Actinobacteria and Acidobcteria have emerged as keystone taxa in blueberry rhizosphere soil [20]. Actinobacteria have the ability to produce diverse antibiotics [48], potentially allowing members of this phylum to take part in protecting plants from pathogenic bacteria. Our results showed that Sphingomonas (Alphaproteobacteria) was the module hub in the P. australis zones network, which could be correlated with signaling molecule regulation (e.g., quorum sensing) [49]. Previous studies showed that microorganisms can use quorum sensing (QS) to control competitive and cooperative behaviors in the plant rhizosphere [18,50,51,52]. In contrast, the module hubs and connectors of bacterial network in the non-P. australis zones were assigned to Anaerolineaceae (Chloroflexi), Ignavibacterium (Ignavibacteriae), and Pelosinus (Firmicutes). These members have been reported to be involved in biogeochemical cycling, such as hydrocarbon degradation, sulfur cycling, and Fe (III) reduction [53,54,55]. In addition, we also observed that the role shifting of keystone species appeared between the two networks, which was the result of changes in keystone species as the surrounding environment changed [18].
4.3. Links among Aquatic Macrophytes, Local Factors, and Bacterial Communities in the Riparian Zone of Lake Taihu
We showed that taxonomic-based dissimilarities in bacterial communities were different in the four assayed zones, which exhibited significant correlations with local factors. Environmental heterogeneity has been suggested to be an important factor driving the distribution of bacterial communities in the riparian zones of lakes [56]. However, P. australis zones were shown to exhibit phylogenetic closeness of the bacterial communities under different hydrologic conditions, indicating the existence of common selective pressures shaping bacterial communities in the rhizosphere [20]. Moreover, the local factors had a lower correlation with bacterial community structures and networks of P. australis zones compared with non-P. australis zones (Table 1 and Table S6). A number of mechanisms could explain why P. australis exerted a strong selection on riparian bacterial communities. Root exudates of the P. australis, such as oxalic, acetic, and succinic acid act as endogenous carbon sources for heterotrophic bacteria [11]. The properties of the soil/sediment close to roots are modified by the plant roots. For example, the roots of P. australis secretes oxygen to oxygenate the anaerobic sediments [57], which can affect the rhizosphere bacterial community composition and diversity. Furthermore, the assembly of rhizosphere bacterial community is based on functional activities related to the nutrient metabolism of plants [58,59]. Similarly, microbes with functional genes encoding transporters were shown to be crucial to the assembly of bacterial rhizosphere communities [60].
Our results showed that the bacterial community structure in the nonrhizosphere zones was correlated with NH4+, DOC, and OM, similar to the results of other studies in different freshwater ecosystems [6,61]. Notably, SWC and particle size correlated with the bacterial community and network structures in both the P. australis and non-P. australis zones. Although P. australis exhibited the ability to select and enrich riparian bacterial community composition and interactions, local factors can also directly or indirectly drive the variation in bacterial communities. Previous studies showed that dynamic riparian hydrology can influence the diffusion of oxygen and organic matter decomposition in the sediment, which can affect bacterial community structure and activity in riparian zones [6,62]. In the context of hydrologic variation, the anaerobic conditions of sediments can prompt the replacement of the rhizomes of P. australis by adventitious roots [2,63], and these changes can indirectly affect rhizosphere bacterial community composition and network structures. Furthermore, the existing literature has shown that differences in bacterial assemblages are associated with different particle sizes in various ecosystems [64,65,66]. Soil particle size can provide different surface properties and microhabitats that strongly contribute to the activity and composition of microbial communities [67,68]. For aquatic macrophytes, the riparian particle size can also affect their growth and root exudates, indirectly regulating the characteristics of rhizosphere bacterial communities.
5. Conclusions
Our results revealed that P. australis roots exerted effects on bacterial community composition and network structures in the riparian zone of Lake Taihu. Although the riparian bacterial communities exhibited differences based on taxonomic dissimilarity in the four assayed riparian zones, the P. australis zones enriched the phylogenetic closeness of the bacterial community. The results showed that two of the bacterial networks were nonrandom networks, and completely different topological properties were observed between the P. australis and non-P. australis zones. The putative keystone taxa of the bacterial network of the P. australis zones were affiliated with Sphingomonas (Alphaproteobacteria), Acidimicrobiales (Actinobacteria), and Gp3 (Acidobcteria), which might be involved in the regulation of bacterial interactions and plant growth. Furthermore, the hydrological regime and particle size of the riparian zone of Lake Taihu were correlated with the bacterial community and network structures. Overall, these results suggest that the distribution of P. australis and local factors are crucial for shaping bacterial communities and interactions in the riparian zone of Lake Taihu.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4441/12/2/432/s1. Figure S1: The relative abundance of different phyla and subphyla in the four zones. Abbreviations: Supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd). Table S1: The properties of local factors in lake riparian. Abbreviations: Supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd); ammonium nitrogen (NH4+); nitrate nitrogen (NO3−); sample water content (SWC); organic matter content (OM); total carbon (TC); total nitrogen (TN); dissolved organic carbon (DOC). Table S2: Estimated OUT richness and alpha diversity indices of the four zones (no significant difference was found for each variable). Abbreviations: Supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd). Table S3: Permutational multivariate analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (ADONIS) based on Bray–Curtis dissimilarity matrix (BC) and Weighted UniFrac dissimilarity matrix (WU) in bacterial community composition among four sampling sites. Abbreviations: P. australis zone (RZ); non-P. australis zone (NZ); supralittoral zone with P. australis (RZa); eulittoral zone with P. australis (RZb); eulittoral zone without P. australis (NZc); infralittoral zone without P. australis (NZd). Table S4: Significance analysis of RDA by Monte Carlo permutation (permutation = 999). Abbreviations: Ammonium nitrogen (NH4+); nitrate nitrogen (NO3−); sample water content (SWC); organic matter content (OM); total carbon (TC); total nitrogen (TN); dissolved organic carbon (DOC). Table S5: Module hubs and connectors in P. australi zone and non-P. australis zone networks. Abbreviations: P. australi zone (RZ); non-P. australis zone (NZ); c, o, f, g represent class, order, family, and genus, respectively. Table S6: The mantel and partial mantel tests on connectivity of networks and local factors. Abbreviations: P. australi zone (RZ); non-P. australis zone (NZ); ammonium nitrogen (NH4+); nitrate nitrogen (NO3−); sample water content (SWC); organic matter content (OM); total carbon (TC); total nitrogen (TN); dissolved organic carbon (DOC). Table S7: Correlations between module eigengenes and local factors. Abbreviations: P. australi zone (RZ); non-P. australis zone (NZ); ammonium nitrogen (NH4+); nitrate nitrogen (NO3−); sample water content (SWC); organic matter content (OM); total carbon (TC); total nitrogen (TN); dissolved organic carbon (DOC).
Author Contributions
Conceptualization, J.Z. and Y.L.; methodology, Y.L. and R.H.; software, Y.L. and R.H.; validation, Y.L., J.Z., and Q.L.W.; formal analysis, Y.L. and R.H.; investigation, Y.L.; writing—original draft preparation, Y.L.; writing—review and editing, J.Z. and Q.L.W.; supervision, J.Z.; funding acquisition, J.Z. and Q.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (41621002, 31730013, 31971478), and the National Key Technology R&D Program (2015BAD13B01).
Acknowledgments
We thank Huibin Lu, Lina Wang, and Huiming Xu for sample collection and data analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McClain, M.E.; Boyer, E.W.; Dent, C.L.; Gergel, S.E.; Grimm, N.B.; Groffman, P.M.; Hart, S.C.; Harvey, J.W.; Johnston, C.A.; Mayorga, E.; et al. Biogeochemical hot spots and hot moments at the interface of terrestrial and aquatic ecosystems. Ecosystems 2003, 6, 301–312. [Google Scholar] [CrossRef]
- Srivastava, J.; Kalra, S.J.S.; Naraian, R. Environmental perspectives of Phragmites australis (Cav.) Trin. Ex. Steudel. Appl. Water Sci. 2014, 4, 193–202. [Google Scholar] [CrossRef]
- Trias, R.; Ruiz-Rueda, O.; Garcia-Lledo, A.; Vilar-Sanz, A.; Lopez-Flores, R.; Quintana, X.D.; Hallin, S.; Baneras, L. Emergent macrophytes act selectively on ammonia-oxidizing bacteria and archaea. Appl. Environ. Microbiol. 2012, 78, 6352–6356. [Google Scholar] [CrossRef] [PubMed]
- Pietrangelo, L.; Bucci, A.; Maiuro, L.; Bulgarelli, D.; Naclerio, G. Unraveling the composition of the root-associated bacterial microbiota of Phragmites australis and Typha latifolia. Front. Microbiol. 2018, 9, 1650. [Google Scholar] [CrossRef]
- Srivastava, J.K.; Chandra, H.; Kalra, S.J.S.; Mishra, P.; Khan, H.; Yadav, P. Plant–microbe interaction in aquatic system and their role in the management of water quality: A review. Appl. Water Sci. 2016, 7, 1079–1090. [Google Scholar] [CrossRef]
- Foulquier, A.; Volat, B.; Neyra, M.; Bornette, G.; Montuelle, B. Long-term impact of hydrological regime on structure and functions of microbial communities in riverine wetland sediments. FEMS Microbiol. Ecol. 2013, 85, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Peralta, A.L.; Matthews, J.W.; Kent, A.D. Habitat specialization along a wetland moisture gradient differs between ammonia-oxidizing and denitrifying microorganisms. Microb. Ecol. 2014, 68, 339–350. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, D.; Wu, J.; Chen, Q.; Long, C.; Li, Y.; Cheng, X. Anti-seasonal submergence dominates the structure and composition of prokaryotic communities in the riparian zone of the Three Gorges Reservoir, China. Sci. Total Environ. 2019, 663, 662–672. [Google Scholar] [CrossRef]
- Laurent, P.; Raaijmakers, J.M.; Philippe, L.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar]
- Haichar, F.e.Z.; Santaella, C.; Heulin, T.; Achouak, W. Root exudates mediated interactions belowground. Soil Biol. Biochem. 2014, 77, 69–80. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.; He, X.; Zhang, S.; Liang, R.; Shen, J. Effects of root exudates on denitrifier gene abundance, community structure and activity in a micro-polluted constructed wetland. Sci. Total Environ. 2017, 598, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Behera, P.; Mahapatra, S.; Mohapatra, M.; Kim, J.Y.; Adhya, T.K.; Raina, V.; Suar, M.; Pattnaik, A.K.; Rastogi, G. Salinity and macrophyte drive the biogeography of the sedimentary bacterial communities in a brackish water tropical coastal lagoon. Sci. Total Environ. 2017, 595, 472–485. [Google Scholar] [CrossRef]
- Bowen, J.L.; Kearns, P.J.; Byrnes, J.E.K.; Wigginton, S.; Allen, W.J.; Greenwood, M.; Tran, K.; Yu, J.; Cronin, J.T.; Meyerson, L.A. Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun 2017, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Zhu, J.N.; Liu, Q.F.; Liu, Y.; Liu, M.; Liu, L.; Zhang, Q. Comparison of the diversity of root-associated bacteria in Phragmites australis and Typha angustifolia L. in artificial wetlands. World J. Microbiol. Biotechnol. 2013, 29, 1499–1508. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Yang, Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2011, 2. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X.; Relman, D.A. Functional Molecular Ecological Networks. mBio 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef]
- Fan, K.; Cardona, C.; Li, Y.; Shi, Y.; Xiang, X.; Shen, C.; Wang, H.; Gilbert, J.A.; Chu, H. Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol. Biochem. 2017, 113, 275–284. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, S.; Li, R.; Zhang, J.; Liu, Y.; Lv, L.; Zhu, H.; Wu, W.; Li, W. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017, 109, 145–155. [Google Scholar] [CrossRef]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2018, 20, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.F.; Zhang, T.; Yang, X.; Wang, S.; Yu, Y.; Dong, L.L.; Guo, Y.D.; Ma, Y.X.; Zang, J.Y. Diversity and composition of bacterial community in soils and lake sediments from an arctic lake area. Front. Microbiol. 2016, 7, 1170. [Google Scholar] [CrossRef]
- Deslippe, J.R.; Jamali, H.; Jha, N.; Saggar, S. Denitrifier community size, structure and activity along a gradient of pasture to riparian soils. Soil Biol. Biochem. 2014, 71, 48–60. [Google Scholar] [CrossRef]
- Ye, C.; Cheng, X.; Zhang, K.; Du, M.; Zhang, Q. Hydrologic pulsing affects denitrification rates and denitrifier communities in a revegetated riparian ecotone. Soil Biol. Biochem. 2017, 115, 137–147. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Z.; Xia, X.; Wang, F. A shallow lake remediation regime with Phragmites australis: Incorporating nutrient removal and water evapotranspiration. Water Res. 2012, 46, 5635–5644. [Google Scholar] [CrossRef] [PubMed]
- Luigimaria, B.; Giovanni, B.; Alessio, M.; Roberto, D.P.; Lorenzo, B. Rhizosphere effect and salinity competing to shape microbial communities in Phragmites australis (Cav.) Trin. ex-Steud. FEMS Microbiol. Lett. 2015, 359, 193–200. [Google Scholar]
- Packer, J.G.; Meyerson, L.A.; Skálová, H.; Pyšek, P.; Kueffer, C. Biological flora of the British Isles: Phragmites australis. J. Ecol. 2017, 105, 1123–1162. [Google Scholar] [CrossRef]
- Hallin, S.; Hellman, M.; Choudhury, M.I.; Ecke, F. Relative importance of plant uptake and plant associated denitrification for removal of nitrogen from mine drainage insub-arctic wetlands. Water Res. 2015, 85, 377–383. [Google Scholar] [CrossRef]
- Kowalski, K.P.; Bacon, C.; Bickford, W.; Braun, H.; Clay, K.; Leduc-Lapierre, M.; Lillard, E.; McCormick, M.K.; Nelson, E.; Torres, M.; et al. Advancing the science of microbial symbiosis to support invasive species management: A case study on Phragmites in the Great Lakes. Front. Microbiol. 2015, 6, 95. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Saunders, A.M.; Schramm, A. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl. Environ. Microbiol. 2009, 75, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The ecology of Acidobacteria: Moving beyond genes and genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Aymé, S.; Omry, K.; Zhao, J.; Susannah Green, T.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Cardinale, M.; Grube, M.; Erlacher, A.; Quehenberger, J.; Berg, G. Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 2015, 17, 239–252. [Google Scholar] [CrossRef]
- Sl, B.S.T.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 1999, 65, 1731–1737. [Google Scholar]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790. [Google Scholar] [CrossRef] [PubMed]
- Debruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Mark, R. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl. Environ. Microbiol. 2011, 77, 6295. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Geetanjali, G. Symbiotic nitrogen fixation in legume nodules: Process and signaling. A review. Agron. Sustain. Dev. 2007, 27, 59–68. [Google Scholar] [CrossRef]
- Ye, W.; Liu, X.; Lin, S.; Tan, J.; Pan, J.; Li, D.; Yang, H. The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu. FEMS Microbiol. Ecol. 2009, 70, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Krause, A.E.; Frank, K.A.; Mason, D.M.; Ulanowicz, R.E.; Taylor, W.W. Compartments revealed in food-web structure. Nature 2003, 426, 282–285. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Morohoshi, T.; Sato, N.; Iizumi, T.; Tanaka, A.; Ikeda, T. Identification and characterization of a novel N-acyl-homoserine lactonase gene in Sphingomonas ursincola isolated from industrial cooling water systems. J. Biosci. Bioeng. 2017, 123, 569–575. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Lindow, S.E.; Firestone, M.K. Bacterial quorum sensing and nitrogen cycling in rhizosphere soil. FEMS Microbiol. Ecol. 2008, 66, 197–207. [Google Scholar] [CrossRef]
- Elasri, M.; Delorme, S.; Lemanceau, P.; Stewart, G.; Laue, B.; Glickmann, E.; Oger, P.M.; Dessaux, Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 2001, 67, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.; Pierson, E.A.; Pierson, L.S.; Stacey, G.; Chatterjee, A. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant. Biol. 2002, 5, 285–290. [Google Scholar] [CrossRef]
- Brown, S.D.; Utturkar, S.M.; Magnuson, T.S.; Ray, A.E.; Poole, F.L.; Lancaster, W.A.; Thorgersen, M.P.; Adams, M.W.; Elias, D.A. Complete genome sequence of Pelosinus sp. strain UFO1 assembled using single-molecule real-time DNA sequencing technology. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.N.; Krumholz, L.R.; Gieg, L.M.; Parisi, V.A.; Suflita, J.M.; Allen, J.; Philp, R.P.; Elshahed, M.S. Biodegradation of low-molecular-weight alkanes under mesophilic, sulfate-reducing conditions: Metabolic intermediates and community patterns. FEMS Microbiol. Ecol. 2010, 72, 485–495. [Google Scholar] [CrossRef]
- Liu, Z.; Frigaard, N.U.; Vogl, K.; Iino, T.; Ohkuma, M.; Overmann, J.; Bryant, D.A. Complete genome of Ignavibacterium album, a metabolically versatile, flagellated, facultative anaerobe from the phylum Chlorobi. Front. Microbiol. 2012, 3, 185. [Google Scholar] [CrossRef]
- Monard, C.; Gantner, S.; Bertilsson, S.; Hallin, S.; Stenlid, J. Habitat generalists and specialists in microbial communities across a terrestrial-freshwater gradient. Sci. Rep. 2016, 6, 37719. [Google Scholar] [CrossRef]
- Tanaka, N.; Yutani, K.; Aye, T.; Jinadasa, K.B.S.N. Effect of broken dead culms of Phragmites australis on radial oxygen loss in relation to radiation and temperature. Hydrobiologia 2007, 583, 165–172. [Google Scholar] [CrossRef]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef]
- Yan, Y.; Kuramae, E.E.; De, H.M.; Klinkhamer, P.G.; van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2016, 11, 56–66. [Google Scholar] [CrossRef]
- Wan, Y.; Ruan, X.; Zhang, Y.; Li, R. Illumina sequencing-based analysis of sediment bacteria community in different trophic status freshwater lakes. Microbiologyopen 2017, 6, e00450. [Google Scholar] [CrossRef] [PubMed]
- Weise, L.; Ulrich, A.; Moreano, M.; Gessler, A.; Kayler, Z.E.; Steger, K.; Zeller, B.; Rudolph, K.; Knezevic-Jaric, J.; Premke, K. Water level changes affect carbon turnover and microbial community composition in lake sediments. FEMS Microbiol. Ecol. 2016, 92, fiw035. [Google Scholar] [CrossRef] [PubMed]
- Ostendorp, W.; Dienst, M.; Schmieder, K. Disturbance and rehabilitation of lakeside Phragmites reeds following an extreme flood in Lake Constance (Germany). Hydrobiologia 2003, 50–509, 687–695. [Google Scholar] [CrossRef]
- Santmire, J.A.; Leff, L.G. The influence of stream sediment particle size on bacterial abundance and community composition. Aquat. Ecol. 2006, 41, 153–160. [Google Scholar] [CrossRef]
- Jackson, C.R.; Weeks, A.Q. Influence of particle size on bacterial community structure in aquatic sediments as revealed by 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 2008, 74, 5237–5240. [Google Scholar] [CrossRef] [PubMed]
- Tkacz, A.; Cheema, J.; Chandra, G.; Grant, A.; Poole, P.S. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 2015, 9, 2349–2359. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Dohrmann, A.B.; Christensen, B.T.; Tebbe, C.C. Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front. Microbiol. 2018, 9, 149. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Christensen, B.T.; Martens, R.; Tebbe, C.C. Soil particle size fractions harbour distinct microbial communities and differ in potential for microbial mineralisation of organic pollutants. Soil Biol. Biochem. 2015, 90, 255–265. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).